Abstract
The knowledge regarding the importance of long non-coding RNAs (lncRNAs), a new class of genes, is very sparse in osteosarcoma. In the present study, we describe the expression profile of lncRNAs in osteosarcomas compared with paired adjacent non-cancerous tissue (n = 7) using microarray analysis. A total of 25,733 lncRNAs were identified in osteosarcoma; 1995 lncRNAs were consistently upregulated and 2226 lncRNAs were consistently under-regulated in all samples analyzed (≥2.0-fold, p < 0.05). We have validated three over-regulated and three under-regulated lncRNAs in patient samples (n = 7). The antisense transcript of SATB2 protein (SATB2-AS1) was identified as one of the upregulated lncRNAs. The SATB2-AS1 is a 3197-bp lncRNA on chromosome 2. This is the first report, where we have documented the increased expression of SATB2-AS1 in osteosarcoma patients and in human osteosarcoma cancer cell lines (U2OS, HOS, MG63). SATB2-AS1 expression was significantly higher in the metastatic tumors compared to non-metastatic tumors. In vitro gain and loss of function approaches demonstrated that SATB2-AS1 regulates cell cycle, cell proliferation, and cell growth. In addition, SATB2-AS1 affects the translational expression of SATB2 gene. Our data demonstrate that an antisense non-coding RNA regulates the expression of its sense gene, and increases the cell growth, therefore pointing the pivotal functions of SATB2-AS1 in osteosarcoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most common type of primary bone cancer [1, 2]. The American cancer statistics (2016) report bone and joint tumors as the common cancer in children and adolescents under the age of 20 [3]. The majority of osteosarcomas originate in the long bones, such as the distal femur and proximal tibia. This cancer is highly aggressive with a metastatic rate of ~20%, with the most common targets being the lung and other bones [2, 4]. Despite recent advancements in multimodal treatments (e.g., wide tumor excision, adjuvant chemotherapy, and radiotherapy), survival expectancies have not improved much over the past 25 years, especially for the patients with metastasis and recurrent osteosarcoma [5]. The exact mechanisms underlying the pathogenesis and progression remain still enigmatic. Therefore, identification of new candidate molecules participating during the pathophysiology becomes pertinent to improve the diagnosis, prevention, and treatment.
Long non-coding RNAs (lncRNAs), members of the non-coding RNA family are a class of RNA over 200 nucleotides in length with no protein-coding potential [6]. It is clear now that this class of RNAs plays crucial roles in the regulation of a wide array of biological processes such as development, growth, apoptosis, and tumorigenesis by regulating gene expression at the transcriptional or post-transcriptional level. Dysregulated expression of lncRNAs has been established in various cancers; therefore, they have recently been of interest to many researchers worldwide [6, 7].
In the present study, microarray analysis revealed a significant dysregulation of lncRNAs in osteosarcoma samples when compared to their paired adjacent non-cancerous tissues. We have validated the expression of three over-regulated (AC017096.1, AL589743.1, AC092484.(1) and three under-regulated (TRHDE-AS1, LOC100505633, AC116614.1) lncRNAs. Looking closely at the lncRNAs which we have validated using qPCR, we observed that lncRNA AC017096.1 is oriented in antisense direction with respect to the SATB2 gene. SATB2 (special AT-rich sequence binding protein (2)) is a multifunctional transcriptional regulator, which promotes osteoblast differentiation, maturation and bone formation, regulates osteosarcoma invasion, and promotes osteosarcoma formation [8, 9].
We found that AC017096.1 is a natural antisense lncRNA to SATB2, and this lncRNA was named as SATB2-AS1. Here, we report that SATB2-AS1, an oncogenic lncRNA, increases cell proliferation, cell growth, and regulates the cell cycle of osteosarcoma cells in vitro. Here, we demonstrated that SATB2-AS1 effects the translational expression of sense gene SATB2. Our findings suggest the altered expression levels of lncRNAs contribute to the occurrence and molecular processes of osteosarcoma.
