Abstract
EMMPRIN (extracellular matrix metalloproteinase inducer, EMN, CD147) is a member of the immunoglobulin superfamily expressed in numerous cell types both as a soluble and a membrane-spanning glycoprotein. It is involved in many physiological processes, as well as in cancer. This study addresses mechanisms of crosstalk between EMN-driven cancer-related cellular responses and the canonical Wnt-pathway in MCF-7 breast carcinoma cells. Genetic knockdown of EMN in MCF-7 resulted in characteristic changes in cellular shape, organization of the actin cytoskeleton and malignancy profile, indicating that EMN expression represses cell motility, but, in contrast, exerts a stimulatory effect on cell proliferation and invasive properties. Increased invasiveness coincided with elevated expression of Wnt-target genes and established invasion driver matrix metalloproteinase MMP14. Activation of the downstream Wnt-pathway by means of heterologous β-catenin and/or TCF-4 expression, through inhibition of GSK-3β by LiCl treatment, or by cell stimulation with insulin-like growth factor-1 (IGF-1) resulted in increased EMN expression. EMN over-expression raised the ratio of the two opposing Wnt pathway-driven transcription factors Sp1 and Sp5, leading to stimulation of the EMN promoter. Furthermore, the EMN promoter was activated by a feed-forward circuit involving an EMN-dependent drop in expression of the repressive signal transducer and activator of transcription 1 (STAT1). Taken together, we show that the influence of EMMPRIN on malignancy-related properties of breast cancer cells is functionally connected to both Wnt- and JAK/STAT pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
EMMPRIN (extracellular matrix metalloproteinase inducer, EMN, basigin, CD147) is a widely expressed glycoprotein belonging to the immunoglobulin superfamily. It exists both as a soluble and a membrane-spanning protein, the latter acting as EMN receptor (EMNR) for soluble EMN in the course of homotypic interactions [1]. As a multifunctional protein, EMN is abundant in various organs and cell types, amongst others in immune cells [2], platelets, and monocytes [3], in fibroblasts [4] or cells of the nervous system [5]. It participates in reproductive [6] and developmental processes like the differentiation of the mammary gland in mice [7] and plays a role in several diseases e.g., human immunodeficiency virus (HIV) infection or rheumatoid arthritis [8].
As EMN was associated with the progression of several cancer types [9, 10] and a poor prognosis for cancer patients like breast cancer patients [11, 12], many studies aimed to clarify the role of EMN in tumor biology. As a stimulator of matrix metalloproteinases like MMP-1, -2, -3, -9, -14, -15 [13,14,15] and associated with cell surface proteins like integrins (α3β1 [16], α6β1 [17]), EMN influences cancer cell proliferation, migration, and invasion. This was also demonstrated by EMN-knockdown studies of our laboratory and others [18,19,20]. While the signaling mechanisms regulating the cellular EMN levels are still not fully understood, diverse factors were identified as stimulators for EMN expression, e.g., epidermal growth factor (EGF) [21], insulin-like growth factor 1 (IGF-I) [22] or transforming growth factor-β1 (TGF-β1) [23]. Studies covering the transcriptional regulation of EMN on the promoter level revealed the involvement of transcription factors Sp1, hypoxia-induced factor-1α (HIF-1α) and c-myc in cancer cells [24]. Sidhu et al. demonstrated an induction of Wnt/β-catenin signaling by overexpression of EMN in lung cancer cells which led to an induction of malignancy parameters like migration and anchorage-independent growth [25].
The Wnt/β-catenin signaling cascade controls various cellular functions including differentiation, proliferation, migration, apoptosis, and cancer development, e.g., of breast cancer [26]. Binding of Wnt ligands to their receptors and co-receptors elicits nuclear translocation and activity of β-catenin together with TCF/LEF transcription factors to regulate target genes like cyclin D1 [27], matrix metalloproteinase 14 (MMP14, MT1-MMP) [28], or the transcription factor Sp5 [29, 30]. In the absence of Wnt ligands the β-catenin destruction complex consisting of APC (Adenomatous polyposis coli), Axin, CK1 (casein kinase 1) and GSK3β binds and phosphorylates β-catenin which in turn is subjected to proteasomal degradation [31].
Furthermore, a positive correlation of EMN and the PI3K/AKT signaling pathway was found in cancer cell lines involving adaption processes of the glucose metabolism in hepatocellular carcinoma cells [28] and the induction of malignancy parameters covering migration and invasion of ovarian carcinoma cells [18].
The aim of our work was to identify relevant signaling molecules and mechanisms involved in EMN regulation and to study how these mechanisms affect malignancy parameters. As EMN is associated with malignancy of breast cancers and is discussed as a marker for progression and prognosis of these cancers [12], we decided to employ the breast cancer model cell line MCF-7 for this purpose. Their epithelial character [32] and well detectable EMN expression levels [20] mark these cells as a convenient cellular model. By using various tools of signaling pathway induction and EMN-knockdown in MCF-7 cells we demonstrate a positive correlation between EMN and MCF-7 malignancy. We obtained insights into EMN regulation by components of the Wnt/β-catenin as well as the JAK/STAT signaling cascade in these cells.
Materials and methods
Cells, cell culture and cell transfection
MCF-7 cells were purchased from the ATCC (American Type Culture Collection). Generation of the stable MCF-7 derivative with EMN-knockdown (MCF-7-shEMN) and the control cell line MCF-7-shscr has been described previously [20]. Cells were routinely cultured in Dulbecco’s Minimum Essential Medium (DMEM) supplemented with phenol red, 4 mM stabilized l-glutamin, 4.5 g/L d-glucose, 10% (v/v) fetal calf serum (FCS) (Biochrom, Berlin, Germany), 1% Gentamycin (Ratiopharm, Ulm, Germany) at 37 °C, and 5% CO2. Media were obtained from Sigma-Aldrich (Steinheim, Germany). Every 2–3 days cells were passaged after incubation for 5 min with trypsin/EDTA (Biochrom) to disrupt the adherent monolayer of cells.
For DNA-mediated transfection, aliquots of 2.5 × 105 cells (for overexpression experiments) or 5 × 104 cells (for reporter gene experiments) were seeded into six well (or 24 well) plates and incubated for 12 h before 200 µl (100 µl) of serum-free DMEM containing 0.25–2 µg of plasmid DNA and 2 µl TurboFect™ reagent (Thermo Fisher Scientific, Schwerte, Germany) per µg DNA were added. After a further incubation for 6 h, medium was changed (DMEM + 10% (v/v) FCS) and after overnight incubation, cells were subjected to analysis.
