Abstract
The pigs have similarities of organ size, immunology and physiology with humans. Porcine-induced pluripotent stem cells (piPSCs) have great potential application in regenerative medicine. Here, we established piPSCs induced from porcine fetal fibroblasts by the retroviral overexpression of Oct4, Sox2, Klf4, and c-Myc. The piPSCs not only express pluripotent markers but also have the capacity for differentiation in vivo and in vitro, including EB and teratoma formation. We supplemented microRNAs during the induction process because miR-302a, miR-302b, and miR-200c have been reported to be highly expressed in human and mouse embryonic stem cells and in iPSCs. In this study, we found that the overexpression of miR-302a, miR-302b, and miR-200c effectively improved the reprogramming efficiency and reduced the induction time for piPSCs in the OSKM and OSK induction systems. Due to the similar induction efficiency of 4F-induced piPSCs or of three factors combined with miR-302a, miR-302b, and miR-200c (3F-miRNA-induced piPSCs), we recommend the addition of miRNAs instead of c-Myc to reduce the tumorigenicity of piPSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine-induced pluripotent stem cells (piPSCs) are a potential resource for application in the studies of heredity and breeding, animal models of human disease and xenotransplantation [1]. The porcine species have similarities to humans in organ morphology and function, and are easier to maintain and breed than monkeys. Despite 20 years of progress, it remains difficult to establish standard porcine ESCs that have the characteristics of true pluripotency [2–4]. Thus, it was difficult to understand the mechanisms of porcine early embryonic development before we were inspired by the first appearance of mouse induced pluripotent stem cells [5]. The retroviral overexpression of Oct4, Sox2, Klf4, and c-Myc (OSKM) is the most classical approach to produce iPSCs, and this approach has been successfully used in mouse, human, monkey, and rat cells [5–8]. In this study, we established porcine-induced pluripotent stem cells through this approach.
MicroRNAs (miRNAs) are small, noncoding RNAs that serve as posttranscriptional regulators of gene expression in higher eukaryotes. The miRNAs can specifically bind to complementary target mRNAs to effect gene silencing via translational repression or target degradation [9, 10]. Previous studies have shown that several microRNAs are uniquely expressed in human and mouse embryonic stem cells and in pluripotent stem cells. With their crucial role in stem cell development, these miRNAs, which primarily belong to four clusters, miR-290, miR-302, miR-371, and miR-200c [11–13], can strongly promote the reprogramming efficiency of iPSCs. However, the study of whether the miRNAs play a similar role in piPSCs has not been performed.
miR-302 and miR200c are predominant miRNA species in human ESCs and iPSCs [14–17, 19]. According to a previous report, miR-302 may increase the efficiency of cell reprogramming in at least three ways. First, miR-302 inhibits NR2F2 [20] while stimulating the expression of Oct4 [21]. Second, miR-302 silences AOF1, AOF2, MECP1-p66, and MECP2 and downregulates DNMT1, which results in genome demethylation and iPSC development. Third, miR-302 inhibits TGFBR2 and RHOC expression, leading to the increased efficiency of reprogramming, in part by inducing a more efficient mesenchymal-to-epithelial transition (MET) [22, 23]. MET has been shown as an important early event in somatic cell reprogramming [22, 24, 25]. A recent report found that miR-200c is directly activated by Oct4 and Sox2 and significantly represses the expression of zinc finger E-box binding homeobox 2 (ZEB2) by directly targeting its 3′ UTR to help fibroblasts overcome the MET barrier and promote iPSC generation [26].
Porcine iPSCs are important to help us better understanding porcine embryonic development, to facilitate studies of heredity and breeding, and to establish animal models of human disease. Here, we established piPSCs by the overexpression of Oct4, Sox2, Klf4, and c-Myc. We found that miR-302a, miR-302b, and miR-200c improved the efficiency of cell reprogramming in the OSKM-induced system. These microRNAs were further shown to play a similar role in the OSK-induced system without c-Myc.
