Abstract
Accumulating data have shown that microRNAs are involved in the pathogenesis of cancer. miR-202 has been confirmed to be downregulated in several types of human cancer. However, the expression and biological role of miR-202 in osteosarcoma (OS) carcinogenesis and progression remain unclear. In this study, we demonstrated that miR-202 expression is significantly decreased in human OS cell lines and specimens. Restoration of miR-202 expression could inhibit OS cell proliferation, induce cell apoptosis, and suppress tumor growth in nude mice models. We subsequently identified the transcription factor Gli2 as a direct target of miR-202. Overexpression of Gli2 blocked the inhibitory function of miR-202. Taken together, our results indicate that miR-202 acts as a novel tumor suppressor to regulate OS cell proliferation and apoptosis through downregulating Gli2 expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteosarcoma (OS) is the most frequent primary malignant bone tumor and accounts for 60 % of all cancer-related death in children and adolescents [1, 2]. Despite recent advance in therapeutic strategies, such as surgery, radiotherapy, and chemotherapy, the 5-year survival rate of OS patients is only 60–70 % [3]. Most patients with OS eventually developed lung metastases, which is the major cause of death [1, 4]. Thus, a better understanding of the molecular mechanism underlying the development and progression of OS is urgently needed to optimize strategies for more effective therapies. At present, emerging evidence has shown that microRNAs (miRNAs) play crucial roles in OS pathogenesis [5–7], which provides novel insights into new biomarker identification, and the development of diagnostic, therapeutic approaches of this disease.
microRNAs are small and endogenous ∼22 nucleotides (nt) non-coding RNAs that play crucial regulatory roles post-transcriptionally through binding to 3′-untranslated region (3′-UTR) of target mRNAs in sequence-specific manner, which results in the silence or degradation of target mRNAs [8, 9]. microRNAs are involved in various biological processes, such as cell cycle, proliferation, differentiation, apoptosis, invasion, and metabolism [10, 11]. Moreover, accumulating evidence has shown that in the pathogenesis and development of numerous human cancers, the dysregulation of miRNAs was found involved in crucially [8, 10–14].
Human miR-202, a member of the let-7 family, located within a chromosomal fragile site in 10q26, has been reported to be involved in various cancers, such as endometrical cancer [15], colorectal carcinoma [16], hepatocellular carcinoma [17], breast cancer [18], multiple myeloma [19], gastric cancer [20], follicular lymphoma [21], and neuroblastoma [22], suggesting common roles of miR-202 in tumorigenesis. However, the specific role and molecular mechanism of miR-202 in OS remain poorly understood.
In the present study, we demonstrated that the expression level of miR-202 is decreased greatly in human OS cell lines and clinical specimens. Restoration of miR-202 expression results in inhibition of OS cell proliferation, induction of apoptosis, and suppression of tumor growth in nude mice models. The transcription factor Gli2 was identified as a direct target of miR-202. Overexpression of Gli2 could abrogate the inhibitory effect of miR-202 in OS cell proliferation and tumor growth. Our study provides new insights that miR-202 acts as a novel tumor suppressor to regulate OS cell proliferation and apoptosis through downregulating Gli2 expression.
Materials and methods
Tissue specimens and cell lines
Fresh OS tissues and matched adjacent non-cancerous tissues were obtained from 16 OS patients in Yantaishan Hospital, China. All tissues are snap frozen in liquid nitrogen immediately after surgery and stored at −80 °C, until total RNA was extracted. For the use of these clinical materials for research purposes, prior patients’ written informed consents were obtained, and this project was approved by the Institutional Ethics Committee. The normal human osteoblastic cell line hFOB1.19 and OS cell lines HOS and MG63 were cultured in Dulbecco’s modified Eagle’s medium in the presence of 10 % FBS, 100 units/ml penicillin, and 100 g/ml streptomyc in a humidified 5 % (v/v) atmosphere of CO 2 at 37 °C incubator.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total mRNA from cultured cells and frozen tissues was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. cDNA was synthesized using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). The expression levels of miR-202 were determined using a TaqMan miRNA assay kit (Applied Biosystems, Foster, CA) according to the manufacturer’s instructions and calculated using RNU6B small nuclear RNA as an endogenous control by 2−ΔΔCT method. All of the reactions were run in triplicate.
