Abstract
Long non-coding RNAs (LncRNAs) have been reported that play important roles in the progression and metastasis of some carcinomas. In the present study, we identified a new LncRNA, FRLnc1, from a microarray analysis in which those LncRNAs were regulated by FOXM1, an oncogene widely studied in most malignancies. Quantitative real-time PCR (qRT-PCR) results in gastric cancer cell lines indicated FRLnc1 expression is positively correlated with FOXM1 level, supporting the microarray data. Furthermore, the RNA level of FRLnc1 is upregulated in 49 % (20/41) of cancer samples compared with neighboring non-cancerous stomach tissues. The in vitro functional analyses demonstrated that FRLnc1 knockdown by RNA interference suppressed cell migration in MGC803 and AGS cells, whereas FRLnc1 overexpression promoted cell migration in BGC823 and SGC7901 cells. Moreover, FRLnc1 could enhance the distant metastasis of SGC7901 cells by tail vein injection approach in mice. We also identified TGFβ1 and Twist as the downstream effectors of FRLnc1 in the regulation of cell migration by qRT-PCR analysis. Taken together, our findings suggest that FRLnc1 is involved in gastric cancer cell migration and for the first time set up the link between FOXM1 and LncRNA in cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A phenomenon discovered by researchers of human genome project indicates that only about 20,000 human genes are responsible for protein coding [1]. Approximately 98 % of the total genes are transcribed into non-coding RNAs (ncRNAs) [2]. Although the genome portions without protein-coding capacity were originally believed as junk DNAs [3], a great deal of evidences support that ncRNAs may play critical roles in physiology and disease [4]. According to the size, ncRNAs are commonly divided into two categories: small ncRNAs and long ncRNAs (LncRNAs) [5]. The former including microRNAs (miRNAs), small interference RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) have been widely studied. The latter, which are composed of more than 200 nucleotides [6, 7], are still not well characterized to date.
According to the current knowledge, LncRNAs are commonly transcribed by RNA polymerase II and are located in either nucleus or cytoplasm [8]. Compared to protein-coding transcripts, LncRNAs have lower levels of expression and weaker conservation and frequently exhibit more tissue specificity [9, 10]. Growing evidences have demonstrated that abnormal expression of LncRNAs is associated with carcinoma [11]. One of the most well-known LncRNA HOX Antisense Intergenic RNA (HOTAIR) is reported to be overexpressed in primary breast cancer [12], and the expression level of HOTAIR is significantly associated with distant metastasis and poor prognosis [13]. Another study reveals that LncRNA CCAT1, which is elevated by c-Myc, has a strong ability to promote primary gastric cancer cells growth and has a close correlation with lymph node metastasis [14, 15]. In a recent report, LncRNA-activated by TGFβ (LncRNA-ATB) as a newly discovered type of LncRNA is overexpressed in metastases of hepatocellular carcinoma and always leads to poor prognosis [16, 17]. These increasing functional LncRNAs are being explored, which might serve as novel biomarkers for cancer diagnosis and prognosis and have the potential to be considered as new targets for therapy [18].
Gastric cancer (GC) is one of the most common types of digestive tumor [19]. It remains a major public-health issue as it leads to the second most frequent cause of cancer-related deaths worldwide [20]. Forkhead box protein M1 (FOXM1), a transcription factor characterized by a 100 amino acid winged-helix DNA binding domain, plays important roles in the regulation of oncogenesis and tumor progression in diverse malignancies [21]. In the context of gastric cancer, FOXM1 is significantly overexpressed and closely correlated with neoplasia and metastasis [22]. Furthermore, FOXM1 overexpression promotes cell growth and angiogenesis [23], whereas FOXM1 inhibition leads to cellular senescence which depends on p27kip1 activity [24].