Materials and method
Patient samples
Subjects were recruited in the present study at the Guangzhou First People’s Hospital, China after obtaining a written informed consent from all the patients. Cancerous tissue and pair-matched non-cancerous tissue were obtained by biopsy. None of the patients underwent chemotherapy or radiotherapy before surgery. This study was performed with the approval from the Ethics Committee of the Guangzhou First People’s Hospital.
Microarray and data analysis
mRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre) was extracted from the samples and submitted to KangChen Bio-tech Company, China for microarray analysis. For microarray analysis, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols. Each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Human LncRNA Array v3.0 (8 × 60 K, Arraystar) and the Agilent Scanner G2505C scanned the arrays.
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, lncRNAs that has at least seven out of 14 samples have flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. Differentially expressed lncRNAs with statistical significance between the two groups were identified through Volcano Plot filtering. Hierarchical Clustering was performed to show the distinguishable lncRNAs expression pattern among samples. Pathway analysis and GO analysis were applied to determine the roles of these differentially expressed RNA.
Cell cultures
The cell lines U2OS, HOS, MG63, and HLF (human fibroblast like) were obtained from ATCC, and were maintained as per ATCC guidelines. Cells were incubated in a humidified 37 °C incubator with a 5% CO2 atmosphere. All cells used in the experiments were at the exponential growth phase.
RNA isolation and qRT-PCR
Total RNA was extracted from tissues and cultured cells using the Trizol reagent (Invitrogen). First-strand cDNA was synthesized using SuperScript™ III Reverse Transcriptase (Invitrogen). qPCR was performed using the SYBR Green PCR Kit (Takara) in the ABI7500 real-time PCR detection system (Applied Biosystems). For each sample, gene expression was normalized to the levels of GAPDH. The primer sequences used are listed in Table 1.
Plasmid constructs, virus production, and infections
SATB2-AS1 overexpression and knockdown of stable cells were established and purchased from Shanghai Gene Chem Co Ltd, China. For loss of function studies, the SATB2-AS1 was knocked down using siRNA in lentivirus system. Briefly, commercially synthesized three different SATB2-AS1 targeting siRNAs were used in the present study: siRNA1-5′-CCUAAUACCUUCUCACUGUTT-3′; siRNA-2 5′-GUAUUGCACUUGCAUCAAATT-3′, and siRNA-3 5′-GCAGAUACCGGCAGGGAATT-3′ (Invitrogen). For gain of function studies, SATB2-AS1 was cloned and overexpressed in cells using lentivirus vector (Vector name: GV303, Component order: Ubi-MCS-SV40-EGFP, Enzyme cutting site: AgeI/NheI). Osteosarcoma cells were transfected with lentivirus (MOI = 10) harboring RNAi sequence and overexpression sequence targeting lncRNA (AC017096.1). Stable cells were selected, and the knockdown and overexpression of lncRNA (AC017096.1) were confirmed using real-time PCR.
Cell proliferation assay
Cells were seeded in triplicate on six-well plates, and cell proliferation over time was measured by counting the number of cells by trypan blue exclusion at the indicated time points.
Measurement of apoptosis
Infected cells were collected by trypsinization, washed twice with PBS, and suspended in staining buffer (5 μl Annexin V-APC/100 μl) (KGA1023, KeyGen BioTECH) for 15 min. Apoptotic cells were sorted by FCM (BD FACSCalibur, USA).
Cell cycle analysis
Cell cycle distribution was analyzed by flow cytometry using Cycle test plus kit (KGA512, KeyGen BioTECH) as per the manufacturer’s instructions.
Cytosolic and nuclear fractionation
Nuclear protein separation kit (P0027, Beyotime) was used to isolate the cytosolic and nuclear fractions from cells as per manufacturer’s protocol.
Wound healing assay
A wound healing assay was used to determine the migratory potential of the cells. Cell motility and migration characteristics were analyzed by wound healing assay. Briefly, the cells were grown to a confluent monolayer on a six-well tissue culture plate. The cell monolayers were “wounded” using a P200 micropipette tip. The wounded monolayers were washed twice with Dulbecco’s phosphate buffered saline (Boster, Wuhan, China) to remove the cell debris, and incubated in culture medium supplemented with 0.1% FBS for 24 h. The migrating cells in the denuded zone were monitored using an inverted microscope equipped with a camera (Leica, Wetzlar, Germany). The cell migration distance was evaluated by measuring the width of the wound.