Antibodies and reagents
EMN (Ig1) goat polyclonal antibody (N19), STAT1 (p84/p91) rabbit polyclonal antibody and horseradish peroxidase (HRP)-conjugated secondary antibody were purchased from Santa Cruz Biotechnologies (Dallas, TX, USA), CD147-PE (phycoerythrin) and -APC (allophycocyanin) conjugated antibodies were from ImmunoTools GmbH (Friesoythe, Germany). Antibodies to cytokeratin and E-cadherin were purchased from Dako (Hamburg, Germany). The anti-GAPDH antibody was purchased from Merck Millipore (Schwalbach, Germany). Secondary goat-anti-mouse-Cy3 antibody was from Dianova (Hamburg, Germany). Phalloidin-Alexa-Fluor® 594 was from Invitrogen (Karlsruhe, Germany).
Plasmids and recombinant DNA work
Expression construct pEGFPN1-EMN encoding full-length EMMPRIN as a fusion with green fluorescent protein (GFP) was a gift from Dr. Andreas Till and has been described [33]. Expression constructs pCS2+-β-Catenin and pCS2+-TCF4 as well as the empty vector pCS2+ have been described previously [34]. pcDNA3.1-STAT1-HA encoding HA-tagged STAT1 [35] was obtained from Prof. Oliver H. Krämer.
pGL-3-pEMN(-254) was constructed by inserting an Nhe I/Hind III restricted PCR-product representing a 254 bp segment upstream of the transcriptional start of the human EMMPRIN gene into luciferase plasmid pGL-3 (Promega). The PCR-product was generated from human genomic DNA using primers pEMN 254 fwd (5′-atcggctagcccgtttcctagcaacgcc-3′) and pEMN 254 rev (5′-atcgaagcttgattcctattcctcgccg-3′) according to published information [36].
Phenotypic characterization of EMN-knockdown and control cells
Phenotypic cell characterisation was performed using the CASY® TT Cell Counter and Analyzer (Roche Diagnostics GmbH, Penzberg, Germany). MCF-7-shEMN and MCF-7-shscr cells were harvested (< 2 × 106 cells) and the cell suspension (60 µl) was resuspended in 6 ml of CASY®-ton (isotonic electrolytic solution). After cleaning with CASY®-clean and calibration with CASY®-ton, the cell mean and peak diameter were detected by quantification of the electrical impulse generated by the cells according to the “pulse area analysis” principle in a 150 µm capillary.
Flow cytometry
MCF-7 cells optionally treated with stimuli or transfected with expression constructs were harvested and washed in phosphate buffered saline (PBS) as described [20]. They were stained with monoclonal antibodies CD147-APC or CD147-PE (10 µg/ml, Ig1-domain) on an orbital shaker. Cells were analyzed in a CyFlo™ Space Cytometer (Partec, Münster, Germany) and data analyses were performed using the Cyflogic Software (CyFlo Ltd, freeware) as detailed previously [20].
Phalloidin-staining of F-actin
Cells were seeded on glass slides and on the following day washed with PBS and fixed in fixation buffer containing 4% (w/v) paraformaldehyde (PFA) and 4% (w/v) glucose for 20 min. For inactivation of PFA, fixed cells were treated with 50 mM NH4Cl. After a washing step with PBS permeabilisation was achieved by treating the cells with 0.5% (v/v) Triton X-100 solution. Blocking of unspecific binding-sites was performed with blocking-buffer (1% (w/v) BSA, 1% (w/v) fish gelatine, 1% (v/v) FCS diluted in PBS) for 25 min at room temperature. Treatment with Phalloidin-Alexa-Fluor® 594 (Invitrogen, Karlsruhe, Germany) in blocking-buffer (1 h in the dark) and Roti-Mount FluorCare with 4′,6-diamidine-2-phenylindol (DAPI) (Carl Roth, Karlsruhe, Germany) allowed for F-actin detection and nuclear staining, respectively.
Immunofluorescence microscopy
Cells (0.3 × 106) were seeded on 8-well chamber slides (BD Transduction Laboratories™, Heidelberg, Germany). On the next day cells were washed with PBS and fixed in ice-cold methanol/acetone (1:1) for 2 min. After drying the slides were stored at − 20 °C. For immunofluorescent staining cells were reconstituted in Tris/HCl-buffered saline (TBS)/0.01% Tween-20 (TBST) pH 7.4. Antibody preparations were made up according to the manufacturer’s instructions in antibody dilutant with background-reducing components solution (Dako), in detail E-cadherin (1:300), β-catenin (1:50), and cytokeratin (1:300).
Cell proliferation assays
Cell proliferation was measured either using the xCELLigence™ system (Roche, Mannheim, Germany) or the CellTiter-Glo™ assay (Promega, Mannheim, Germany). xCELLigence™ is based on the measurement of impedance triggered by cell attachment. The dimension-less cell index (CI) quantifies cell attachment resulting from cell growth and proliferation. The relative cell index (rCI) represents the ratio between the CI at a specific time point and the CI determined at the start of the measurement (t0). Samples of 1.5 × 104 MCF-7 cells were seeded into each well of an E-plate (Roche) and grown overnight in an xCELLigenceTM/RTCA DP instrument (Roche). Stimulus was optionally added to wells diluted in fresh 200 µl DMEM/10% (v/v) FCS per well on the next day, thus defining t0.
The CellTiter-Glo™ assay (Promega) was performed following instructions of the manufacturer. MCF-7 cells were seeded into 96-well-plates (1000 cells per well). Measurements were performed according to the manufacturer’s instructions by adding 40 µl of the assay reagent directly to the cells for 10 min followed by quantification of luminescence using a Glo-Max™ 96 Microplate Luminometer (Promega).
Cell migration assays
Cell motility was quantified by wound-healing (scratch) assays and the xCELLigence™ system (Roche). The wound-healing assay is based on monitoring the closing of a wound scratched into a monolayer of MCF-7-shEMN and control MCF-7-shscr cells. Samples of 2 × 105 cells/well were grown in 24 well-plates for 24 h before generating cross-shaped scratches with a pipet tip. The degree of wound-healing was determined after 24 h incubation by taking microscopic pictures. Photographs were analyzed for the percentage of scratch closure using the TScratch software developed by Tobias Geback and Martin Schulz and provided as freeware by the Computational Science and Engineering Laboratory of the Eidgenössische Technische Hochschule (ETH), Zurich, Switzerland.