A number of downstream target genes of FOXM1 associated with tumors have been reported [25]. These genes include Cyclin B1, Cyclin D1, Slug, MMP-2, VEGF, Skp2, uPA, Caveolin-1 and so on. However, to date, no report is found whether LncRNAs mediate FOXM1 function in tumor. To investigate the correlation between FOXM1 and LncRNAs, we performed a LncRNA microarray assay in gastric cancer cell line MGC803 in which FOXM1 was knocked down by RNA interference (unpublished data). Among the LncRNAs downregulated with FOXM1 knockdown, we focused on a novel LncRNA, designated as FRLnc1 (FOXM1-related LncRNA 1), that its role in tumor is still unclear. FRLnc1 is a long intergenic non-coding RNA (LincRNA) with the length of 1126 nucleotides. It is located on chromosome 17 without any intron (GeneSymbol: RP11-721K1.1). Quantitative RT-PCR analysis indicated that FRLnc1 expression is changed with FOXM1 increase or decrease, supporting the previous microarray data. Biological functions analysis revealed that FRLnc1 is required for cell migration in vitro and in vivo and this effect was mediated in part by Twist and TGFβ1. We also provided the expression pattern of FRLnc1 in 41 paired of clinical samples. Our findings suggest that FRLnc1 is a FOXM1 regulated LncRNA and involved in gastric cancer cell migration.
Materials and methods
Cell lines and culture conditions
Four human gastric cancer cell lines (MGC803, AGS, BGC823, and SGC7901) were obtained from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China) and cultured in MEM medium supplemented with 10 % fetal bovine serum (FBS, Hyclone) as well as 100 U/ml penicillin and 100 μg/ml streptomycin. All above cells were maintained in a humidified incubator at 37 °C with 5 % CO2.
Tissue samples
Fresh gastric cancer tissue samples and neighboring non-cancerous stomach tissue samples were obtained from 41 patients who underwent primary surgical resection of gastric cancer at Shanghai East Hospital affiliated to Tongji University between October 2013 and May 2014. Non-cancerous tissues were taken from a distance of 5–10 cm from the primary tumors. After resection, all the samples were snap-frozen in liquid nitrogen and stored at −80 °C prior to RNA extraction. Patients who received preoperative treatment were excluded from the study. All patients in this study provided written informed consent. The protocols involving human samples were conducted in conformity with the ethical principles of research and approved by the Human Resources Ethics Committee of Shanghai East Hospital affiliated to Tongji University.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from tissue samples and cultured cells using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The concentration of extracted RNA was measured by NanoDrop ND-1000 Spectrophotometer (Agilent, CA, USA). RNA was reversely transcribed into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara, China). Quantitative real-time PCR (qRT-PCR) was performed using the SYBR PrimeScript RT-PCR kit (Takara, Japan) and ABI 7500 System (Applied Biosystems, USA). β-actin was measured as an internal control for cells and tissues. Primers used in quantitative real-time PCR analysis were designed as follows: For FRLnc1, 5-ATGCGTGATTGCAGTCTCTG-3 (Forward), 5-TCTTGCAATATTTCCTGTGA-3 (Reverse); For FOXM1, 5-GGGCGCACGGCGGAAGATGAA-3 (Forward), 5-CCACTCTTCCAAGGGAGGGCTC-3 (Reverse); For TGF-β1, 5-GGAGAGCCCTGGATACCAAC-3 (Forward), 5-GCAGGGTCCCAGACAGAAG-3 (Reverse); For Twist, 5-CAAGTCTGCAGCTCTCGCCA-3 (Forward), 5-CCAACGGCTGGCGCACAC-3 (Reverse); For β-actin, 5-CCTGGCACCCAGCACAATG-3 (Forward), 5- GGGCCGGACTCGTCATACT-3(Reverse). The differential expression level was calculated using 2−ΔΔCt formula. All the experiments were conducted for at least three times.