Results
Characterization of piPSCs reprogrammed from fibroblasts
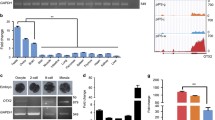
Porcine fetal fibroblasts (PFFs) were infected with retroviral vectors designed to express either four or three reprogramming genes (Oct4, Klf4, Sox2, and c-Myc; or Oct4, Klf4, and Sox2) (Fig. 1b). The reprogramming protocol is shown as a schematic diagram (Fig. 1a). The initial time of infection was designated as D0; the cell morphology began to change at D5, and progressed until ES-like colonies appeared at D12 (Fig. 1c). Cell staining by DAPI indicated that these colonies had well-defined borders and a high nuclear: cytoplasmic ratio (Fig. 1e) and were positive for alkaline phosphatase (AP) (Fig. 1f). The karyotype analysis of these piPSCs showed that no chromosomal abnormalities were present during cell reprogramming (Fig. 1g). These cells could be routinely passaged on feeders without losing their characteristics, but splitting without feeders induced rapid differentiation.
Porcine iPSC lines derived from PFFs. a Schematic diagram of reprogramming protocol in general, the miRNAs were added in medium at D2 and D4. b Vector maps of retroviral constructs used for reprogramming. c Cell morphology at D5, D8, and D12 during infection. d Typical ES-like colonies appeared at D14. e High nuclear/cytoplasmic ratio of piPSC colonies using DAPI stain. f The piPSC colonies were AP positive. g Karyotype analysis of piPSCs
piPSCs express pluripotency markers and can differentiate both in vivo and in vitro
The piPSCs were positive for the protein expression of pluripotency markers REX1, OCT4, NANOG, and SSEA4 by immunocytochemical staining (Fig. 2a). Porcine iPSCs then underwent 10 days of EB differentiation to assess their ability to differentiate into three germ layers (Fig. 2b). Each of the germ layer markers—the endoderm gene (AFP, alpgaAT), the mesoderm gene (BMP4, Enolase), and the ectoderm gene (GFAP, Neurod)—were expressed in EBs (Fig. 2c). The reprogramming of the PFFs with exogenous factors resulted in the reactivation of endogenous porcine Oct4, Sox2, Klf4, and Nanog, which were not expressed in PFFs (Fig. 2d). Semiquantitative RT-PCR indicated that mRNA products for the exogenous transgenes were not silenced (Fig. 2d). To ascertain their multilineage differentiation, piPSCs were injected subcutaneously into NOD/SCID mice and, after 2.5–4 weeks, resulted in teratomas (Fig. 2f) containing tissues derived from the three germ layers, including ectoderm-derived neural epithelium, mesoderm-derived striated muscle, and endoderm-derived crypt-like structures (Fig. 2g). We also analyzed the methylation of the Oct4 promoter in piPSCs and PFFs and found obvious demethylation in piPSCs compared to PFFs (Fig. 2e).