Lentivirus production and transduction
The primary transcript genomic region of miR-202 was cloned into the downstream of the CMV promoter in the FUGW vector, and none insert vectors used as negative control. The miR-202-expressing or negative control lentiviral plasmid along with delta 8.9 and VSVG (packaging plasmids) were co-transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen) to generate lentivirus. HOS and MG63 cells were infected with the recombinant lentivirus-transducing units in the presence of 6 μg/ml of Polybrene. Cells were cultured for 48 h and analyzed by qRT-PCR.
Vector construction
The plasmid pcDNA-Gli2-3′UTR was constructed by inserting the Gli2 cDNA into the pcDNA3.1(+) vector (Invitrogen). pcDNA-Gli2 contained the full-length Gli2 coding region lacking the 3′-UTR. Transfection of these plasmids was performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. To construct a luciferase reporter plasmid, the wild-type 3′-UTR of Gli2 containing a putative miR-202 binding site was cloned into psiCHECK2. Meanwhile, a mutant 3′-UTR of Gli2, which contains the point mutant in targeting region of miR-202 seed sequences, was also inserted to psiCHECK2. These plasmids were confirmed by direct sequencing.
Cell proliferation and apoptosis analysis
Cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). Cells (2 × 103) were seeded into 96-well plates in a final volume of 100 µl and cultured for 48 and 96 h. Cell Counting Kit-8 solution (10 μl) was added into each well, and the optical density at 450 nm was measured to calculate the number of viable cells. Three parallel wells were carried out for each group. For apoptosis detection, cells were seeded into six-well plates (4 × 105/well). At 48 h after transfection, cells were stained with AnnexinV/PI double staining kit (BD biosciences, Bedford, MA) according to the manufacturer’s protocol. Apoptotic cells were examined by flow cytometry on a FACScan (Beckman Instruments, Fullerton, CA). These experiments were conducted in triplicates and repeated three times independently.
Luciferase reporter assay
HOS and MG63 cells were seeded in 24-well plates and allowed to settle for 24 h. A mixture of 100 ng reporter plasmid, 200 ng of FUGW or FUGW-miR-202 and 10 ng pRL-TK Renilla plasmid (Promega, Madison, WI) was transfected into these cells using Lipofectamine 2000. After 48 h, Firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay Kit (Promega) according to a protocol provided by the manufacturer.
Western blotting
Cells were lysed in RIPA buffer along with Protease Inhibitor, and then electrophoresed in 10 % SDS-PAGE gel. After transfer to nitrocellulose membrane (Bio-Rad, Hercules, CA), 5 % milk was used to block the member. Then, the member was incubated with anti-Gli2 primary antibody (Cell Signaling, Danvers, MA) overnight. The corresponding horseradish peroxidase (HRP)-conjugated immunoglobulin G was incubated at room temperature for 1 h. Finally, signals were visualized by enhanced chemiluminescence. GAPDH was used as a loading control.
In vivo tumorigenesis assay
HOS cells (1 × 106) stably expressing miR-202 or vector control were suspended in 100 µl PBS and injected subcutaneously into the flank of BALB/c nude mice (4 weeks of age, male). Tumor volume (V) was monitored every week and calculated using the formula: V = length × width2 × 0.5. Mice were sacrificed after 6 weeks and tumor weights were assessed. All experiments were carried out under the approval of the Committee for the Use and Care of Experimental Animals of the Yantaishan Hospital.