The xCELLigence™ system was employed along with CIM-plates. Cells (2 × 104) were seeded into the upper chamber and migration through a porous membrane (8 µm pores) to the lower chamber was monitored as dimension-less cell index (CI).
Colony formation assay (CFA)
Quantification of cellular spreading into an agarose layer was used as a malignancy marker for MCF-7-shEMN and MCF-7-shscr cells. Cells were suspended in an agarose (0.3%)/DMEM mixture to a dilution of 2.5 × 103/ml, 1 ml of the cell suspension was seeded into an agarose (0.6%)/DMEM-coated 6-well plate and cultivated overnight at 37 °C. On the next day, wells were covered with 2 ml of DMEM containing 100 µg/ml G418 and quantitative detection of the grown colonies was performed after 21 days of cultivation at 37 °C via photography and Clono-counter freeware [37].
Reporter gene assay
MCF-7 cells were (co-)transfected with 0.25 µg of luciferase reporter gene construct, 0.5 µg of the Renilla luciferase reference construct pRL-TK (Promega) and, optionally, specific expression construct(s) as described above employing the TurboFect™ reagent (Thermo Scientific). Luciferase activity was quantified by chemiluminescence measurement using a Glo-Max™ 96 microplate luminometer (Promega) and normalized for constitutive Renilla luciferase activity as described previously [20].
Analysis of DNA sequences for transcription factor binding-sites
The DNA sequence upstream the EMN transcriptional start site was analyzed for potential transcription factor recognition sites using the ALLGEN-PROMO-Software [38].
RNA extraction, cDNA synthesis and RT-PCR
Extraction of total mRNA was performed using an RNA isolation kit from Macherey-Nagel (Düren, Germany) following the manufacturer’s protocol. 5 ng of RNA was reverse transcribed with RevertAid™ reverse transcriptase from (Thermo Scientific) employing random hexamer primers and oligo-(dT)18 primers (Thermo Scientific). Q-Real-Time-PCR was performed by using the Universal ProbeLibrary (UPL)™ technology (Roche). Target-specific probes and primers were identified employing the Universal ProbeLibrary Assay Design Center (Roche). Primer-probe combinations used in this study were: GAPDH sense (5′-agccacatcgctcagacac-3′), GAPDH antisense (5′-gcccaatacgaccaaatcc-3′), probe #60; EMN sense (5′-ccttggctccaagatactcct-3′), EMN antisense (5′-cgtcggagtccaccttga-3′), probe #68; MMP14 sense (5′ gccttggactgtcaggaatg-3′), MMP14 antisense (5′-aggggtcactggaatgctc-3′), probe #37, Sp5 sense (5′-ggggagactcagcagacg-3′), Sp5 antisense (5′-tgggtccctatgtccgaag-3′), probe #36. Sp1 sense (5′-ctatagcaaatgccccaggt-3′), Sp1 antisense (5′- tccacctgctgtgtcatcat-3′), probe #9, STAT1 sense (5′-ttggcacctaacgtgctg-3′), STAT1 antisense (5- ttcgtaccactgagacatcctg-3′), probe #68. PCR products were generated by 40 cycles of 15 s, 95 °C—60 s, 60 °C—90 s, 72 °C in MicroAmp™ optical 96-well reaction plates using the StepOne™ real time PCR procedure (Applied Biosystems, Darmstadt, Germany). Relative fold-changes were calculated by the comparative Ct method. Gene expression levels of EMN-kd (shRNA) samples were related to control (scrambled RNA) samples (control = 1).
SDS-polyacrylamide gel electrophoresis and Western blot
SDS-PAGE and Western blot analyses were performed as described previously [20]. Briefly, whole cell extracts were solubilized in gel loading buffer (62.5 mM Tris/ HCl pH 6.8; 2% (w/v) SDS; 25% (v/v) glycerol; 1‰ (w/v) phenol blue; 5% (v/v) β-mercaptoethanol), boiled for 5 min and separated through 12% acrylamide SDS-polyacrylamide gels. Proteins were silver- or Coomassie-stained or blotted onto nitrocellulose membranes (GE Healthcare, Munich, Germany). Non-specific binding-sites on blot membranes were blocked with 5% (w/v) milk protein in Tris/HCl-buffered saline (TBS)/0.01% (v/v) Tween-20 pH 7.4. Primary antibodies for detection were dissolved in TBST to final concentrations of 0.4 µg/ml for polyclonal EMMPRIN (N19), 5 µg/ml anti-β-catenin and anti-GAPDH, and 1 µg/ml anti-STAT1 antibodies. Bound primary antibodies were detected using HRP-conjugated secondary antibodies (dilution 1:10,000). Visualization was performed using Lumi-Light™ Western blot substrate (Roche).
Statistics
Distribution of values was analyzed for normality. All values were normally distributed and determination of statistical significance was performed using analysis of equality of variances (Levene’s Test) and t test (SPSS-Software). Values of p < 0.05 were considered as statistically significant.
Results
EMN determines the cellular phenotype of breast cancer cells
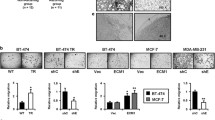
To assess the role of EMN in the control of malignancy parameters, an EMN-knockdown (EMN-kd) was induced in MCF-7 breast cancer cells by means of an EMN-targeting shRNA (a scrambled shRNA construct was employed as a specificity control), resulting in the two stable cell lines MCF-7 shEMN and MCF-7 shscr [20]. We first compared EMN expression in both cell lines and determined strongly decreased EMN mRNA (Fig. 1a) and EMN protein levels (Fig. 1b).