Characteristics and generational abilities of piPSCs in vivo and in vitro. a Immunocytochemical staining confirmed the expression of REX1, OCT4, NANOG, and SSEA4 in piPSCs. b Semiquantitative RT-PCR demonstrated the integration of the four transgenes into the genome of piPSC lines, and the robustly increased expression of endogenous Oct4, Nanog, Klf4, and Sox2. c Embryoid bodies derived from porcine iPS cells. d The positive expression of marker genes of the three germ layers was detected from the embryoid bodies: the endoderm gene (AFP, alpgaAT), the mesoderm gene (BMP4, Enolase), and the ectoderm gene (GFAP, Neurod). e Bisulfite genomic sequencing of the promoter regions of Oct4. The open and closed circles indicate unmethylated and methylated CpGs, respectively. f NOD/SCID mouse injected with 4F-iPSCs after 20days. g Teratomas derived from piPSCs revealed a contribution to all three germ layers
miRNAs promote the induction of pluripotent stem cells
To test the role of microRNAs in cell reprogramming, we selected miR-302a, miR-302b, and miR-200c, which have crucial role in maintaining the characteristics of embryonic stem cells and in reprogramming mouse and human iPSCs. We separately transfected 20 pmol of each group of miRNAs into the infected cells on D2 and D4 after the introduction of OKSM and then scored the AP-positive colonies at D15 (Fig. 3a). The induction efficiency with four factors was ~0.02–0.04 %; however, the efficiency of four factors combined with miR-302a and miR-302b, or four factors combined with miR-200c was obviously increased to 0.2–0.3 % (Fig. 3b). The time until the appearance of colonies was shortened from an average of 12 to 10 days (Fig. 3c). The results indicated that miR-302a, miR-302b, and miR-200c could efficiently promote the induction of OSKM-mediated piPSCs. Next, we introduced the combination of miR-302a, miR-302b, and miR200c into the OSK induction system and found that the induction efficiency increased from ~0.003 to 0.02 % (Fig. 3b); the time of appearance of colonies was shortened from an average of 14.5 to 10 days (Fig. 3c). These results suggested that miR-302a, miR-302b, and miR-200c could efficiently improve the induction of both OSKM- and OSK-mediated piPSCs.
miRNA302a, 302b and 200c promote the induction of piPSCs. a AP staining of 4F-induced piPSCs; 4F+miR-302a+miR-302b induction; 4F+miR-200c induction; 3F induction; 3F+miR-302a+miR-302b+miR-200c induction (4F:Oct4, Klf4, Sox2, c-Myc;3F:Oct4, Klf4, Sox2). b Induced efficiency of 4F-induced piPSCs; 4F+miR-302a+miR-302b induction; 4F+miR-200c induction; 3F induction; 3F+miR-302a+miR-302b+miR-200c induction (4F:Oct4, Klf4, Sox2, c-Myc;3F:Oct4, Klf4, Sox2). c Days of piPSC induction, 4F induction; 4F+miR-302a+miR-302b induction; 4F+miR-200c induction; 3F induction; 3F+miR-302a+miR-302b+miR-200c induction (4F: Oct4, Klf4, Sox2, c-Myc; 3F: Oct4, Klf4, Sox2)
3F-miRNA-induced piPSCs are pluripotent and can differentiate both in vivo and in vitro
To further investigate whether the miR-piPSCs induced by three factors reach a pluripotent state, the expression of pluripotent markers was analyzed by semiquantitative RT-PCR (Fig. 4a). These cells were all positive for the protein expression of pluripotency markers REX1, OCT4, NANOG, and SSEA4 by immunocytochemical staining (Fig. 4b). The analysis of embryonic bodies (Fig. 4c) and teratomas (Fig. 4d, e) indicated the miR-piPSCs have differentiation capacity both in vitro and in vivo. Further studies showed that miR-302a, miR-302b, and miR-200c induced 3F-piPSCs have similar characteristics as OSKM-mediated piPSCs.
Characteristics of 3F-miRNA-induced piPSCs in vivo and in vitro. a 3F-miRNA-induced piPSC colonies immunocytochemically stained positive for the human stage-specific embryonic antigen SSEA4 and the transcription factors OCT4, NANOG, and REX1. b Semiquantitative RT-PCR demonstrated the integration of the four transgenes into the genome of 3F-miRNA-induced piPSCs lines (P2iPS1201, P2iPS1202) and 4F-induced piPSCs (P1iPS0501, P1iPS0801), and the robustly increased expression of endogenous Oct4, Nanog, Lin28, and Sox2. c Embryoid bodies derived from 3F-miRNA-induced piPSCs. d NOD/SCID mouse injected with 3F-miRNA-iPSCs after 27 days. e Teratomas derived from 3F-miRNA-induced piPSCs revealed a contribution to all three germ layers
Discussion
The generation of piPSCs has been reported using the traditional four factors Oct4, Sox2, Klf4, and c-Myc by retroviral vectors [27–29] and by lentiviruses [11], and 4 factors with the addition of Nanog and Lin28 has been reported by DOX-inducible lentiviral vectors [12]. However, low efficiency is still a barrier to obtaining piPSCs.