Statistical analysis
All data analyzed as the mean ± SD are from at least three separate experiments. The relationship between the miR-202 and Gli2 expression levels was detected using the Pearson’s correlation analysis. Student’s t test or one-way ANOVA test was used to determine the statistical significance, and P < 0.05 in all cases was considered significant.
Results
miR-202 is downregulated in OS
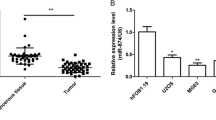
First, we detected the expression levels of miR-202 by qRT-PCR in human normal osteoblastic cell line hFOB1.19 and OS cell lines HOS and MG63 to investigate the role of miR-202 in human OS development, and found that miR-202 expression was downregulated significantly in OS cell lines compared with normal osteoblastic cell line (Fig. 1a). Similarly, the down expression of miR-202 was also observed in 16 pairs of clinical human OS tissues when compared with adjacent normal tissues (Fig. 1b). These results suggested that miR-202 is downregulated in human OS cells, implicating a potential regulating role of miR-202 in OS.
miR-202 is downregulated in OS cell lines and clinical specimens. a The expression levels of miR-202 were measured by qRT-PCR in human normal osteoblastic cell line hFOB1.19 and OS cell lines HOS and MG63. b The expression levels of miR-202 in 16 pairs of human OS tissues and adjacent normal tissues. All data were normalized by RNU6B. *P < 0.05
miR-202 reduces cell proliferation, promotes cell apoptosis, and suppresses tumorigenicity
To study the biological function of miR-202 in OS, we established miR-202 stably expressing cell lines of HOS and MG63 by lentivirus infection, and qRT-PCR analysis confirmed successful overexpression of mature miR-202 in these cell lines (Fig. 2a). We found that overexpression of miR-202 markedly suppressed cell proliferation in HOS and MG63 cells (Fig. 2b). Furthermore, we explored the effect of miR-202 on apoptosis by FACS. As shown in Fig. 2c, both OS cells expressing miR-202 displayed a significant increase in the frequency of apoptosis compared with their corresponding controls.
Overexpression of miR-202 inhibits proliferation and tumor growth of OS cells. Human OS cell lines (HOS and MG63) were infected with miR-202 lentivirus and control lentivirus, respectively. a miR-202 level of the indicated cells was measured by qRT-PCR. b Cell proliferation of the indicated cells was analyzed using CCK-8. c Apoptosis of the indicated cells was analyzed using flow cytometry. d HOS cells infected with miR-202 lentivirus or control lentivirus was injected subcutaneously into the flank of nude mice. Tumor volumes were measured on the indicated days. At the experimental endpoint, tumors were dissected, photographed, and weighed. *P < 0.05
Next, we identify the effect of miR-202 on tumorigenicity in mouse models. When injected with HOS cells stably expressing miR-202 in nude mice, the tumors grew more slowly than that of miR-NC, and the gap expanded until the endpoint of 6 weeks test (Fig. 2d). Consistently, the miR-202 overexpressed tumors were smaller in size and weight compared with the control tumors (Fig. 2d). Together, our data indicated that miR-202 not only reduces cell proliferation and promotes cell apoptosis in vitro, but also suppresses tumorigenicity in vivo.
Gli2 is a direct target of miR-202
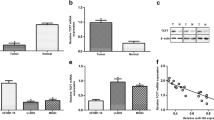
We further analyzed the candidate genes regulated by miR-202 using two most-used publicly bioinformatic algorithms, TargetScan and miRanda. The results showed that Gli2, a transcription factor in hedgehog-GLI signaling pathway, which involved in various stages of embryonic development and tumorigenesis [23, 24], was a potential target gene of miR-202 theoretically. To determine whether Gli2 is a direct target of miR-202, we constructed luciferase reporter plasmids containing the 3′-UTR region of Gli2 gene carrying either a wild-type or mutant putative miR-202 binding site (Fig. 3a). After overexpression of miR-202 in HOS and MG63 cells, the relative luciferase activity of the construct carrying the wild-type 3′-UTR of Gli2 decreased significantly, but the similar effect was not observed when the three nucleotides in the miR-202 seed binding site of the Gli2 3′-UTR were mutated (Fig. 3b). Furthermore, Western blotting analysis demonstrated that miR-202 overexpression significantly suppressed endogenous Gli2 expression in HOS and MG63 cells (Fig. 3c). These results suggest that miR-202 downregulates Gli2 expression by directly targeting its 3′-UTR.