Recombinant plasmid construction and RNA interference
The full-length cDNA of FRLnc1 (GeneSymbol: RP11-721K1.1) was amplified from human gastric cancer cell MGC803 cDNA library and cloned into the pCDH-CMV-EF1-copGFP expression vector to generate pCDH-FRLnc1 expression plasmid. Accurate reading frame insertion was verified by DNA sequencing. The siRNAs targeting FRLnc1 were designed and the sequences were as follows: siFRLnc1-1 (sense 5-CAAGAUUAAAUGCCUAAGAdTdT-3); siFRLnc1-2 (sense 5-CCAAUAUCAUCCUUUACGUdTdT-3). The siRNA targeting FOXM1 was designed as siFOXM1 (sense 5-CUCUUCUCCCUCAGAUAUAdTdT-3). The irrelevant nucleotides not targeting any annotated human genes were used as negative control: siNC (sense 5-UUCUCCGAACGUGUCACGUdTdT-3). All the siRNAs were chemically synthesized by GenePharma, Shanghai. Cell transfection with plasmids or siRNAs was conducted using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. The lentivirus overexpressing FOXM1 or FRLnc1 (LV-FOXM1, LV-FRLnc1) and knocking down FOXM1 or FRLnc1 (LV-shFOXM1, LV-shFRLnc1) were packaged and purchased from GenePharma, Shanghai using above corresponding sequences.
Cell migration assay in vitro
5 × 104 transfected gastric cancer cells in serum-free media were placed into the upper part of a transwell chamber in a 24-well format with 8 mm diameters (Corning, USA). In the bottom chamber, 600 µl of normal MEM medium containing 10 % FBS was added as a chemoattractant and the chambers were incubated for 24–48 h at 37 °C and 5 % CO2. The cells on the upper part of the transwell chamber were removed by the use of a cotton swap, and the cells that had migrated through the membrane were stained with 0.05 % crystal violet for 2 h. Finally, the migrated cells were counted in five random fields under a microscope and the average number of five fields was calculated. All assays were performed in triplicate and repeated three times.
Wound healing assay
For the wound healing assay, 3.5 × 105 of examined gastric cancer cells were seeded into six-well plates to achieve 90 % confluence. Wounds were created in confluent monolayer cells using a plastic tip, cell debris were removed using PBS, and 0.5 % FBS-containing MEM was added. Then the scratched cells were incubated at 37 °C with 5 % CO2. The initial scratched gap breadth (0 h) and the residual scratched gap breadth (48 h) were measured using the light microscope (Nikon, Japan).
Animal studies
The protocol for animal experiments was viewed and approved by the Institutional Animal Care and Use Committee of Tongji University. Male athymic nude mice (4 weeks old) were obtained from Animal Center of the Chinese Academy of Science (Shanghai, China) and maintained in sterile laminar flow cabinets. At the age of 6 weeks old, the mice were randomly divided into four groups and injected with 1.5 × 106 of MGC803-shNC, MGC803-shFRLnc1 or SGC7901-Vector, SGC7901-FRLnc1 through tail vein, respectively. After 5 weeks, the group received SGC7901 cells were euthanized, while the group received MGC803 were euthanized 8 weeks after injection. The lungs were removed and photographed, the obvious lung metastatic tumor on the surface were calculated. In addition, the lung tissues were fixed in 4 % paraformaldehyde and store them in 70 % ethanol, treat them with paraffin-embedding, sectioning, H&E staining and histopathological examination.
Statistical analysis
Quantitative data were presented as mean ± SEM. Parameters of two-tailed, 95 % CI were used for statistical analysis. Only a P value of less than 0.05 was considered significant. “*” indicates P < 0.05; “**” indicates P < 0.01.
Results
FRLnc1 expression is regulated by FOXM1 in gastric cancer cells
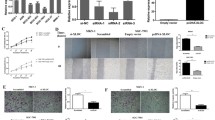
To verify whether FRLnc1 expression is regulated by FOXM1, qRT-PCR analysis was performed to detect the FRLnc1 RNA level in a few of gastric cancer cell lines in which FOXM1 expression was stably upregulated or downregulated. These stable cell lines were achieved through infection with lentivirus expressing FOXM1 coding sequence or short hairpin interference RNA against FOXM1, respectively. The final data showed that FRLnc1 was decreased with FOXM1 knockdown in MGC803, AGS and SGC7901 cells (Fig. 1a), while FRLnc1 was increased with FOXM1 overexpression in AGS and SGC7901 cells (Fig. 1b). These results demonstrated that FRLnc1 level is positively correlated with FOXM1 expression, suggesting FOXM1 may be a transcriptional activator of FRLnc1.