Molecular and cellular effects of EMN-knockdown in MCF-7 cells. a Comparison of EMN mRNA abundance in MCF-7 cells stably transfected with EMN shRNA (shEMN) and a scrambled shRNA control (shscr). Samples of 2 × 105 cells were subjected to mRNA isolation and subsequent quantification of EMN mRNA by qRT-PCR as described in “Materials and methods” section. Data are derived from three independent experiments. Error bars represent standard deviation, asterisks indicate significance of differences (***p < 0.001). b Effect of EMN shRNA on EMN protein abundance in MCF-7 cells. Lysates from MCF-7 cells stably transfected with EMN shRNA (shEMN) and a scrambled shRNA control (shscr) were subjected to SDS-PAGE (12% polyacrylamide), Western blot and probing with a goat polyclonal anti-EMN antibody. Bands representing highly (HG) and low-glycosylated (LG) EMN are indicated. Probing with a specific antibody to α-tubulin (bottom panel) served as loading control. c Phenotypical comparison of EMN-kd MCF-7 cells (shEMN) and control cells transfected with scrambled shRNA (shscr) by immunofluorescence microscopy. Cells were seeded onto glass slides and stained with a mouse antibody to cytokeratin. Visualization was achieved using a Cy3-coupled goat anti-mouse antibody as described in “Materials and methods” section. Cell nuclei were stained with DAPI. d Effect of EMN-knockdown on the size of MCF-7 cells. Samples of roughly 107 MCF-7 cells transfected with EMN shRNA (shEMN) or scrambled shRNA (shscr) were analyzed for peak and average size by pulse area analysis employing a CASYR Cell Counter and Analyzer. Standard deviation was derived from the results of eight independent measurements. Significance of differences (**p < 0.01, ***p < 0.001) was determined by Student’s t test
Notably, both cell lines differed in their phenotype. Compared to control cells, MCF shEMN cells showed a more expanded, flattened shape (Fig. 1c) and an increased average cell size (Fig. 1d) by immunofluorescence microscopy and automated cell sizing, respectively.
EMN stimulates proliferation but represses motility of breast cancer cells
Effects of EMN-kd on proliferation, migration, adhesion-independent growth, and cellular invasiveness were determined in MCF-7 cells. While EMN-kd cells showed a significant decrease in proliferation rate compared to control cells (Fig. 2a), migratory activity appeared clearly enhanced (Fig. 2b). These results indicate that EMN acts in favor of proliferation but reduces migration of MCF-7 breast cancer cells.
Influence of EMN on proliferative and migratory properties of MCF-7 cells. a Proliferative behavior of EMN-kd MCF-7 cells compared to control cells. Proliferation of cells stably transfected with shEMN (squares) and a scrambled shRNA control (shscr—triangles) was followed for a period of 120 h. At the indicated time points, aliquots of 1000 cells were subjected to proliferation tests by the CellTiter-Glo™ assay as detailed in the “Materials and methods” section. Data points represent relative increase of proliferation-related luminescence as determined for respective samples after the indicated periods of time. Readings at the start of the incubation were set 1. Three independent experiments, each with triplicate measurements were performed. Error bars show standard deviations, statistical significance of differences is indicated by an asterisk (*p < 0.05). b Effect of EMN-kd on migration of MCF-7 cells. MCF-7-shEMN (black bars) and MCF-7-shscr (grey bars) cells were subjected to a wound-healing assays upon introduction of scratches into cell monolayers as detailed in the “Materials and methods” section. The closed wound area is indicated as percentage of scratch closure after 24 h of incubation. Data are derived from three independent experiments, each performed in triplicate and were processed using the TScratch software. Error bars show standard deviations, asterisks indicate statistical significance of differences (**p < 0.01)
EMN enhances invasive cell properties
Next, the effect of EMN-kd on the invasive potential of MCF-7 cells was assessed. In comparative invasion assays, cells proficient for EMN (MCF-7-shscr) showed a profoundly stronger rate of passing through a matrigel layer, i.e., an artificial basal membrane equivalent (Fig. 3a).
Influence of EMN on invasiveness-related properties of MCF-7 cells. a Invasive behavior of MCF-7-shEMN cells (squares) in comparison to MCF-7-shscr cells transfected with a scrambled control shRNA (triangles). Invasion was determined by a transwell invasion assay combined with electrical impedance measurement using an xCELLigence Real Time Cell Analyzer (RTCA) device (Roche). Cells were placed on CIM-plates-16 coated with a MatrigelR layer as described in “Materials and methods” section. Quantification of migrated (invasive) cells was performed by repeated determination of impedance (proportional to invasive cells) over a time period of 48 h. Values (relative invasion rate) were related to the readings obtained after 3 h of incubation which was arbitrarily set 1. Data represent mean values of three independent experiments. b Effect of EMN-kd on F-actin distribution in MCF-7 cells. Fixed MCF-7-shEMN cells and control MCF-7-shscr cells transfected with a scrambled control shRNA were seeded onto glass slides and stained for F-actin with phalloidin-Alexa-Fluor® 594 and for nuclei with DAPI prior to visualization by fluorescence microscopy as detailed in “Materials and methods” section. Arrows indicate noticeable actin assemblies in protrusions in EMN-kd cells. c Effect of EMN-kd on the expression of Wnt-target gene MMP14. MCF-7-shEMN and MCF-7-shscr cells were subjected to mRNA isolation, cDNA synthesis and comparative mRNA quantification as detailed in the “Materials and methods” section. Normalized mRNA levels in EMN-kd cells (black bars) were related to those in shscr control cells (grey bars) which were arbitrarily set 1. Error bars represent standard deviation, asterisks indicate significance of changes relative to sh scr cells (***p < 0.001)
We then aimed at correlating EMN-dependent alterations in invasiveness and motility behavior of MCF-7 cells with structural and signaling determinants. Cell staining for F-actin revealed notable effects of the EMN-kd on the shape of the cytoskeleton. An evident assembly of F-actin in protrusions, presumably forming lamellipodia was observable at the edges of MCF-7-shEMN cells, whereas such structures were absent in control MCF-shscr cells (Fig. 3b).
In a previous study we have shown that soluble EMN can drive both invasiveness and activation of β-catenin-mediated gene transcription in breast cancer cells, presumably via homotypic interaction with the membrane-bound EMN receptor [20]. We, hence, hypothesized that EMN-related enhancement of cell invasion is connected to Wnt-signaling and therefore tested the effect of EMN-kd on a Wnt-target gene with an established role in invasiveness. The Wnt-target gene MMP-14 is known to be upregulated in invasive cancer cells [28]. We compared its mRNA level in MCF-7-shEMN and MCF-7-shsrc cells and observed significantly reduced expression in EMN-depleted cells (Fig. 3c).
The downstream Wnt-pathway upregulates EMNR on breast cancer cells
We aimed at characterizing the functional overlap and interconnection of EMN-driven cellular responses and Wnt-signaling. Previously, we had observed that soluble EMN can stimulate both β-catenin-mediated gene regulation and enhanced expression of EMN [20]. To characterize underlying mechanisms, we activated the downstream Wnt pathway in MCF-7 cells by inhibiting GSK3β-mediated phosphorylation of β-catenin by treatment with LiCl. Interestingly, this led to an increase in EMNR surface presentation in a dose-dependent manner (Fig. 4a). We then tested if components of the malignancy-related Wnt pathway influence EMNR expression. MCF-7 cells were transfected with expression constructs for β-catenin and T-cell factor 4 (TCF4) and subsequently assayed cytometrically for surface EMN (EMNR) expression. Both Wnt effectors enhanced EMNR surface expression in an additive manner (Fig. 4b). This effect was paralleled by a similar activation pattern of a minimal EMN promoter in a reporter gene assay (Fig. 4c) and by an increase of the transcriptional level of the endogenous EMN gene as determined by real time qRT-PCR (Fig. 4d).