Protein extraction and western blot analysis
Cell lysates were prepared in lysis buffer (Beyotime) with protease and phosphatase cocktail inhibitor (ThermoFisher Scientific). Cellular debris was cleared from lysates by centrifugation, and protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad). Samples were separated on precast 4–15% TGX-PAGE (Bio-Rad), transferred to a PVDF membrane (Bio-Rad), blotted with the indicated antibodies [SATB2 (Abcam-EPNCIR130A) and GAPDH (Proteintech-10494-1-AP)], and visualized with enhanced chemiluminescence substrate (Tiangen Biotech, Beijing, China).
Statistical analysis
The results are reported as the means ± SDs. Quantification of Western blots was performed using ImageJ (NIH Software). Statistical analysis (two-tailed t test or ANOVA) was based on a minimum of three replicates using Prism 5 (version 5, Graphpad Software). The differences were considered significant if p ≤ 0.05.
Results
Differentially expressed lncRNAs in osteosarcomas
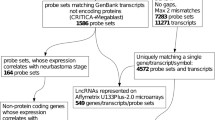
A significant and differential expression of lncRNAs was observed in normal bone tissue and osteosarcoma (Fig. 1a, b). Microarray analysis revealed the presence of a total of 25,733 lncRNAs in osteosarcoma. We identified 1995 over-regulated lncRNAs and 2226 under-regulated lncRNAs that were significantly differentially expressed in these human primary osteosarcomas (≥2.0-folds) (Data not shown).
a, b The profiles of differentially expressed lncRNAs in osteosarcoma when compared to the paired non-cancerous tissues. a Heat map and Hierarchical Clustering for “Differentially Expressed LncRNAs in T vs. C”. “Red” indicates high relative expression, and “blue” indicates low relative expression. b The scatter plot demonstrating the variations of the lncRNAs. The values of X and Y axes in the Scatter Plot are the normalized signal values of the samples (log2 scaled) or the averaged normalized signal values of groups of samples (log2 scaled). The green lines are Fold Change Lines (The default fold change value given is 2.0). The LncRNAs above the top green line and below the bottom green line indicated more than 2.0-fold change of LncRNAs between the two compared samples or the two compared groups of samples. c Six of the differential expression of lncRNAs were validated by qRT-PCR (three over-regulated and three under-regulated) in osteosarcoma and their paired non-tumor tissues (n = 7). d GO pathway analysis. e. Schematic outlining the genomic organization of SATB2-AS1 and SATB2. f SATB2-AS1 expression significantly upregulated in the metastatic osteosarcoma (n = 15) samples. ***p ≤ 0.001. (Color figure online)
Following this data, we compared the lncRNA expression levels between seven human primary osteosarcomas and their paired adjacent non-cancerous tissues. We have validated the expression of three over-regulated (AC017096.1, AL589743.1, AC092484.1) and three under-regulated (TRHDE-AS1, LOC100505633, AC116614.1) lncRNAs in these samples (Fig. 1c).
Construction of the coding-non-coding gene co-expression network
Cytoscape (http://www.cytoscape.org/) was used to draw coding and non-coding expression network of the six validated lncRNAs and observed that 246 coding genes have a relationship with these six validated lncRNAs (Data not shown). GO analysis showed that 180 of these genes relate to protein binding, and continue to subdivide these 180 genes, and found one-third of these genes relate to binding and catalytic activity, respectively (Fig. 1d). These analyses suggested that these lncRNAs play an important role in protein binding and catalytic activities. Future studies are needed to explore this aspect of the osteosarcoma-related lncRNAs in the pathophysiology of disease.
The high expression of SATB2-AS1 related to osteosarcomas metastasis
In the present study, we chose to characterize and study the functional role of an overregulated lncRNA AC017096.1. Our analysis reveled that this lncRNA is located at chromosome 2, and is oriented in antisense direction with respect to the SATB2 gene and is complimentary for about 3197 nucleotides (Fig. 1e). Finding it as a natural antisense transcript further in text lncRNA AC017096.1 will be addressed as SATB2-AS1. As shown in Fig. 1c, SATB2-AS1 (AC017096.1) is increased in the osteosarcoma compared to paired non-tumor tissues. Further, we observed that SATB2-AS1 expression is significantly high in the metastatic tumors compared to non-metastatic tumors (n = 15) (Fig. 1f).