FRLnc1 RNA is positively correlated with the FOXM1 levels in gastric cancer cells. a FOXM1 relative expression in MGC803, AGS and SGC7901 cells stably infected with LV-shFOXM1 or LV-shNC. The expression levels of FRLnc1 were declined after FOMX1 knockdown. b FOXM1 relative expression in AGS and SGC7901 cells stably infected with LV-FOXM1 or LV-Vector. The expression levels of FRLnc1 were elevated after FOXM1 overexpression
FRLnc1 is frequently increased in gastric caner
To determine whether FRLnc1 is upregulated in gastric cancer, we explored the relative expression of FRLnc1 in 41 pairs of gastric cancer tissues and neighboring non-cancerous stomach tissues via qRT-PCR analysis. The data demonstrated that in 20 of 41 (49 %) cancer samples, FRLnc1 was upregulated at least 1.5-fold than neighboring non-cancerous stomach tissues. About 14 of 41 (34 %) cancer samples were downregulated, and the other seven paired samples showed no significant difference in FRLnc1 expression. The expression pattern of FRLnc1 in gastric cancer samples is shown in Fig. 2a, b. We also investigated FOXM1 expression in nine paired of gastric cancer samples in which express high levels of FRLnc1. As shown in Fig. 2c, FOXM1 expression was increased in seven of nine paired of cancer samples compared with their non-cancerous counterparts. This implicates the expression relevance of the two genes in clinical samples. However, we did not find any correlation between FRLnc1 expression and gastric cancer metastasis, tumor size, tumor grade, or other clinicopathological characteristics. This is possibly attributed to small amount of clinical samples.
FRLnc1 expression pattern in gastric cancer samples. a FRLnc1 level was examined by quantitative RT-PCR in 41 paired gastric cancer tissues (C) and neighboring non-cancerous tissues (N). β-actin was used as an internal loading control. Data were shown as the upregulated fold change (C/N) of FRLnc1 RNA level. b the percentage of FRLnc1 expression alteration in 41 paired gastric cancer samples with “≥1.5” indicating upregulation, “≤0.66” indicating downregulation, and “>0.66 but <1.5” indicating no significant variation. c the expression of FRLnc1 and FOXM1 by semiquantitative PCR analysis in nine paired of representative gastric cancer samples
FRLnc1 exhibits a significant effect on gastric cancer cell migration in vitro
Cancer cell migration is a critical aspect of cancer progression. To investigate whether FRLnc1 had a functional role in promoting gastric cancer cell migration, two specific small interference RNAs, siFRLnc1-1, and siFRLnc1-2 or negative control siNC were transfected into MGC803 and AGS cells, respectively, the efficiency of knockdown is shown in Fig. 3a. Transwell migration assays indicated that FRLnc1 knockdown led to a significant decrease in MGC803 and AGS cells migration (Fig. 3b). Consistent with transwell assay results, MGC803 cells in which FRLnc1 was knocked down exhibited more slowly migration rate compared with controls in wound healing assays (Fig. 3c). These data demonstrated that FRLnc1 downregulation decreases the ability of gastric cancer cell migration.