Gli2 is a direct target of miR-202 in OS cells. a Bioinformatic analysis identifed Gli2 as a potential target for miR-202. Gli2 wild and Gli2 mut luciferase report plasmids were constructed. b OS cells were co-transfected with Lv-miR-NC or Lv-miR-202 with a wild-type or a mutant Gli2 3′-UTR. Relative luciferase activity was analyzed. c The Gli2 protein levels in OS cell lines infected with Lv-miR-NC or Lv-miR-202. d The Gli2 protein levels in human normal osteoblastic hFOB 1.19 cell line versus OS HOS and MG63 cell lines. e Pearson’s correlation analysis of miR-202 and Gli2 protein in 16 human OS samples. The expression of miR-202 was examined using qRT-PCR analysis. The expression of Gli2 was quantified by Western blotting with mean optical density and normalized to GAPDH expression. *P < 0.05
We subsequently compared Gli2 protein levels in hFOB 1.19 cells and OS cell lines HOS and MG63, as miR-202 was observed to be downregulated in these OS cells. As shown in Fig. 3d, Gli2 protein levels were significantly upregulated in OS cells. In addition, the correlation analysis between the relative protein levels of Gli2 and miR-202 expression was performed in 16 human clinical OS tissues, and the results revealed that the expression of miR-202 and Gli2 was negatively correlated (Fig. 3e). These results demonstrated that Gli2 is a direct target of miR-202.
The anti-tumor function of miR-202 is mediated by downregulating Gli2 expression
To identify whether the anti-tumor function of miR-202 in OS cells was due to downregulating Gli2 expression, the effect of Gli2 (without 3′-UTR) and Gli2-3′-UTR (with 3′-UTR) was examined in the miR-202 expressing cells. As shown in Fig. 4a, Gli2 expression was markedly increased in these cells transfected with Gli2 plasmid, but not in those transfected with Gli2-3′-UTR. Cell Counting Kit-8 assay showed that overexpression of Gli2 significantly promoted the proliferation of miR-202 expressing HOS and MG63 cells, whereas Gli2-3′-UTR overexpression had no significant impact on the proliferation of these cells (Fig. 4a and b). Moreover, the apoptosis rate was decreased obviously after Gli2 transfection in the miR-202 expressing cells (Fig. 4c). These results suggest that Gli2 is a functional downstream target of miR-202.
Re-introduction of Gli2 rescues the miR-202-induced proliferation inhibition. a Western blotting analysis of Gli2 in HOS and MG63 cells and the indicated cells transfected with vector, Gli2 (without 3′-UTR) and Gli2-3′-UTR (with 3′-UTR). b Cell proliferation assay. c Cell apoptosis analysis. *P < 0.05 versus miR-NC group, & P < 0.05 versus Vector group
Discussion
Osteosarcoma is the most common human primary malignant bone tumor and always results in an aggressive clinical course [1, 25]. However, the molecular mechanism under OS carcinogenesis and progression remains to be explored. Recent evidence has shown that dysgrugulation of miRNAs was involved in the development of OS, such as miR-21, miR-140, miR-34, and miR-143 [26].
microRNAs negatively regulate their target genes in a sequence-specific manner [10]. Emerging evidence has shown that miRNAs play crucial roles in many biological processes including tumorigenesis [27]. The dysregulation of miR-202 was found in many cancers [15–22]. Functional interactions among identified targets suggested that miR-202 is involved in follicular lymphomagenesis and can be a potential tumor suppressor in follicular lymphoma [21]. Moreover, miR-202 was found to inhibit cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma [16]. In addition, miR-202 suppresses cell proliferation in human hepatocellular carcinoma by downregulating LRP6 post-transcriptionally [17]. However, it remains unclear whether dysregulation of miR-202 was associated with the development of OS.
