Abstract
Signaling transduction pathways are established by interactions between growth factors, protein kinases, and transcription factors, and they play a crucial role in tooth development. Precursor cells of the dental follicle (DFCs) are used for in vitro studies about molecular mechanisms during periodontal development. Previous studies have already shown that the growth factor BMP2 and the transcription factor EGR1 are involved in the osteogenic differentiation in DFCs while interactions with protein kinase-based pathways remain elusive. In this current study, we investigated the role of the AKT kinase signaling pathway for the osteogenic differentiation in DFCs. The AKT signaling pathway was activated in DFCs after the induction of the osteogenic differentiation by BMP2. The inhibition of AKT in DFCs repressed the differentiation and the expression of the transcription factor EGR1. Interestingly, EGR1 bound to the phosphorylated form of SMAD1/5 (pSMAD). The binding of pSMAD to EGR1 was increased after the induction with BMP2. Moreover, the overexpression EGR1 increased the osteogenic differentiation of DFCs. Our results suggest that the AKT signaling pathway submits the BMP2-dependent osteogenic differentiation in DFCs via the expression of the transcription factor EGR1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Signaling pathways such as the BMP pathway control the spatial and temporal regulation of the development of mineralized tissues [1,2]. Cultivated cells from the dental follicle (DFCs) contain precursor cells for alveolar osteoblasts and cementoblasts and cell cultures with DFCs are valuable for investigations under in vitro conditions about molecular mechanisms during the development of mineralizing periodontal tissues [3,4].
Recently, we reported that the BMP2/DLX3 pathway control partially the osteogenic differentiation in human DFCs [5]. Previous studies have shown that DLX3, which is an essential transcription factor for skeletal development, is induced not only by BMP2 but also through post-transcriptional modifications such as phosphorylation by the serine/threonine protein kinase AKT1 [5-7]. The phosphorylation of DLX3 by AKT1 enhances the function of DLX3 for the osteogenic differentiation by supporting protein stability and transcriptional activity [7]. AKT1 is part of the phosphatidylinositol 3 kinase (PI3K)/AKT signaling pathway and similar to DLX3 this pathway regulated the proliferation and differentiation of mineralizing cells [5,8].
The relevance of multipathway kinase signatures for the osteogenic differentiation was recently shown in bone marrow-derived multipotent stromal cells (MSCs) [9] and particular the activation of the (PI3K)/AKT signaling pathway is required for differentiation into osteoblast [10]. Here, BMP2 induces the (PI3K)/AKT signaling pathway that regulates the BMP2-specific SMAD function during osteoblast differentiation [10].
Recently, we showed that DLX3 induces the expression of the transcription factor early growth response gen (EGR)1 in DFCs [5]. Interestingly, this transcription factor is also regulated through diverse signaling pathways such as the mitogen-activated protein (MAP) kinase and AKT [11,12] and participates in multiple biological processes including cell proliferation and differentiation [13,14]. Moreover, we have already shown that EGR1 supports the osteogenic differentiation of dental stem cells [15], but an interaction between BMP2, AKT, and EGR1 has not been shown for the osteogenic differentiation of dental stem cells. In this study, we showed for the first time an interaction between BMP2, AKT, and EGR1, and the relevance of this signal for the osteogenic differentiation of dental stem cells.
Materials and methods
Cell culture
Human dental follicle cells (DFCs) were isolated as described previously [5,15]. Briefly, the dental follicle was minced and digested in a solution of collagenase type I, hyaluronidase (Sigma-Aldrich, Munich, Germany), and DNase I (Roche, Mannheim, Germany). The digested tissue suspension was seeded into cell culture flasks in standard culture medium (Sigma-Aldrich, Munich, Germany) at 37 °C in 5 % CO2. The standard culture medium (DMEM) consists of Dulbecco’s Modified Eagle Medium (Sigma-Aldrich) supplemented with 10 % fetal bovine serum, FBS (Sigma-Aldrich), and 100 μg/mL Penicillin/Streptomycin. For experiments, DFCs were used at cell passage 6.
Osteogenic differentiation
DFCs were cultivated until subconfluence (>80 %) in standard cell culture medium before they were stimulated with BMP2 (10 ng/ml) comprised Dulbecco’s Modified Eagle Medium (Sigma-Aldrich) supplemented with 1 % fetal bovine serum (Sigma-Aldrich), 100 μmol/l ascorbic acid 2-phosphate, 10 mmol/l KH2PO4, 1 × 10−8 mol/L dexamethasone sodium phosphate (Sigma-Aldrich), HEPES (20 mmol/l), and 100 μg/ml Penicillin/Streptomycin. For inhibition of AKT the AKT inhibitor, IV (Calbiochem) was added to DMEM or BMP2 (10 ng/ml) at a final concentration of 1.25 µM, respectively.