FRLnc1 knockdown weakens the migratory capacity of gastric cancer cells in vitro. a Efficiency of FRLnc1 knockdown with si-FRLnc1-1 and si-FRLnc1-2 in MGC803 and AGS cells by qRT-PCR. b Transwell chamber assays were performed to investigate the change of migratory ability of gastric cancer cells indicated. c Wound healing assays were performed to investigate the mobility of gastric cancer cells indicated
On the other hand, we generated two stable cell lines BGC823-FRLnc1 and SGC7901-FRLnc1 by infecting FRLnc1 overexpressing lentivirus. qRT-PCR data showed FRLnc1 was successfully overexpressed in the two cell lines (Fig. 4a). Transwell assays revealed that the migratory ability was dramatically increased in FRLnc1 overexpressing cells (Fig. 4b), and would healing assays indicated obvious migration of SGC7901-FRLnc1 cells toward the wound blank than that of SGC7901-Vector cells (Fig. 4c). Collectively, these findings suggested that overexpression of FRLnc1 promotes cell migration of gastric cancer cells.
FRLnc1 overexpression enhances the migratory capacity of gastric cancer cells in vitro. a Efficiency of FRLnc1 overexpression in BGC823 and SGC7901 cells by qRT-PCR. b Transwell migration assays were performed to investigate the change of migratory ability of gastric cancer cells indicated. c Wound healing assays were performed to investigate the mobility of gastric cancer cells indicated
FRLnc1 promotes the formation of pulmonary metastasis in vivo
To further determine the role of FRLnc1 in formation of tumor metastasis in vivo, 1.5 × 106 of SGC7901-FRLnc1 stable cells were injected directly into tail veins of nude mice. 5 weeks after injection, the mice were euthanized, lungs were removed and photographed, and visible lung metastatic tumors on the surface were calculated. As shown in Fig. 5a, b, the tumor on the lung surface from mice receiving SGC7901-FRLnc1 cells were significantly more than that of the control cell SGC7901-Vector. Histological analysis confirmed the presence of metastatic tumors in the lungs of these mice (Fig. 5c). Furthermore, we established a subclone of MGC803 cell line, which was stably infected by a lentivirus expressing shRNA-FRLnc-1 for FRLnc1 knockdown. However, we did not observed any tumor on the surface of lungs, whether from mice receiving MGC803-shFRLnc1 cells or control cells (data not shown). Our overexpression experiment suggested that FRLnc1 could promote pulmonary metastasis in vivo.
FRLnc1 promotes gastric cancer cells metastasis in vivo. a Lung metastatic tumor nodules observed in lung surface of nude mice received tail vein injection with SGC7901-Vec or SGC7901-LFRLnc1 cells. b the mean number of tumor nodules from each experimental group (n = 5). c the hematoxylin and eosin (HE) stained lung sections
FRLnc1 expression levels are associated with TGF-β1 and Twist
To address the possible mechanism by which FRLnc1 could induce cell migration and promote gastric cancer metastasis in vivo, we examined a serious of molecule markers involved in epithelial mesenchymal transition (EMT) or tumor distant metastasis by qRT-PCR analysis. Among these genes, we found that the expression level of TGFβ1 and Twist were changed with FRLnc1 increase or decrease. As shown in Fig. 6a, when FRLnc1 was knocked down in AGS cells, the two genes were reduced as well; in contrast, when FRLnc1 was overexpressed in SGC7901 cells, the two genes were also increased (Fig. 6b). These results demonstrated that Twist and TGFβ1 might mediate the FRLnc1 function in cell migration.
Discussion
FOXM1 functions as an oncogene in diverse of malignancies [26]. Deregulation of FOXM1 affects tumor initiation, progression, EMT, metastasis, angiogenesis, and drug resistance [27]. The underlying mechanisms by which FOXM1 promotes these processes are being explored. Reports have revealed some proteins and signaling transduction factors are downstream targets of FOXM1 [28]. However, there is no report about the correlation between FOXM1 and LncRNAs to date. LncRNAs, initially considered as the transcription noise, are playing pivotal roles in carcinogenesis [29]. In the present study, we identified a new LncRNA as downstream target of FOXM1 for the first time. We designated it as FRLnc1 (FOXM1-related LncRNA 1). By evaluating the expression levels of FRLnc1 in FOXM1 upregulating or downregulating gastric cancer cells via qRT-PCR, we confirmed FRLnc1 is regulated by FOXM1. This regulation appears to be indirect for no FOXM1 binding site is found in the promoter region of FRLnc1. As for the clinicopathological significance of FRLnc1 expression in gastric cancer, we should gather more clinical samples of gastric cancer in future since in the present samples we did not find any clinical correlation although FRLnc1 is upregulated in 49 % cancer samples.