In the present study, we found that miR-202 was downregulated in OS cell lines and clinical OS specimens. Overexpression of miR-202 reduces cell proliferation and promotes cell apoptosis in vitro, also suppresses tumorigenicity in vivo, indicating miR-202 as a tumor suppressor. Next, we tried to explore the mechanism by which miR-202 exerts influence on the development of OS. Gli2 was predicted to be the theoretical target gene of miR-202 using two publicly bioinformatic algorithms in combination. We found that Gli2 expression is increased in OS cell lines compared with human normal osteoblastic cell line. Overexpression of miR-202 significantly downregulated the expression of Gli2 protein. In addition, luciferase reporter assay showed that miR-202 could directly target the 3′-UTR of Gli2 mRNA.
Gli2 is a transcription factor in hedgehog-Gli signaling pathway, which involved in various stages of embryonic development and tumorigenesis [23, 24]. Gerhard and colleagues showed that Gli2, along with Gli1, was involved in the positive feedback mechanism in human basal cell carcinoma [28]. Moreover, Hiroko and co-workers found that Gli2 was significantly upregulated in human OS tissues, and Gli2 knockdown by RNA interferences prevented OS growth under the mechamism of promoting the arrest of OS cells at G1 phase [29]. Our study revealed that Gli2 is a direct and functional target of miR-202 in OS. Re-introduction of Gli2 greatly abrogated miR-202-induced proliferation inhibition of OS cells. Furthermore, there is a negatively correlation between the expression of miR-202 and Gli2 in OS tissues. These results demonstrate that miR-202 inhibits the proliferation of OS cells by downregulating the expression of Gli2 through directly targeting its 3′-UTR.
In conclusion, our data suggest that miR-202 is downregulated in OS, and functions as a novel tumor suppressor to regulate OS cell proliferation and apoptosis through downregulating Gli2 expression. Thus, miR-202 and Gli2 could potentially serve as targets for OS treatment.
References
Ando K, Heymann MF, Stresing V, Mori K, Redini F, Heymann D (2013) Current therapeutic strategies and novel approaches in osteosarcoma. Cancers (Basel) 5:591–616. doi:10.3390/cancers5020591
Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF (2011) The molecular pathogenesis of osteosarcoma: a review. Sarcoma 2011:959248. doi:10.1155/2011/959248
Gorlick R (2009) Current concepts on the molecular biology of osteosarcoma. Cancer Treat Res 152:467–478. doi:10.1007/978-1-4419-0284-9_27
Marina N, Gebhardt M, Teot L, Gorlick R (2004) Biology and therapeutic advances for pediatric osteosarcoma. Oncologist 9:422–441
Lulla RR, Costa FF, Bischof JM, Chou PM, de F BM, Vanin EF, Soares MB (2011) Identification of differentially expressed microRNAs in osteosarcoma. Sarcoma 2011:732690. doi: 10.1155/2011/732690
Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T, Ito H, Oshimura M, Ochiya T (2011) MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther 19:1123–1130. doi:10.1038/mt.2011.53
Jones KB, Salah Z, Del MS, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, Volinia S, Stein GS, Croce CM, Lian JB, Aqeilan RI (2012) miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res 72:1865–1877. doi:10.1158/0008-5472.CAN-11-2663
Garzon R, Calin GA, Croce CM (2009) MicroRNAs in Cancer. Annu Rev Med 60:167–179. doi:10.1146/annurev.med.59.053006.104707
Liu J (2008) Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol 20:214–221. doi:10.1016/j.ceb.2008.01.006