There is a series of roadblocks during the epigenetic transformation from somatic cells to iPSCs, which is well known as the MET [30]. During the reprogramming, microRNA networks regulate MET [31] and have a crucial role in iPSC development.
According to previous reports, the miR-302 cluster enhances the reprogramming of human fibroblasts and hair follicle cells (hHFCs) through multiple targets including cell cycle regulators, epigenetic modifiers, and MET regulators [23, 32]. Other homologous miRNAs have also been found in hESCs, including miR-17/92, 371, 372, and 294; however, these miRNAs are not as highly expressed as miR-302 in undifferentiated embryonic stem cells [32].
A recent report demonstrated that the miR-200 family promotes the reprogramming of fibroblasts by activating Oct4 and Sox2 to overcome the MET barrier [26]. The combination of miR-302, miR-200c, and miR-369 has also been reported to induce pluripotency in mouse and human somatic cells [33]. Our results have shown that miR-302 and miR-200c could also enhance the reprogramming efficiency of porcine OSKM-induced iPSCs.
We further investigated whether these miRNAs could promote piPSCs generation with the OSK system because c-Myc is broadly implicated in tumorigenesis [34] and found to be nonessential for iPSC generation, although the speed of reprogramming is twice as slow without c-Myc [35]. miR-291-3p, 294, 295, and 302d were reported to enhance mouse iPSCs generation with OSK factors [36], and the expression of these miRNAs is high in ES cells [37] and is controlled by c-Myc [38]. These miRNAs directly target multiple cell cycle regulators, including p21, to accelerate the G1/S transition [39]; they simultaneously increase the expression of pluripotent transcription factors [40]. Moreover, microRNA clusters 17-92, 106b-25, 106a-363, and 302-367 also enhance mouse somatic cell reprogramming using either three or four factors [41, 42]. In this study, we produced piPSCs without using c-Myc and found that the combination of miR-302a, miR-302b, miR-200c, and OSK factors could efficiently enhance the generation of colonies compared to OSK alone to a similar efficiency as OSKM.
In conclusion, we successfully established piPSCs reprogrammed from PFFs with four or three factors (Oct4, Klf4, Sox2, and c-Myc; or Oct4, Klf4, and Sox2) and validated them both in vivo and in vitro to have similar characteristics as embryonic stem cells. Furthermore, we found that miR-302a, miR-302b, and miR-200c could not only enhance the formation of piPSCs colonies but also reduce the induction cycles in both OSKM- and OSK-induced systems.
Materials and methods
Reagents and animals
Chemicals, media, and cell culture reagents were purchased from Hyclone (US) unless otherwise stated. Pregnant Yorkshire pigs were purchased from the Jilin University Pig Farm. Specific SPF level ICR mice used for feeder cells and nonobese diabetic/severe combined immune deficient (NOD/SCID) mice used for teratoma formation were purchased from Vital River Laboratories (Beijing, China). All experiments involving animals were approved by and complied with the experimental practices and standards of the Animal Welfare and Research Ethics Committee at Jilin University (Approval ID: 20101008-2).
Cell preparation and culture
Porcine fetal fibroblasts (PFFs) were isolated from Yorkshire pigs at E25-30. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 1 mM GlutaMAX (Invitrogen, USA), and 50 units/ml penicillin–streptomycin (Sigma, USA). Only early four passages of PFFs were used for piPSCs induction. Mouse embryonic fibroblasts (MEFs) were isolated from ICR mice at E12.5. MEFs were used for feeder layer cells after treatment with mitomycin C (10 μg/ml) for 3 h. HEK293GP cells were maintained in high-glucose DMEM supplemented with 10 % FBS. The piPSCs were grown and passaged on feeder cells in ES medium with knockout DMEM supplemented with 20 % knockout serum, 1 mM GlutaMAX, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol (Sigma, USA), 100 U/ml leukemia inhibitory factor (LIF) (Millipore, USA), and 50 units/ml penicillin–streptomycin (Invitrogen, USA). The medium was changed every other day.