Transfection experiments
Two days before transfection, DFCs were seeded at a cell density of 7 × 103 cm2. For transfection, the cells were grown until subconfluence and cultivated in DMEM-containing antibiotics. DFCs were transiently transfected with the EGR1 expression plasmid, pCMV6-EGR1 (pEGR1) (OriGen, Rockville, MD). The transfection was performed using the FUGEN HD Transfection Reagent (Roche) according to the manufacturer’s instructions. Reaction complexes were formed from the combination of FUGEN HD and the plasmid diluted in DMEM in a ratio of 5:1.5 (5 µl FUGEN HD per 1.5 µg DNA).
As control, the empty vector without an insert, pCDNA3.1 (+ ) (Invitrogen) (pEV) was used. The expression of EGR1 in DFCs was determined by quantitative reverse transcription polymerase chain reaction (RT-qPCR) after 48 h.
Measurement of alkaline phosphatase (ALP) activity
For evaluation of the osteogenic differentiation, potential of DFCs was quantified the ALP activity after 10 days of cultivation in BMP2 (10 ng/ml) or standard culture medium (DMEM). DFCs were washed with PBS buffer, disrupted by freezing −80 °C, and thereafter 60 µl of each sample was mixed with 60 µl of 1.5 M alkaline buffer (Sigma) and 100 µl of 100 mM p-nitrophenylphosphate (Sigma). The reaction was stopped after 60 min incubation at 37 °C by adding 300 μl of 0.3 M NaOH. The liberated p-nitrophenol was measured spectrophotometrically at 405 nm. ALP activity values were normalized to total DNA concentration determined by Quant-iT PicoGreen dsDNA Assay (Invitrogen). At least three biological replicates were analyzed for each condition, and the data were presented as the means ± S.E. (σ/√n).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cell using RNA isolation kit (Qiagen). The cDNA synthesis was performed using 500 ng total RNA and QuantiTect Reverse Transcriptase Kit (Qiagen). Quantitative RT-PCR (RT-qPCR) was conducted with the Fast Start DNA Master SYBR® Green I kit (Roche) or the Light Cycler TaqMan kit (Roche) and the Light Cycler PCR System (LightCycler 480 II, Roche). For normalization was used the mean of the control group gene expression and as the housekeeper control was used GAPDH. Primer pairs used for RT-qPCR are listed in (Table 1).
Immunoblotting
DFCs were washed with PBS at indicated points in time and harvested by scraping after 5 min incubation on ice with lysis buffer (20 mM Tris, 48 mM NaF, 150 mM NaCl, 2 mM Na-Orthovanadate, 1 % NP-40 and 10 % Glycerol) and freshly added protease (Roche) and phosphatase (Sigma) inhibitor cocktail. An aliquot of 20 µg protein extract was denatured and reduced by boiling in lithium dodecyl sulfate (LDS) sample buffer containing DTT, separated by SDS–polyacrylamide electrophoresis in NuPAGE® Novex® 4–12 % Bis–Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were blocked with skimmed milk and incubated for 60 min at room temperature followed by incubation with primary antibody at 4 °C overnight. The used antibodies are as follows: EGR1 (15F7), phosphoAKT(S473) (D9E), phosphoSMAD1/5 (41D10), LaminA/C, pmTOR(S2481) (Cell Signaling, Beverly, MA), and β-Actin (AC-15) (Novus Biologicals).
After washing, the membranes were incubated either with a biotinylated anti-mouse IgG or a biotinylated anti-rabbit IgG for 60 min followed by incubation with avidin-conjugated horseradish peroxidase (HRP). Detection was performed by chemiluminescence (Pierce). For quantitation, X-ray films were scanned and the program ImageJ version 1.47 (public domain; http://imagej.nih.gov/ij/) was used for the evaluation of protein band intensities. For calculation of relative pixel intensities of protein bands, the measured intensities were normalized to the intensities of the housekeeping protein β-Actin.
For nuclear and cytoplasmic protein partition, the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) were used according to the manufacturer's instructions. Solutions for protein extraction contain Protease Inhibitor tablets (complete mini, Roche) to minimize protein degradations.