Upregulation of EMN expression by the activated Wnt-signaling pathway in MCF-7 cells. a Effect of LiCl on EMN surface expression (EMNR). MCF-7 cells were treated with the indicated concentrations of LiCl for 24 h. They were then incubated with APC-conjugated anti-CD147 (EMN) antibody for 1 h before EMN was detected by flow cytometry. b Effect of β-catenin and/or TCF4 overexpression on EMN surface expression. MCF-7 cells were transfected with 1 µg each of expression constructs for β-catenin and T-cell factor 4 (TCF4) individually or in combination as indicated and subsequently assayed for surface EMN (EMNR) expression essentially as in (a), but employing a PE-conjugated anti-CD147 (EMN) antibody. c Effect of β-catenin and/or TCF4 over-expression on EMN promoter activity. A firefly luciferase reporter gene construct encompassing 254 bp upstream from the transcriptional start site of the human EMN gene (pGL3-EMN-254) was transfected together with a Renilla luciferase reference construct and the indicated β-catenin or/and TCF4 expression constructs (pCS2+-β-Catenin, pCS2+-TCF4) into MCF-7 cells. After 18 h cells were lysed and subjected to quantification of luciferase activity. Data represent means of three independent experiments with standard deviation. The asterisks indicate significance of change relative to non-stimulated cells (*p < 0.05, **p < 0.01, ***p < 0.001). d Effect of β-catenin and/or TCF4 over-expression in MCF-7 cells on expression of the endogenous EMN gene. Cells were transfected with β-catenin or/and TCF4 expression constructs as in (c). After 24 h, mRNA was isolated and quantified by RT-PCR as described in the “Materials and methods” section. Normalized mRNA levels were related to those in non-transfected control cells (arbitrarily set 1). Error bars and statistics are analogous to Figs. 1, 2 and 3
In line with these findings, EMN depletion resulted in a reduced expression level of Wnt pathway effector protein cyclin D1, and increased expression of tumor suppressor p53 as analyzed by RT-PCR (Fig. 5a, b). To further solidify the notion that downstream components of the Wnt pathway are operative in upregulating EMNR expression, we made use of the instance that the IGF-1 receptor is a known activator of the PI3K/AKT kinase cascade. As shown in Fig. 5c, stimulation of MCF-7 cells with IGF-1 enhanced EMNR surface expression.
Influence of EMN expression on the activity of the Wnt-signaling pathway. a RT-PCR-based quantitative determination of Wnt-target gene cyclin D1 expression in dependence of EMN-ld in MCF-7 cells. mRNA levels in MCF-7-shEMN (shEMN) and MCF-7-shscr (shscr) cells were compared and represented as in Fig. 3c. b Influence of EMN-kd on p53 expression. The experiment was performed and represented in analogy to (a). c Effect of Wnt-pathway stimulation by IGF-1 receptor activation on EMN surface expression. Samples of 2.5 × 105 MCF-7 cells were treated with 20 ng/ml IGF-1 in a volume of 200 µl for 3 h or left untreated. They were then stained with an antibody to EMNR and a PE-conjugated secondary antibody, and subsequently analyzed cytometrically as in Fig. 4a, b
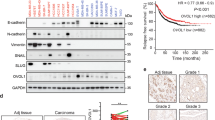
The EMN promoter is regulated by both the downstream Wnt pathway and a feed-forward circuit involving EMN and STAT1
Next we sought to further characterize mechanisms determining the regulation of the EMN promoter under the influence of signaling mediated by the EMN- GSK3β-Wnt pathway axis. The genomic DNA sequence upstream of the EMN transcriptional start site was analyzed for potential transcription factor recognition sites. As depicted in Fig. 6a, within a 320 bp promoter segment, cognate elements for TCF, STAT1, the estrogen receptor α (ERα) and, interestingly, for Sp1/Sp5 were identified. Because Sp1 and Sp5 have been described as Wnt pathway-driven transcriptional regulators with opposing activities [30], we studied the consequence of EMN-kd on the respective expression levels of the two factors by qRT-PCR. Of note, the presence of EMN in control cells coincides with stimulation of Sp1 expression and with profound repression of Sp5 expression (Fig. 6b). From this finding we draw the conclusion that one reason for the enhancing effect of EMN on EMN transcription is that it alters the Sp1/Sp5 ratio.
Feed-forward regulation of EMN expression through Wnt-pathway- and STAT1-dependent transcriptional control. a Schematic representation of potential transcription factor binding-sites within the proximal 320 bp of the human EMN promoter. Numbers indicate distances upstream from the transcriptional start site (arrow), sequences represent consensus recognition sites for the indicated transcription factors. Not to scale. b Effect of EMN depletion on mRNA levels of Wnt-pathway-dependent transcription factors Sp1 (left) and Sp5 (right). mRNA from MCF-7-shEMN (shEMN) and MCF-7-shscr (shscr) cells was quantitatively analyzed for the levels of Sp1 (left) and Sp5 transcripts (right) by qRT-PCR using specific primers given in Material and methods. Procedure and representation was as in Fig. 5a, b. c Effect of EMN-kd on the expression of STAT1 in MCF-7 cells. Lysates from MCF-7-shEMN (shEMN) or MCF-7-shscr cells (shscr) were probed with an antibody to STAT1 (top) and reprobed with an antiserum to EMN (N19) to confirm the EMN-kd (middle panel) and to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (bottom panel) to test for equal loading. d Dose-dependent repressive influence of STAT1 on EMN promoter activity. EMN promoter luciferase reporter gene construct pGL-3-pEMN(-254) was transfected together with a Renilla luciferase reference construct and the indicated amounts of STAT1 expression plasmid pcDNA3.1-STAT1-HA into MCF-7 cells. Luciferase activity was determined and presented as in Fig. 4c. e Repression of EMN surface expression on MCF-7 cells by heterologous expression of STAT1. MCF-7 cells were transfected with the indicated amounts of STAT1 expression plasmid pcDNA3.1-STAT1-HA and subsequently assayed for surface EMN (EMNR) expression as in Fig. 4a, b. f, g Effect of EMN over-expression on the expression of transcription factors STAT1 and Sp5. 2.5 × 105 MCF-7 cells were transfected with 1 µg of EMN expression plasmid pEGFPN1-EMN or empty vector pEGFPN1 as indicated. Cells were lysed 24 h post transfection and subjected to mRNA isolation, cDNA synthesis and comparative mRNA quantification for STAT1 (f) and Sp5 (g) by qRT-PCR employing specific primer pairs as detailed in the “Materials and methods” section. Normalized mRNA levels in EMN-transfected cells (black bars) were related to those in control cells tranfected with the empty vector cells (grey bars) which were arbitrarily set 1. Error bars represent standard deviation, asterisks indicate significance of changes relative to MCF-7-shscr cells (***p < 0.001)
Since a potential binding-site for STAT1 resides within the EMN-promoter sequence, we studied the role of STAT1 in EMN-governed cellular processes. The abundance of cellular STAT1 protein was found enhanced in EMN-kd cells, i.e., STAT1 expression is negatively correlated with EMN expression (Fig. 6c). This suggests that EMN triggers a feed-forward loop involving STAT1 to activate its own expression. To address this in more detail, we overexpressed STAT1 in MCF-7 cells to determine how, in turn, STAT1 affects EMN function. Reporter gene experiments and cytometric analyses revealed that heterologous STAT1 expression dose-dependently decreased both EMN-promoter activity and EMN surface expression (Fig. 6d, e). These results indicate that, on the one hand, STAT1 is repressive for EMN expression and that, on the other hand, EMN-dependent reduction of STAT1 expression has a stimulatory effect on the EMN promoter, consistent with a feed-forward regulatory mechanism.