Retroviral production and generation of piPSCs from PFFs
Retroviral plasmids containing mouse Oct4, Sox2, Klf4, and c-Myc were kindly provided by Professor Qi Zhou, Institute of Zoology, Chinese Academy of Sciences. HEK293GP cells (1 × 107) were plated in 100 mm culture dishes (Corning, USA). Plasmids were transfected on the following day. A quantity of 10 ml OptiMEM (Gibco, USA) was added to each culture dish when cells reached 80 % confluence. HEK293GP cells were then transduced separately with pCMV-VSV-G plasmid and single-factor retroviral plasmids using Lipofectamine 2000 (Invitrogen, USA). After 6 h, the medium was changed to 10 ml fresh HEK293GP medium. Viruses were collected 48 and 72 h later and filtered (0.45 μm pore size) before added onto PFFs.
PFFs were seeded at a density of 106 cells per well in 6-well plates (Corning, USA) and maintained in 5 % CO2, and 95 % air at 37.8 °C. After 24 h, the cells were infected with cocktails of the previously packaged retroviruses. After 5 days, the PFFs were digested by 0.25 % trypsin (Gibco, USA) and moved onto feeder layer cells in ES media. When ESC-like colonies appeared, they were harvested and maintained in ES media (Fig. 1a). The media were changed every other day until the colonies were large enough to be isolated.
miRNA mimic composition and induction
The miRNA, FAM positive control, and standard negative control miRNA mimics were purchased from Genepharma Co., Ltd. (Shanghai, China). Mature miRNAs are conserved between different species, so we used human mature microRNAs of miR-302a, miR-302b, and miR200c. The sequences were as follows:
-
Has-miR-302a-3p: sence(5′–3′): UAAGUGCUUCCAUGUUUUGGUGA
-
antisense(5′-3′): ACCAAAACAUGGAAGCACUUAUU
-
Has-miR-302b-3p: sence(5′-3′): UAAGUGCUUCCAUGUUUUAGUAG
-
antisense(5′-3′): ACUCAAACAUGGAAGCACUUAUU
-
Has-miR-200c-3p: sence(5′-3′): UAAUACUGCCGGGUAAUGAUGGA
-
antisense(5′-3′): CAUCAUUACCCGGCAGUAUUAUU
A quantity of 20 pmol miRNAs was separately transfected into the infected cells with Lipofectamine 2000 on D2 and D4 after the introduction of OSKM or OSK factors (Fig. 1a); AP-positive colonies were scored at D15 (Fig. 3a).
Reverse transcription and semi-quantitative PCR analyses
Total RNA was isolated with Trizol reagent (Invitrogen, USA). The synthesis of the cDNA was performed by Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA). Semiquantitative PCR reactions were performed in a 25 μl volume containing 1 μl of cDNA, 12.5 μl of rTaq premix (TaKaRa, Japan), 10.5 μl of RNase-free water, and 0.5 μl each of the forward and reverse primers (10 pmol) (primers shown in Table 1) for each gene. All the products were resolved on ethidium bromide-stained agarose gels. PCRs were repeated at least twice, and the mean relative expression level was calculated. The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The PCR primers are listed in Table 2.
Alkaline phosphatase staining and immunocytochemical staining
The piPSCs were washed twice in PBS (with Mg2+ and Ca2+) and fixed with 4 % paraformaldehyde before staining. The AP staining was performed using AP substrate kit (Chemicon, USA). The piPSCs were immunocytochemically stained with OCT4 (1:100, Abcam, ab18976), SOX2 (1:100, Sigma, AV38232), NANOG (1:100, Abcam, ab80892), REX1 (1:100, Abcam, ab50828), and SSEA4 (1:100, Abcam, ab16287) antibodies. The cells were incubated with secondary antibodies (Alexa Fluor 488 goat anti-rabbit, 1:400, Invitrogen; Alexa Fluor 568 goat anti-mouse, 1:400, Invitrogen). Photomicrographs were taken with a fluorescent microscope (TE2000-U Nikon/C-SHG1, Japan) and a light microscope (AE20 Motic, German).