The activation of the AKT signaling pathway was assayed using the PathScan AKT Signaling Antibody Array Kit (Cell Signaling), chemiluminescent signals were detected with the imaging system, and ChemiDoc (PEQLAB) and the spot intensities were quantified using the ImageJ software.
Immunoprecipitation
Immunoprecipitation assays were performed using the Protein G Agarose Immunoprecipitation Kit (Abnova, Walnut, USA) and following the protocol described by the manufacturers. DFCs were lysed in buffer containing 150 mM NaCl, 50 mM Tris, 1 % tritonX-100 together with freshly added protease Inhibitor (Roche), and phosphatase inhibitor cocktail (Sigma-Aldrich). Cleared cell lysates were incubated overnight at 8 °C with a specific antibody against EGR1 (Cell Signaling). To capture antibody protein complex, protein G beads were added and incubated by shaking the tubes for 1 h at 8 °C. After washing four times in lysis buffer, immunoprecipitates were eluted by boiling the beads with 1X SDS-PAGE loading buffer for 5 min and assayed by Western blotting as above.
Results
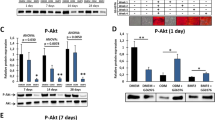
AKT pathway array. Protein lysates of the DFCs after 3 days of osteogenic induction with BMP2 or in DMEM for control were assayed using the PathScan AKT Signaling Antibody Array Kit. The pixel density of the control (DFCs in DMEM) was used for calibration. All values are means plus standard error (σ/√n) of three biological replicates. Significant differences are shown with the Student’s t test (**p < 0.0125 and ***p < 0.005)
Influence of AKT inhibitor on the phosphorylation of AKT, SMAD1/5, and mTOR and the expression of the transcription factor EGR1. A As indicated by immunoblotting, a specific AKT inhibitor (AKT Inh) hampers the phosphorylation of AKT at Serin 473 after 6 h supplementation; for quantitation supplemental Fig. 2. B Western blot analyses of DFCs nuclear extracts using specific antibodies for phospho-mTOR (S2481), EGR1, phospho-SMAD1/5, β-Actin as whole protein loading control and Laminin A/C for nuclear loading control after 3 days osteogenic differentiation induced with BMP2 or BMP2 and AKT Inh or in DMEM; for quantitation supplemental Fig. 3. C RT-qPCR of EGR1 in DFCs treated with BMP2 (Ctr.) or BMP2 and AKT inhibitor (AKT Inh) at day three of osteogenic differentiation. All values are means of three biological replicates ± SEM
AKT influences alkaline phosphatase (ALP) expression and activity. A RT-qPCR of ALP in DFCs treated with BMP2 or BMP2 and AKT inhibitor (AKT Inh) at day three of osteogenic differentiation. All values are means of three biological replicates ± SEM. B Alkaline phosphatase (ALP) activity after 10 days osteogenic differentiation with BMP2 or BMP2 and AKT Inh of DFCs. For calibration, DFCs in DMEM were used. All values are means ± SEM of at least four biological replicates. Significant differences are shown with the Student’s t test (***p < 0.005)
EGR1 interacts with the phosphorylated form of SMAD1/5. Immunoprecipitation (IP) assay with an EGR1 antibody with cell lysates from DFCs in DMEM or DFCs induced three days with BMP2. Immunoprecipitates were analyzed with specific antibodies against phospho-SMAD1/5 and EGR1. IPs with an unspecific rabbit IgG were used as negative control. The Diagram shows relative pixel intensities of pSMAD1/5 after EGR1 immunoprecipitation. Protein band pixel intensities of pSMAD1 were normalized to pixel intensities of EGR1 protein bands
EGR1 overexpression induces the alkaline phosphatase (ALP) activity in DFCs. A Western blot analyses with whole lysates of DFCs after 72 h transfection with the EGR1 plasmid (pEGR1) or an empty vector (pEV) in DFCs and specific antibodies against EGR1 and β-Actin, which was used as loading control. Diagram shows the relative pixel intensities of EGR1 normalized to the loading control. B Alkaline phosphatase (ALP) activity of DFCs after 48 h transfection with the EGR1 plasmid (pEGR1) or an empty vector (pEV) and 10 days osteogenic differentiation with BMP2. For calibration, pEV-transfected DFCs in BMP2 were used. All values are means ± SEM of at least four biological replicates
Analyses with the PathScan AKT Signaling Antibody Array Kit and Western blots discovered an increased phosphorylation (activation) level of proteins of the AKT signaling pathway after the induction of the osteogenic differentiation with BMP2 (Fig. 1, Fig. S1). Furthermore, the phosphorylation/activation of proteins such as AKT and SMAD1/5 was decreased using a specific inhibitor for AKT (AKT Inh) (Fig. 2). The AKT inhibitor repressed the BMP2-induced expression of EGR1 and the activation (phosphorylation) of mammalian target of rapamycin mTOR (S481) after 3 days induction with BMP2 (Fig. 2B). The AKT inhibitor inhibited the expression of EGR1 in DFCs (Fig. 2C) and the osteogenic differentiation of DFCs (Fig. 3). Interestingly, the interaction of the phosphorylated form of SMAD1/5 with the transcription factor EGR1 increased after the induction of the osteogenic differentiation with BMP2 (Fig. 4). These results lead to the assumption that the AKT signaling pathway, which is induced by BMP2, supports the osteogenic differentiation in DFCs via the transcription factor EGR1. To evaluate the influence of EGR1 on the osteogenic differentiation in DFCs, this transcription factor was overexpressed (Fig. 5A). The overexpression of EGR1 induced significantly the ALP activity in DFCs (Fig. 5B).