SATB2-AS1 promotes cell proliferation and cell migration of osteosarcoma cells in vitro
To further investigate whether SATB2-AS1 could modulate the proliferation and migration of osteosarcoma cells, we tested three different human osteosarcoma cell lines (MG63, HOS, and U2OS) which express the SATB2-AS1 (Fig. 2a), and chose the two cell lines which had highest expression of SATB2-AS1 in our hands (U2OS and HOS). Silencing and overexpression of SATB2 were confirmed by qPCR (Fig. 2b–d). In the present study, we used three different siRNA constructs. SiRNA-2 was found to be more efficient, compared to siRNA1 and siRNA3; therefore, all experiments were done using siRNA2. Silencing of SATB2-AS1 demonstrated a significant decrease in the cell proliferation as determined by cell counting assay in both the cell line (Fig. 3a, b). Also, overexpression of SATB2-AS1 leads to hyperproliferation of the cells (Fig. 3c, d). In corroboration, silencing of SATB2-AS1 inhibited the wound closure significantly (Fig. 3e–h). These in vitro results demonstrated significance of SATB2-AS1 in cell proliferation and cell growth.
a Expression of SATB2-AS1 in different cells lines as determined by qPCR. GAPDH was used as a reference control. b, c Confirmation of silencing of SATB2-AS1 in osteosarcoma cell lines (U2OS and HOS) as determined by qPCR. Three different siRNA viral constructs were used, and scrambled RNA (scrRNA) was used as a control. Expression was calculated compared to mock cells. d SATB2-AS1 was overexpressed and the overexpression was determined by qPCR in osteosarcoma cells. For all panels, data are shown as mean ± SD based on at least three independent experiments
SATB2-AS1 promotes cell proliferation cell migration of osteosarcoma cells in vitro. a, b Knock down of SATB2-AS1 downregulates the cell proliferation of osteosarcoma cells as determined by cell counting assay. c, d Overexpression of SATB2-AS1 upregulates the cell proliferation as determined by cell counting assay. Knock down of SATB2-AS1 decreases migration of osteosarcoma cells as determined by wound healing assay. e, f Representative pictures of the wound healing migration assay. g, h Quantitative measurements of the wound closure in the different cells. Panels A, C, E, and G represent results for the U2OS, and panels B, D, F, and H represent the results for HOS cells. For all panels, data are shown as mean ± SD based on at least three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
Silencing of SATB2-AS1 promotes G1 arrest and causes apoptosis
To determine the potential mechanism of SATB2-AS1 in the osteosarcoma cells proliferation, we measured the distribution of cell cycle phases of U2OS and HOS cells in the absence of SATB2-AS1. As shown in Fig. 4a, b, the percentage of cells in G0/G1 phase increased significantly, while that of cells at S phase decreased markedly after SATB2-AS1 knockdown. In addition, the percentage of early apoptotic cells is significantly increased with the silencing of the SATB2-AS1 (Fig. 4c, d, Supplemental Figs. 1, 2). Together, these results suggest silencing of SATB2-AS1 inhibits osteosarcoma cells proliferation through inhibiting cell cycle progression and promoting the cell apoptosis.
Silencing of SATB2-AS1 promotes G1 arrest and causes apoptosis. a, b A significant increase in the G0/G1 phase of the cells in osteosarcoma cells. c, d Significant increase in the early apoptotic cells with the silencing of the SATB2-AS1. Results are compared to the mock cells. For all panels, data are shown as mean ± SD based on at least three independent experiments. *p ≤ 0.05
Cellular localization of SATB2-AS1
We examined the localization of SATB2-AS1 lncRNA in two osteosarcoma cell lines and found that SATB2-AS1 is present in both the cytoplasmic and nuclear fractions. However, the higher abundance of SATB2-AS1 was found to be in the cytoplasmic fractions in both the cells (Fig. 5).