In vitro and in vivo differentiation of piPSCs
Porcine iPSCs were harvested by Dispase (Gibco, USA), and were incubated twice on 0.1 % gelatin-coated plates for 30 min to exclude residue feeders. After incubation, 1 × 106 cells were injected into nonobese diabetic/severe combined immune deficient (NOD/SCID) mice per site. After 2.5–4 weeks, the sections of tumors embedded in paraffin were stained with hematoxylin/eosin (HE). Porcine iPSCs were digested by 0.25 % trypsin and were incubated twice on 0.1 % gelatin-coated plates for 30 min to exclude residue feeders. Every liquid drop with 3 × 105 single cells were cultured in nonadherent bacterial culture plates in KSR-based media without LIF. Embryoid bodies (EBs) were collected after six days. Total RNA derived from plated embryoid bodies on day six was used for RT-PCR analysis.
Bisulfite genomic sequencing and karyotype analysis
Treatment for piPSCs was performed with a CpGenome modification kit (Chemicon, USA). The amplified PCR products were cloned into the T-Simple vector (T-Simple; Takara, Japan), and at least ten random clones were sequenced (primers shown in Table 1). Karyotype analysis was performed for G-banding straining [42] and suggested the piPSCs had a normal karyotype.
Data analyses
Data are shown as the mean and SD. All statistical analyses were performed with Excel 2010 (Microsoft). Differences were considered statistically significant at *p < 0.05.
References
Hall V (2008) Porcine embryonic stem cells: a possible source for cell replacement therapy. Stem Cell Rev 4:275–282. doi:10.1007/s12015-008-9040-2
Strojek RM, Reed MA, Hoover JL, Wagner TE (1990) A method for cultivating morphologically undifferentiated embryonic stem cells from porcine blastocysts. Theriogenology 33:901–913
Li M, Zhang D, Hou Y, Jiao L, Zheng X, Wang WH (2003) Isolation and culture of embryonic stem cells from porcine blastocysts. Mol Reprod Dev 65:429–434. doi:10.1002/mrd.10301
Blomberg LA, Schreier LL, Talbot NC (2008) Expression analysis of pluripotency factors in the undifferentiated porcine inner cell mass and epiblast during in vitro culture. Mol Reprod Dev 75:450–463. doi:10.1002/mrd.20780
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. doi:10.1016/j.cell.2007.11.019
Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317. doi:10.1038/nature05934
Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H (2008) Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3:587–590. doi:10.1016/j.stem.2008.10.014
Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S (2009) Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4:16–19. doi:10.1016/j.stem.2008.11.014
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi:10.1016/j.cell.2009.01.002
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. doi:10.1038/nature02871
Ezashi T, Telugu BP, Alexenko AP, Sachdev S, Sinha S, Roberts RM (2009) Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA 106:10993–10998. doi:10.1073/pnas.0905284106
Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, Dai H, Zhu H, Teng X, Cheng L, Xiao L (2009) Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol 1:46–54. doi:10.1093/jmcb/mjp003
Banito A, Gil J (2010) Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep 11:353–359. doi:10.1038/embor.2010.47
Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS (2004) Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270:488–498. doi:10.1016/j.ydbio.2004.02.019
Zovoilis A, Nolte J, Drusenheimer N, Zechner U, Hada H, Guan K, Hasenfuss G, Nayernia K, Engel W (2008) Multipotent adult germline stem cells and embryonic stem cells have similar microRNA profiles. Mol Hum Reprod 14:521–529. doi:10.1093/molehr/gan044
Ciaudo C, Servant N, Cognat V, Sarazin A, Kieffer E, Viville S, Colot V, Barillot E, Heard E, Voinnet O (2009) Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet 5:e1000620. doi:10.1371/journal.pgen.1000620
Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC (2009) MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 18:749–758. doi:10.1089/scd.2008.0247