To further confirm the down-regulatory influence of EMN on the abundance of both Sp5 and STAT1 in MCF-7 cells, we analyzed the effect of EMN over-expression on the expression levels of both transcription factors. As shown in Fig. 6f, g, cells transfected with an EMN expression construct showed profoundly reduced mRNA levels of both factors.
Discussion
Here, we show that EMN exerts a profound and multi-faceted influence on malignancy properties of breast carcinoma cells and that EMN-based signaling and the oncogenic Wnt-pathway are functionally connected with each other. Knockdown of EMN resulted in an alteration of the malignancy parameters profile of MCF-7 breast carcinoma cells which is virtually inverse to the one we have recently characterized upon treatment with the soluble extracellular domain of EMN [20]. In this earlier study, upregulation of EMN mRNA in response to cell stimulation with soluble EMN was strongly hinting at the existence of a positive EMN feedback loop which involves transcriptional regulation via β-catenin/TCF.
The present study underscores the view that enhanced EMN surface expression favors cell proliferation. While invasiveness is also promoted by EMN-dependent signaling (Fig. 3a), cellular motility, in contrast, drops in response to soluble EMN and is stronger in EMN-knockdown cells (Fig. 2b). These findings are consistent with an earlier report on a comparable role of EMN in cervical carcinoma cells [39]. Reports suggested that EMN surface expression strengthens cell contacts via homotypic interactions as well as by contacting the β1 subunit of integrins α3β1 and α6β1 [17, 40], which, in consequence, may reduce cellular motility. We also assume that in accordance with earlier findings [41], lowered stabilization of integrin clusters in EMN-kd cells is functionally connected with reduced signaling by focal adhesion kinases (FAK). In addition, reduced MMP-14 expression in EMN-depleted MCF-7 cells (Fig. 3c) hints at lower integrin convertase activity, which is likely to affect cell adhesion and invasion processes as well as FAK-associated signal transduction [42].
Earlier studies on MCF-7 cells showed that overexpression or activation of IGF-1 favored interaction of the IGF-1R with the E-cadherin complex and ZO tight junction proteins, resulting in stabilized cell–cell contacts and enhanced cellular growth rate [43, 44]. Interestingly, ZO-2 was reported to exert regulatory functions in the cell nucleus and to reduce cyclin D1 expression as well as cell proliferation [45, 46]. Since we observed reduced IGF-R tyrosine phosphorylation in EMN-kd cells (data not shown), we hypothesize that EMN-mediated activation of these IGF-1R functions contributes to the observed EMN-related positive effect on proliferation and negative effect on motility.
We did not observe expression changes in markers for epithelial–mesenchymal transition (EMT) such as E-cadherin and vimentin upon EMN-kd (data not shown). Since MCF-7 cells show an islet-like growth pattern, we attribute enhanced migration of EMN-kd cells to a collective manner of movement of “cell sheets” as it has been described during the development of the mammary gland [47].
EMN-triggered signaling and the Wnt-pathway are obviously interconnected in a complex and malignancy-relevant manner. Our results suggest that the Wnt-pathway and EMN signal equidirectionally in favor of cell proliferation through increased abundance of cyclin D1 or lowered levels of the tumor suppressor p53.
Interestingly, TCF4 can stimulate transcriptional activity of a minimal EMN promoter which does not contain the consensus TCF binding-site at around 300 bp upstream of the transcriptional start site (Fig. 4c), even to a stronger extent than it stimulates a 1055 bp promoter version containing a bona fide TCF cognate element (data not shown). It is likely that TCF4 is involved in transcriptional regulation of EMN expression in both a direct and an indirect manner, the latter mediated by complex joint activities with other transcription factors. It is established that TCF4 can regulate the expression of transcriptional repressor Sp5 via multiple binding-sites within its promoter [30]. Our results suggest that TCF/LEF transcription factors can act in a dual manner on EMN promotor activity. A direct positive influence takes place by binding to their specific binding-site in the EMN promotor sequence. An additional indirect effect probably relies on down-regulation of Sp5 expression, thereby favoring Sp1 interaction with the EMN promotor in competition with Sp5, thereby upregulating EMN expression.
The EMN promoter contains a STAT1-binding DNA element. With regard to EMN regulation, STAT1 overexpression showed a negative influence on both EMN promoter activity and EMN surface expression in this study. The mechanism behind the latter phenomenon is unclear, but in the light of the simultaneous increase in cytoplasmic EMN levels it can be speculated that STAT1 acts in favor of EMN internalization. Interestingly, in EMN-knockdown cells elevated STAT1 levels were observed. It appears reasonable to assume that this effect contributes to the reduced proliferation rate of EMN-kd cells, since growth-inhibitory influences of STAT1 are well established [48]. Notable in the context of breast cancer is a recent report which associated reduced STAT1 levels with enhanced development of EGFR-positive breast tumors in a mouse disease model [49]. Our findings indicate that STAT1 is repressive for EMN expression and that EMN-dependent reduction of STAT1 expression has an enhancing effect on the EMN promoter, resulting in a feed-forward regulation.