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Zhao Y, Srivastava D (2007) A developmental view of microRNA function. Trends Biochem Sci 32:189–197
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10:704–714. doi:10.1038/nrg2634
Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I (2008) MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 26:462–469. doi:10.1038/nbt1392
Shenouda SK, Alahari SK (2009) MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev 28:369–378. doi:10.1007/s10555-009-9188-5
Peiffer SL, Herzog TJ, Tribune DJ, Mutch DG, Gersell DJ, Goodfellow PJ (1995) Allelic loss of sequences from the long arm of chromosome 10 and replication errors in endometrial cancers. Cancer Res 55:1922–1926
Wang Q, Huang Z, Guo W, Ni S, Xiao X, Wang L, Huang D, Tan C, Xu Q, Zha R, Zhang J, Sheng W, He X, Du X (2014) microRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Clin Cancer Res 20:1146–1157. doi:10.1158/1078-0432.CCR-13-1023
Zhang Y, Zheng D, Xiong Y, Xue C, Chen G, Yan B, Ye Q (2014) miR-202 suppresses cell proliferation in human hepatocellular carcinoma by downregulating LRP6 post-transcriptionally. FEBS Lett 588:1913–1920. doi:10.1016/j.febslet.2014.03.030
Schrauder MG, Strick R, Schulz-Wendtland R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A, Hein A, Bayer CM, Bani MR, Richter S, Adamietz BR, Wenkel E, Rauh C, Beckmann MW, Fasching PA (2012) Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One 7:e29770. doi:10.1371/journal.pone.0029770
Yu J, Qiu X, Shen X, Shi W, Wu X, Gu G, Zhu B, Ju S (2014) miR-202 expression concentration and its clinical significance in the serum of multiple myeloma patients. Ann Clin Biochem 51:543–549
Zhao Y, Li C, Wang M, Su L, Qu Y, Li J, Yu B, Yan M, Yu Y, Liu B, Zhu Z (2013) Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS One 8:e69756. doi:10.1371/journal.pone.0069756
Hoffman AE, Liu R, Fu A, Zheng T, Slack F, Zhu Y (2013) Targetome profiling, pathway analysis and genetic association study implicate miR-202 in lymphomagenesis. Cancer Epidemiol Biomarkers Prev 22:327–336. doi:10.1158/1055-9965
Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, Einvik C (2011) Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer 105:296–303. doi:10.1038/bjc.2011.220
Yang W, Liu X, Choy E, Mankin H, Hornicek FJ, Duan Z (2013) Targeting hedgehog-GLI-2 pathway in osteosarcoma. J Orthop Res 31:502–509. doi:10.1002/jor.22230
Scales SJ, de Sauvage FJ (2009) Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 30:303–312. doi:10.1016/j.tips.2009.03.007
Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D, Li C, Tang H (2013) MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone 56:220–226. doi:10.1016/j.bone.2013.05.020
Kobayashi E, Hornicek FJ, Duan Z (2012) MicroRNA involvement in osteosarcoma. Sarcoma 2012:359739. doi:10.1155/2012/359739
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Regl G, Neill GW, Eichberger T, Kasper M, Ikram MS, Koller J, Hintner H, Quinn AG, Frischauf AM, Aberger F (2002) Human GLI2 and GLI1 are part of a positive feedback mechanism in Basal Cell Carcinoma. Oncogene 21:5529–5539
Nagao H, Ijiri K, Hirotsu M, Ishidou Y, Yamamoto T, Nagano S, Takizawa T, Nakashima K, Komiya S, Setoguchi T (2011) Role of GLI2 in the growth of human osteosarcoma. J Pathol 224:169–179. doi:10.1002/path.2880
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, Z., Zhang, T., Hong, H. et al. miR-202 suppresses proliferation and induces apoptosis of osteosarcoma cells by downregulating Gli2. Mol Cell Biochem 397, 277–283 (2014). https://doi.org/10.1007/s11010-014-2195-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-014-2195-z