Discussion
Intracellular protein kinase-based signaling pathways such as the FAK/ERK pathway play important roles in the regulation of the osteogenic differentiation of DFCs [16,17]. Although the regulation of other protein kinases such as AKT was reported for the osteoblastic differentiation [8,10], almost nothing was known about the role of AKT during the osteogenic differentiation of dental stem cells.
We showed that DFCs express a number of phosphorylated proteins of the AKT signaling pathway. The phosphorylation level of AKT pathway-associated proteins was increased after the induction of the osteogenic differentiation. Previous studies have already shown that the activation of the AKT kinase was required for the BMP2-induced differentiation of osteoblasts and the activation of mTOR downstream from AKT was associated also with the osteogenic differentiation of human MSCs [10,18,19]. In our study, the inhibition of AKT with a specific inhibitor (AKT inh) prevents the activity of ALP in DFCs. Moreover, we could show that inhibition of AKT prevents the phosphorylation of SMAD1/5 after the induction with BMP2. Similar to this observation, Ghosh-Choudhury and co-workers reported that a dominant-negative AKT inhibited both the BMP/SMAD pathway and the osteogenic differentiation of human MSCs [10].
EGR1 is involved in the osteogenic differentiation of dental stem cells and induced by BMP2 at early time points of the osteogenic differentiation in DFCs [15]. Our new results showed that an EGR1 overexpression increased the ALP activity of DFCs, which is in agreement with our previous results after the inhibition of EGR1 in DFCs with a specific siRNA [15]. Here, however, the overexpression of EGR1 in stem cells from the dental apical papilla did not increase the expression of ALP [15]. Additional studies are required to investigate the role of EGR1 during the osteogenic differentiation of DFCs, but EGR1 is probably involved in the regulation of genes, which are induced by BMP2 [5, 15, 20].
Our results propose that BMP2 induces the osteogenic differentiation in DFCs by the induction of the AKT kinase and the expression of EGR1 downstream from AKT. Our results indicate furthermore that the AKT signaling pathway supports the BMP2-induced activation of SMAD, which is not only required for the induction of transcription factors including EGR1 but also for the osteogenic differentiation of DFCs [15, 21]. Our assumption that a pathway of BMP2/AKT does partly control the differentiation of DFCs via transcription factor EGR1 is in coherence with the results of previous studies with human MSCs [10,18], although further studies have to investigate the activation of PI3K by BMP2. Interestingly, Gu et al. have shown that the PI3K/AKT signaling pathway inhibits the expression of osteogenic differentiation markers such as ALP in MC3T3-E1 preosteoblast [8]. Nevertheless, further studies must investigate the regulation of transcription factors such as DLX3 downstream from the PI3K/AKT pathway.