Taken together, this work emphasizes the influence of EMN surface expression on the malignancy profile of MCF-7 breast cancer cells. It demonstrates that combined signaling involving downstream components of Wnt- and JAK/STAT pathways are operative in a feed- forward mechanism which enhances EMN expression in EMN-driven manner.
The above considerations and findings from the present and a previous study [20] have been integrated into a working model shown in Fig. 7. It depicts mechanisms underlying upregulation of EMN expression and activity in MCF-7 cells as a result of signals emanating from downstream sections of Wnt- and JAK/STAT pathways and EMN-associated effects. Our observations led us to the postulation of a feed-forward circuit entailing elevated EMN promoter activity to result in additional stimulation of EMN-mediated signaling.
Model of connections between EMN function and the downstream Wnt-pathway in MCF-7 breast carcinoma cells, resulting in a feed-forward signaling circuit with malignancy-enhancing consequences. Triangles with color gradients represent directional changes from lower (blue) to higher (red) expression or/and activity. “Wnt” in brackets along with the dotted arrow indicates that to date no Wnt-type ligand eliciting the depicted reactions in breast cancer cells has been identified. For further details see text. (Color figure online)
References
Muramatsu T (2016) Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem 159:481–490
Hu J, Dang N, Yao H, Li Y, Zhang H, Yang X, Xu J, Bian H, Xing J, Zhu P, Chen Z (2010) Involvement of HAb18G/CD147 in T cell activation and immunological synapse formation. J Cell Mol Med 14:2132–2143
Schmidt R, Bultmann A, Fischel S, Gillitzer A, Cullen P, Walch A, Jost P, Ungerer M, Tolley ND, Lindemann S, Gawaz M, Schomig A, May AE (2008) Extracellular matrix metalloproteinase inducer (CD147) is a novel receptor on platelets, activates platelets, and augments nuclear factor kappaB-dependent inflammation in monocytes. Circ Res 102:302–309
Biswas C, Zhang Y, DeCastro R, Guo H, Nakamura T, Kataoka H, Nabeshima K (1995) The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res 55:434–439
Fan QW, Yuasa S, Kuno N, Senda T, Kobayashi M, Muramatsu T, Kadomatsu K (1998) Expression of basigin, a member of the immunoglobulin superfamily, in the mouse central nervous system. Neurosci Res 30:53–63
Chen H, Lam Fok K, Jiang X, Chan HC (2012) New insights into germ cell migration and survival/apoptosis in spermatogenesis: lessons from CD147. Spermatogenesis 2:264–272
Szymanowska M, Hendry KA, Robinson C, Kolb AF (2009) EMMPRIN (basigin/CD147) expression is not correlated with MMP activity during adult mouse mammary gland development. J Cell Biochem 106:52–62
Iacono KT, Brown AL, Greene MI, Saouaf SJ (2007) CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol 83:283–295
Weidle UH, Scheuer W, Eggle D, Klostermann S, Stockinger H (2010) Cancer-related issues of CD147. Cancer Genom Proteom 7:157–169
Riethdorf S, Reimers N, Assmann V, Kornfeld JW, Terracciano L, Sauter G, Pantel K (2006) High incidence of EMMPRIN expression in human tumors. Int J Cancer 119:1800–1810
Liu F, Cui L, Zhang Y, Chen L, Wang Y, Fan Y, Lei T, Gu F, Lang R, Pringle GA, Zhang X, Chen Z, Fu L (2010) Expression of HAb18G is associated with tumor progression and prognosis of breast carcinoma. Breast Cancer Res Treat 124:677–688
Zhao S, Ma W, Zhang M, Tang D, Shi Q, Xu S, Zhang X, Liu Y, Song Y, Liu L, Zhang Q (2013) High expression of CD147 and MMP-9 is correlated with poor prognosis of triple-negative breast cancer (TNBC) patients. Med Oncol 30:335
Kataoka H, DeCastro R, Zucker S, Biswas C (1993) Tumor cell-derived collagenase-stimulatory factor increases expression of interstitial collagenase, stromelysin, and 72-kDa gelatinase. Cancer Res 53:3154–3158
Sameshima T, Nabeshima K, Toole BP, Yokogami K, Okada Y, Goya T, Koono M, Wakisaka S (2000) Glioma cell extracellular matrix metalloproteinase inducer (EMMPRIN) (CD147) stimulates production of membrane-type matrix metalloproteinases and activated gelatinase A in co-cultures with brain-derived fibroblasts. Cancer Lett 157:177–184
Guo H, Zucker S, Gordon MK, Toole BP, Biswas C (1997) Stimulation of matrix metalloproteinase production by recombinant extracellular matrix metalloproteinase inducer from transfected Chinese hamster ovary cells. J Biol Chem 272:24–27
Berditchevski F, Chang S, Bodorova J, Hemler ME (1997) Generation of monoclonal antibodies to integrin-associated proteins. Evidence that alpha3beta1 complexes with EMMPRIN/basigin/OX47/M6. J Biol Chem 272:29174–29180
Dai JY, Dou KF, Wang CH, Zhao P, Lau WB, Tao L, Wu YM, Tang J, Jiang JL, Chen ZN (2009) The interaction of HAb18G/CD147 with integrin alpha6beta1 and its implications for the invasion potential of human hepatoma cells. BMC Cancer 9:337
Zhao Y, Chen S, Gou WF, Niu ZF, Zhao S, Xiao LJ, Takano Y, Zheng HC (2013) The role of EMMPRIN expression in ovarian epithelial carcinomas. Cell Cycle 12:2899–29913
Wang B, Xu YF, He BS, Pan YQ, Zhang LR, Zhu C, Qu LL, Wang SK (2010) RNAi-mediated silencing of CD147 inhibits tumor cell proliferation, invasion and increases chemosensitivity to cisplatin in SGC7901 cells in vitro. J Exp Clin Cancer Res 29:61
Knutti N, Kuepper M, Friedrich K (2015) Soluble extracellular matrix metalloproteinase inducer (EMMPRIN, EMN) regulates cancer-related cellular functions by homotypic interactions with surface CD147. FEBS J 282:4187–4200
Menashi S, Serova M, Ma L, Vignot S, Mourah S, Calvo F (2003) Regulation of extracellular matrix metalloproteinase inducer and matrix metalloproteinase expression by amphiregulin in transformed human breast epithelial cells. Cancer Res 63:7575–7580