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, Ruzzo WL, Ware C, Radich JP, Gentleman R, Ruohola-Baker H, Tewari M (2008) MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26:2496–2505. doi:10.1634/stemcells.2008-0356
Rosa A, Brivanlou AH (2011) A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J 30:237–248. doi:10.1038/emboj.2010.319
Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28:6426–6438. doi:10.1128/MCB.00359-08
Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63. doi:10.1016/j.stem.2010.04.014
Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29:443–448. doi:10.1038/nbt.1862
Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7:64–77. doi:10.1016/j.stem.2010.04.015
Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, Qin D, Pan G, Chen J, Pei D, Zheng H (2013) Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT–MET mechanism for optimal reprogramming. Nat Cell Biol 15:829–838. doi:10.1038/ncb2765
Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, Gao L, Ye D, Zhou Y, Chen J, Wang J, Wu M, Liu H, Kang J (2013) Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci USA 110:2858–2863. doi:10.1073/pnas.1212769110
Roberts RM, Telugu BP, Ezashi T (2009) Induced pluripotent stem cells from swine (Sus scrofa): why they may prove to be important. Cell Cycle 8:3078–3081
Telugu BP, Ezashi T, Roberts RM (2010) Porcine induced pluripotent stem cells analogous to naive and primed embryonic stem cells of the mouse. Int J Dev Biol 54:1703–1711. doi:10.1387/ijdb.103200bt00bt
Esteban MA, Xu J, Yang J, Peng M, Qin D, Li W, Jiang Z, Chen J, Deng K, Zhong M, Cai J, Lai L, Pei D (2009) Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem 284:17634–17640. doi:10.1074/jbc.M109.008938
Esteban MA, Bao X, Zhuang Q, Zhou T, Qin B, Pei D (2012) The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev 22:423–428. doi:10.1016/j.gde.2012.09.004
Bao X, Zhu X, Liao B, Benda C, Zhuang Q, Pei D, Qin B, Esteban MA (2013) MicroRNAs in somatic cell reprogramming. Curr Opin Cell Biol 25:208–214. doi:10.1016/j.ceb.2012.12.004
Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT (2011) Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 39:1054–1065. doi:10.1093/nar/gkq850
Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8:633–638. doi:10.1016/j.stem.2011.05.001
Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448:445–451. doi:10.1038/nature05953
Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26:101–106. doi:10.1038/nbt1374
Judson RL, Babiarz JE, Venere M, Blelloch R (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27:459–461. doi:10.1038/nbt.1535
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533. doi:10.1016/j.cell.2008.07.020
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117. doi:10.1016/j.cell.2008.04.043
Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40:1478–1483. doi:10.1038/ng.250
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463:621–626. doi:10.1038/nature08725
Li Z, Yang CS, Nakashima K, Rana TM (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J 30:823–834. doi:10.1038/emboj.2011.2
Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B, Zhang R, Wu J, Lai L, Teng M, Niu L, Zhang B, Esteban MA, Pei D (2011) MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 286:17359–17364. doi:10.1074/jbc.C111.235960
Gustavsson I (1988) Standard karyotype of the domestic pig. Committee for the Standardized Karyotype of the Domestic Pig. Hereditas 109:151–157
Acknowledgments
This work was supported by the National Natural Science Foundation (No. 31271591), the National Basic Research Program (No. 2011CBA01003) and the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT1248) in China.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Kuiying Ma and Guangqi Song have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ma, K., Song, G., An, X. et al. miRNAs promote generation of porcine-induced pluripotent stem cells. Mol Cell Biochem 389, 209–218 (2014). https://doi.org/10.1007/s11010-013-1942-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-013-1942-x