References
Sodek J, McKee MD (2000) Molecular and cellular biology of alveolar bone. Periodontol 24:99–126
Chai Y, Maxson RE Jr (2006) Recent advances in craniofacial morphogenesis. Dev Dyn Off Publ Am Assoc Anat 235:2353–2375. doi:10.1002/dvdy.20833
Ten Cate AR (1997) The development of the periodontium—a largely ectomesenchymally derived unit. Periodontol 2000(13):9–19
Morsczeck C, Götz W, Schierholz J et al (2005) Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol J Int Soc Matrix Biol 24:155–165. doi:10.1016/j.matbio.2004.12.004
Viale-Bouroncle S, Felthaus O, Schmalz G et al (2012) The transcription factor DLX3 regulates the osteogenic differentiation of human dental follicle precursor cells. Stem Cells Dev 21:1936–1947. doi:10.1089/scd.2011.0422
Depew MJ, Simpson CA, Morasso M, Rubenstein JLR (2005) Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat 207:501–561. doi:10.1111/j.1469-7580.2005.00487.x
Choi YH, Choi H-J, Lee K-Y, Oh J-W (2012) Akt1 regulates phosphorylation and osteogenic activity of Dlx3. Biochem Biophys Res Commun 425:800–805. doi:10.1016/j.bbrc.2012.07.155
Gu Y-X, Du J, Si M-S et al (2013) The roles of PI3K/Akt signaling pathway in regulating MC3T3-E1 preosteoblast proliferation and differentiation on SLA and SLActive titanium surfaces. J Biomed Mater Res A 101A:748–754. doi:10.1002/jbm.a.34377
Platt MO, Wilder CL, Wells A et al (2009) Multipathway kinase signatures of multipotent stromal cells are predictive for osteogenic differentiation: tissue-specific stem cells. Stem Cells Dayt Ohio 27:2804–2814. doi:10.1002/stem.215
Ghosh-Choudhury N, Abboud SL, Nishimura R et al (2002) Requirement of BMP-2-induced phosphatidylinositol 3-kinase and Akt serine/threonine kinase in osteoblast differentiation and Smad-dependent BMP-2 gene transcription. J Biol Chem 277:33361–33368. doi:10.1074/jbc.M205053200
Yu J, Zhang SS, Saito K et al (2009) PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J 28:21–33. doi:10.1038/emboj.2008.238
Chu L, Wang T, Hu Y et al (2013) Activation of Egr-1 in human lung epithelial cells exposed to silica through MAPKs signaling pathways. PLoS ONE 8:e68943. doi:10.1371/journal.pone.0068943
Sukhatme VP, Cao XM, Chang LC et al (1988) A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 53:37–43
Hofer G, Grimmer C, Sukhatme VP et al (1996) Transcription factor Egr-1 regulates glomerular mesangial cell proliferation. J Biol Chem 271:28306–28310
Press T, Viale-Bouroncle S, Felthaus O et al (2014) EGR1 supports the osteogenic differentiation of dental stem cells. Int Endod J. doi:10.1111/iej.12299
Viale-Bouroncle S, Gosau M, Morsczeck C (2014) Laminin regulates the osteogenic differentiation of dental follicle cells via integrin-α2/-β1 and the activation of the FAK/ERK signaling pathway. Cell Tissue Res. doi:10.1007/s00441-014-1869-x
Viale-Bouroncle S, Gosau M, Morsczeck C (2014) Collagen I induces the expression of alkaline phosphatase and osteopontin via independent activations of FAK and ERK signalling pathways. Arch Oral Biol 59:1249–1255. doi:10.1016/j.archoralbio.2014.07.013
Pantovic A, Krstic A, Janjetovic K et al (2013) Coordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cells. Bone 52:524–531. doi:10.1016/j.bone.2012.10.024
Mukherjee A, Wilson EM, Rotwein P (2010) Selective signaling by Akt2 promotes bone morphogenetic Protein 2-mediated osteoblast differentiation. Mol Cell Biol 30:1018–1027. doi:10.1128/MCB.01401-09
Viale-Bouroncle S, Klingelhöffer C, Ettl T et al (2014) A protein kinase A (PKA)/β-catenin pathway sustains the BMP2/DLX3-induced osteogenic differentiation in dental follicle cells (DFCs). Cell Signal. doi:10.1016/j.cellsig.2014.12.008
Saugspier M, Felthaus O, Viale-Bouroncle S et al (2010) The differentiation and gene expression profile of human dental follicle cells. Stem Cells Dev 19:707–717. doi:10.1089/scd.2009.0027
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Viale-Bouroncle, S., Klingelhöffer, C., Ettl, T. et al. The AKT signaling pathway sustains the osteogenic differentiation in human dental follicle cells. Mol Cell Biochem 406, 199–204 (2015). https://doi.org/10.1007/s11010-015-2437-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2437-8