Chen Y, Gou X, Ke X, Cui H, Chen Z (2012) Human tumor cells induce angiogenesis through positive feedback between CD147 and insulin-like growth factor-I. PLoS ONE 7:e40965
Wu J, Ru NY, Zhang Y, Li Y, Wei D, Ren Z, Huang XF, Chen ZN, Bian H (2011) HAb18G/CD147 promotes epithelial–mesenchymal transition through TGF-beta signaling and is transcriptionally regulated by Slug. Oncogene 30:4410–4427
Ke X, Fei F, Chen Y, Xu L, Zhang Z, Huang Q, Zhang H, Yang H, Chen Z, Xing J (2012) Hypoxia upregulates CD147 through a combined effect of HIF-1alpha and Sp1 to promote glycolysis and tumor progression in epithelial solid tumors. Carcinogenesis 33:1598–1607
Sidhu SS, Nawroth R, Retz M, Lemjabbar-Alaoui H, Dasari V, Basbaum C (2010) EMMPRIN regulates the canonical Wnt/beta-catenin signaling pathway, a potential role in accelerating lung tumorigenesis. Oncogene 29:4145–4156
Howe LR, Brown AM (2004) Wnt signaling and breast cancer. Cancer Biol Ther 3:36–41
Tetsu O, McCormick F (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422–426
Huang Q, Li J, Xing J, Li W, Li H, Ke X, Zhang J, Ren T, Shang Y, Yang H, Jiang J, Chen Z (2014) CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol 61:859–866
Takahashi M, Nakamura Y, Obama K, Furukawa Y (2005) Identification of SP5 as a downstream gene of the beta-catenin/Tcf pathway and its enhanced expression in human colon cancer. Int J Oncol 27:1483–1487
Fujimura N, Vacik T, Machon O, Vlcek C, Scalabrin S, Speth M, Diep D, Krauss S, Kozmik Z (2007) Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem 282:1225–1237
Aberle H, Bauer A, Stappert J, Kispert A. Kemler R (1997) beta-catenin is a target for the ubiquitin–proteasome pathway. EMBO J 16:3797–3804
Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83:249–289
Till A, Rosenstiel P, Brautigam K, Sina C, Jacobs G, Oberg HH, Seegert D, Chakraborty T, Schreiber S (2008) A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. J Cell Sci 121:487–495
Weiske J, Albring KF, Huber O (2007) The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA 104:20344–20349
Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T (2006) Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev 20:473–485
Kong LM, Liao CG, Fei F, Guo X, Xing JL, Chen ZN (2010) Transcription factor Sp1 regulates expression of cancer-associated molecule CD147 in human lung cancer. Cancer Sci 101:1463–1470
Niyazi M, Niyazi I, Belka C (2007) Counting colonies of clonogenic assays by using densitometric software. Radiat Oncol 2:4
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334
Sato T, Watanabe M, Hashimoto K, Ota T, Akimoto N, Imada K, Nomizu M, Ito A (2012) A novel functional site of extracellular matrix metalloproteinase inducer (EMMPRIN) that limits the migration of human uterine cervical carcinoma cells. Int J Oncol 40:236–242
Li Y, Wu J, Song F, Tang J, Wang SJ, Yu XL, Chen ZN, Jiang JL (2012) Extracellular membrane-proximal domain of HAb18G/CD147 binds to metal ion-dependent adhesion site (MIDAS) motif of integrin beta1 to modulate malignant properties of hepatoma cells. J Biol Chem 287:4759–4772
Desgrosellier JS, Cheresh DA (2010) Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 10:9–22
Ratnikov BI, Rozanov DV, Postnova TI, Baciu PG, Zhang H, DiScipio G, Chestukhina GG, Smith JW, Deryugina EI, Strongin AY (2002) An alternative processing of integrin alpha(v) subunit in tumor cells by membrane type-1 matrix metalloproteinase. J Biol Chem 277:7377–7385
Guvakova MA, Surmacz E (1997) Overexpressed IGF-I receptors reduce estrogen growth requirements, enhance survival, and promote E-cadherin-mediated cell-cell adhesion in human breast cancer cells. Exp Cell Res 231:149–162
Mauro L, Bartucci M, Morelli C, Ando S, Surmacz E (2001) IGF-I receptor-induced cell-cell adhesion of MCF-7 breast cancer cells requires the expression of junction protein ZO-1. J Biol Chem 276:39892–39897
Tapia R, Huerta M, Islas S, Avila-Flores A, Lopez-Bayghen E, Weiske J, Huber O, Gonzalez-Mariscal L (2009) Zona occludens-2 inhibits cyclin D1 expression and cell proliferation and exhibits changes in localization along the cell cycle. Mol Biol Cell 20:1102–1117
Gonzalez-Mariscal L, Bautista P, Lechuga S, Quiros M (2012) ZO-2, a tight junction scaffold protein involved in the regulation of cell proliferation and apoptosis. Ann N Y Acad Sci 1257:133–141
Friedl P, Gilmour D (2009) Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10:445–457
Schneckenleithner C, Bago-Horvath Z, Dolznig H, Neugebauer N, Kollmann K, Kolbe T, Decker T, Kerjaschki D, Wagner KU, Muller M, Stoiber D, Sexl V (2011) Putting the brakes on mammary tumorigenesis: loss of STAT1 predisposes to intraepithelial neoplasias. Oncotarget 2:1043–1054
Bailey SG, Cragg MS, Townsend PA (2012) Role of STAT1 in the breast. JAKSTAT 1:197–199
Acknowledgements
This work was supported by the Thüringer Aufbaubank (European Regional Development Fund Grants FE 9034 and FE 9053) and the Interdisziplinäres Zentrum für klinische Forschung (IZKF), Jena. We are grateful to Andreas Till for EMN cDNA and the EMN-knockdown constructs, to Oliver Krämer for the STAT1 expression construct and to Sonnhild Mittag for valuable discussions.
Author information
Authors and Affiliations
Contributions
NK performed and documented all experiments, OH designed, supervised and interpreted experiments concerned with Wnt pathway analysis, KF designed and supervised the study and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Knutti, N., Huber, O. & Friedrich, K. CD147 (EMMPRIN) controls malignant properties of breast cancer cells by interdependent signaling of Wnt and JAK/STAT pathways. Mol Cell Biochem 451, 197–209 (2019). https://doi.org/10.1007/s11010-018-3406-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-018-3406-9