Next, we examined the potential role of FRLnc1 in gastric cancer cell migration by transwell assay and wound healing assay. The results demonstrate that FRLnc1 promotes gastric cancer cell migration in vitro, in accordance with the phenotypes of FOXM1 reported. Interestingly, FRLnc1 did not affect cell growth in four gastric cancer cell lines examined (data not shown). Considering FRLnc1 is regulated by FOXM1, our results suggest that FRLnc1 might be a mediator of FOXM1 in the regulation of cell migration, but not cell growth. Furthermore, tail vein injection with gastric cancer cells was performed on nude mice. The resulting data indicate that FRLnc1 could facilitate lung metastasis of tumor cells in vivo, supporting the conclusion in vitro.
Finally, we attempted to explore the potential mechanism by which FRLnc1 promotes cell migration. A number of genes associated with cell migration or tumor metastasis were selected and analyzed by qRT-PCR. These genes include Slug, Cav-1, Acp5, Snail, uPA, uPAR, TGFβ1, ZEB1, ZEB2, Twist, Vimentin, and E-cadherin. Two molecules, TGFβ1 and Twist, were screened out in the end, of which expression was regulated by FRLnc1. The two factors are both well-known responsible for cancer metastasis. TGFβ1 is a regulator involved in late stages of tumorigenesis [30]. More than one study has revealed that TGFβ1 suppression can significantly prevent metastasis in different cancer types [31, 32]. Twist has been considered as an important inducer for metastatic progression by promoting EMT in many cell systems [33]. The findings that TGFβ1 and Twist are regulated by FRLnc1 support FRLnc1 involvement in cancer cell migration from molecular level.
Taken together, FRLnc1 is a FOXM1-related LncRNA which is increased in gastric cancer tissues. It has a potential to promote gastric cancer cell migration and may function as a propeller of tumor distant metastasis. Our findings support that FRLnc1 plays a significant role in the progression of gastric cancer and has an opportunity to be a new biomarker for its prognosis. Further studies are needed to expound the detailed underlying mechanism through which FRLnc1 contributes to the cell migration in gastric cancer.
References
Tanpakushitsu HM, Koso K et al (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931–945. doi:10.1038/nature03001
Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM (2011) Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 10:141. doi:10.1186/1476-4598-10-141
Ponting CP, Belgard TG (2010) Transcribed dark matter: meaning or myth? Hum Mol Genet 19:R162–R168. doi:10.1093/hmg/ddq362
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159. doi:10.1038/nrg2521
Brosnan CA, Voinnet O (2009) The long and the short of noncoding RNAs. Curr Opin Cell Biol 21:416–425. doi:10.1016/j.ceb.2009.04.001
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. doi:10.1146/annurev-biochem-051410-092902
Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5:e1000459. doi:10.1371/journal.pgen.1000459
Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, Gonzales C, Sarre A, Alexanian M, Blow MJ, May D, Johnson R, Dauvillier J, Pennacchio LA, Pedrazzini T (2014) Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol 76:55–70. doi:10.1016/j.yjmcc.2014.08.009
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi:10.1101/gr.132159.111
Marques AC, Ponting CP (2009) Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol 10:R124. doi:10.1186/gb-2009-10-11-r124
Yang G, Lu X, Yuan L (2014) LncRNA: a link between RNA and cancer. Biochim Biophys Acta 1839:1097–1109. doi:10.1016/j.bbagrm.2014.08.012
Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, Benedetto C, Yu H (2012) Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat 136:875–883. doi:10.1007/s10549-012-2314-z
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi:10.1038/nature08975
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G (2013) Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol 139:437–445. doi:10.1007/s00432-012-1324-x
He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z (2014) C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol 35:12181–12188. doi:10.1007/s13277-014-2526-4