SATB2-AS1 affects the translation expression of SATB2 gene not the transcriptional expression
Long non-coding RNAs with an antisense orientation toward known protein-coding genes are demonstrated to significantly contribute to the repertoire of regulatory mechanisms that are used by mammalian cells to modulate gene expression [6]. As stated earlier that SATB2-AS1 is an antisense lncRNA to the gene SATB2, in literature, many reports have shown that antisense transcripts regulate the expression of sense genes, especially for non-coding RNAs as antisense transcripts [6]. Therefore, as a natural antisense transcript, we were interested to know the association of these two at the transcriptional and translational level. We hypothesize that SATB2-AS1 can regulate the SATB2 expression and then contribute to osteosarcoma development. To verify this hypothesis, firstly we analyzed the correlations of the expression levels of SATB2-AS1 and SATB2 in osteosarcoma biopsies and their paired adjacent peri-tumoral samples. Surprisingly, the levels of mRNA expression of SATB2 in osteosarcoma did not differ when compared to their paired non-cancerous tissues (Fig. 6). To confirm this, we checked the expression SATB2 in SATB2-AS1 knockdown and overexpressed cells. We observed that the silencing of SATB2-AS1 in both the cellular models (U2OS and HOS), mRNA expression of SATB2, remains unaffected (Fig. 7a, b, d, e); however, the protein expression of SATB2 was found to be directly correlated to the SATB2-AS1 expression (Fig. 7c, f).
SATB2-AS1 effects the translation expression of SATB2 gene, not the transcriptional expression. a, b Knockdown and d, e overexpression of the SATB2-AS1 do not affect the transcriptional expression of the SATB2 gene in osteosarcoma cells as measured by the qRT-PCR. c Knockdown and f overexpression of the SATB2-AS1, respectively, downregulate and upregulate the translational expression of the SATB2 as determined by the western blot (representative blots are shown). For all panels, data are shown as mean ± SD based on at least three independent experiments. ***p ≤ 0.001
Discussion
Newer technologies have led to the identification of thousands of lncRNAs across wide array of cells and is now well accepted that lncRNAs play critical role as epigenetic regulators, as scaffolds for protein binding partners or modulate the activity of other coding and non-coding RNAs [6]. Despite these advancements, the involvement of lncRNAs in osteosarcoma pathophysiology is not widely studied.
In the present study, using microarray analysis, we found that lncRNA expression profiles were altered in the osteosarcoma compared to adjacent non-cancerous tissues. Previously, Li et al. have also described the dysregulated profile of lncRNAs in the osteosarcoma [10]. Using the similar statistical parameters, we have found almost five times over-regulated (1995 vs. 403) and three times under-regulated (2226 vs. 798) lncRNAs compared to the previous report by Li et al. [10]. These results suggest that there is sample-to-sample variation, and more studies are needed to reach the point where we can draw a firm conclusion. Therefore, caution must be exercised in extrapolating data based on small study samples. Comparing and thoroughly analyzing the results from various studies reported worldwide are needed to provide a direction for future research. Many lncRNAs including MALAT-1 [11], the lncRNA SNHG12 (Small Nucleolar RNA Host Gene 12) [12], and HULC (Highly Upregulated in Liver Cancer) [13], are upregulated in osteosarcoma patients, and lncRNA TUSC7 (Tumor Suppressor Candidate 7) [14] was downregulated in osteosarcoma, and in vitro studies show that they play important roles in the disease.
Mechanistically, lncRNAs regulate expression of neighboring protein-coding genes and also regulate gene expression via a trans-acting mechanism by associating with protein complexes at the levels of transcription, post-transcriptional modification, and even translation [6]. The results of the gain and loss of function experiments revealed that SATB2-AS1 did not affect the transcriptional expression of the SATB2 gene; however, the translational expression is affected. We do not know the exact reason and mechanism for this, and more studies in future can be carried out in this direction. Biologically, the discrepancies between mRNA and protein expression can be caused by post-transcriptional regulation, as well as differences in mRNA and protein turnover rates [15, 16]. However, the degree of validity of this key assumption has seldom been answered with certainty. Further insights into the other functional aspects and clinical implications of this lncRNA may facilitate the identification of novel diagnostic or prognostic biomarkers and various drug targets for osteosarcoma.