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH (2014) A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25:666–681. doi:10.1016/j.ccr.2014.03.010
Li W, Kang Y (2014) A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-beta. Cancer Cell 25:557–559. doi:10.1016/j.ccr.2014.04.014
Qi P, Du X (2013) The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 26:155–165. doi:10.1038/modpathol.2012.160
Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K, Yamashita M, Taniguchi H, Nosho K, Suzuki H, Yasuda H, Shinomura Y, Itoh F (2014) An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol 20:3927–3937. doi:10.3748/wjg.v20.i14.3927
Smith MG, Hold GL, Tahara E, El-Omar EM (2006) Cellular and molecular aspects of gastric cancer. World J Gastroenterol 12:2979–2990
Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K, Miyata H, Yamasaki M, Kurokawa Y, Mori M, Doki Y (2013) Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol 20:1035–1043. doi:10.1245/s10434-012-2680-0
Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K (2009) Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res 69:3501–3509. doi:10.1158/0008-5472.can-08-3045
Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, Huang S (2008) FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res 68:8733–8742. doi:10.1158/0008-5472.can-08-1968
Zeng J, Wang L, Li Q, Li W, Bjorkholm M, Jia J, Xu D (2009) FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J Pathol 218:419–427. doi:10.1002/path.2530
Huang C, Du J, Xie K (2014) FOXM1 and its oncogenic signaling in pancreatic cancer pathogenesis. Biochim Biophys Acta 1845:104–116. doi:10.1016/j.bbcan.2014.01.002
Bella L, Zona S, de Moraes GN, Lam EW (2014) FOXM1: a key oncofetal transcription factor in health and disease. Semin Cancer Biol 29c:32–39. doi:10.1016/j.semcancer.2014.07.008
Lam EW, Brosens JJ, Gomes AR, Koo CY (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 13:482–495. doi:10.1038/nrc3539
Behren A, Muhlen S, Acuna Sanhueza GA, Schwager C, Plinkert PK, Huber PE, Abdollahi A, Simon C (2010) Phenotype-assisted transcriptome analysis identifies FOXM1 downstream from Ras-MKK3-p38 to regulate in vitro cellular invasion. Oncogene 29:1519–1530. doi:10.1038/onc.2009.436
Fang XY, Pan HF, Leng RX, Ye DQ (2015) Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett 356:357–366. doi:10.1016/j.canlet.2014.11.005
Bartscht T, Lehnert H, Gieseler F, Ungefroren H (2012) The Src family kinase inhibitors PP2 and PP1 effectively block TGF-beta1-induced cell migration and invasion in both established and primary carcinoma cells. Cancer Chemother Pharmacol 70:221–230. doi:10.1007/s00280-012-1904-0
Ge R, Rajeev V, Ray P, Lattime E, Rittling S, Medicherla S, Protter A, Murphy A, Chakravarty J, Dugar S, Schreiner G, Barnard N, Reiss M (2006) Inhibition of growth and metastasis of mouse mammary carcinoma by selective inhibitor of transforming growth factor-beta type I receptor kinase in vivo. Clin Cancer Res 12:4315–4330. doi:10.1158/1078-0432.ccr-06-0162
Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA (2011) TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res 71:175–184. doi:10.1158/0008-5472.can-10-2651
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939. doi:10.1016/j.cell.2004.06.006
Acknowledgments
This work was supported in part by grants from National Natural Science Foundation of China (81272917 and 81272241), Shanghai medical key specialty (ZK2012A26), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2014-04) and Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (PWRl2013-02).
Conflict of interest
The authors declare that no conflicts of interest exist.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hui Cai and Jingde Chen have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cai, H., Chen, J., He, B. et al. A FOXM1 related long non-coding RNA contributes to gastric cancer cell migration. Mol Cell Biochem 406, 31–41 (2015). https://doi.org/10.1007/s11010-015-2421-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2421-3