It is suggested that lncRNAs may exert their functions through interactions with coding transcripts and proteins in osteosarcoma. An investigation into the connections between signal pathways and lncRNAs is critical for developing novel strategies for the early diagnosis and treatment of osteosarcoma. Since, SATB2 plays a crucial role in the regulation of bone development and differentiation of osteoblasts [8], future studies to explore the mechanistic association of SATB2 and SATB2-AS1 is needed to understand the molecular mechanisms and biological functions of lncRNAs in osteosarcoma.
With respect to osteosarcoma, research area on functional aspects of non-coding RNAs is still in its infancy and we predict that outcome will significantly increase over the coming years, which will provide us important knowledge. In conclusion, we for the first time established that SATB2-AS1 is an oncogenic non-coding RNA which increases cell proliferation and cell growth of osteosarcoma cells.
References
Siclari VA, Qin L (2010) Targeting the osteosarcoma cancer stem cell. J Orthop Surg Res 5:78. doi:10.1186/1749-799X-5-78
Yan GN, Lv YF, Guo QN (2016) Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett 370:268–274. doi:10.1016/j.canlet.2015.11.003
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66:7–30. doi:10.3322/caac.21332
Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G (2006) Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treat Rev 32:423–436. doi:10.1016/j.ctrv.2006.05.005
Kager L, Zoubek A, Potschger U, Kastner U, Flege S, Kempf-Bielack B, Branscheid D, Kotz R, Salzer-Kuntschik M, Winkelmann W, Jundt G, Kabisch H, Reichardt P, Jurgens H, Gadner H, Bielack SS and Cooperative German-Austrian-Swiss Osteosarcoma Study G (2003) Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J Clin Oncol 21:2011–8. doi:10.1200/JCO.2003.08.132
Bartonicek N, Maag JL, Dinger ME (2016) Long non-coding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer 15:43. doi:10.1186/s12943-016-0530-6
Huynh NP, Anderson B, Guilak F, McAlinden A (2016) Emerging roles for long non-coding RNAs in skeletal biology and disease. Connect Tissue Res. doi:10.1080/03008207.2016.1194406
Zhao X, Qu Z, Tickner J, Xu J, Dai K, Zhang X (2014) The role of SATB2 in skeletogenesis and human disease. Cytokine Growth Factor Rev 25:35–44. doi:10.1016/j.cytogfr.2013.12.010
Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125:971–986. doi:10.1016/j.cell.2006.05.012
Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T (2013) Microarray expression profile of long non-coding RNAs in human osteosarcoma. Biochem Biophys Res Commun 433:200–206. doi:10.1016/j.bbrc.2013.02.083
Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA (2012) A triple helix stabilizes the 3′ ends of long non-coding RNAs that lack poly(A) tails. Genes Dev 26:2392–2407. doi:10.1101/gad.204438.112
Ruan W, Wang P, Feng S, Xue Y, Li Y (2016) Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol 37:4065–4073. doi:10.1007/s13277-015-4256-7
Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC (2015) Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol 8:2994–3000
Cong M, Li J, Jing R, Li Z (2016) Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. doi:10.1007/s13277-015-4414-y
Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, Jiang H, Tan L, Xie J, Zhu X, Liang S, Deng H (2008) How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin (Shanghai) 40:426–436
Mahajan N, Bisht D, Dhawan V, Singh S, Minz RW (2010) Transcriptional expression and gelatinolytic activity of matrix metalloproteinases in Henoch-Schonlein purpura. Acta Paediatr 99:1248–1252. doi:10.1111/j.1651-2227.2010.01781.x
Acknowledgements
The National Nature Science Foundation of China (Nos. 31601031, 81272941, 81201864), and The Guangdong Planned Project of Science and Technology (2014A020212009) financially supported the present study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, SH., Zhu, JW., Xu, HH. et al. A novel antisense long non-coding RNA SATB2-AS1 overexpresses in osteosarcoma and increases cell proliferation and growth. Mol Cell Biochem 430, 47–56 (2017). https://doi.org/10.1007/s11010-017-2953-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-017-2953-9