Abstract
Oncogenic KRAS, an important target for antitumor therapy, contributes to pancreatic cancer tumorigenesis, progression and maintenance. However, intracellular compensation regulation can help cells to resist the targeted therapy. Gene knockdown method such as RNAi may help to understand this intracellular regulatory system and discover novel therapeutic approach. In this study, two stable transfected cell lines were constructed through lentivirus-mediated shRNA targeting KRAS of PANC-1 cells, to investigate cell response to the knockdown of KRAS. Human whole genome microarray was then used to compare the gene expression profile. As a result, ribosomal proteins L26 and L29 (RPL26 and RPL29) were dramatically upregulated by KRAS-shRNA specifically. To identify whether RPL26 or RPL29 was critical for PANC-1 cells, siRNAs against RPL26 and RPL29 were designed and transfected in vitro. The results showed that knockdown of RPL26 or RPL29 expression significantly suppressed cell proliferation, induced cell arrest at G0/G1 phase and enhanced cell apoptosis. Reactive oxygen species (ROS) assay indicated that silencing of RPL26 or RPL29 markedly decreased the intracellular ROS generation. Our findings imply that siRNA interference against RPL26 and RPL29 is of potential value for intervention of pancreatic cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RAS genes are frequently mutated in human cancers. The proteins encoded by the RAS genes (K-RAS, H-RAS and N-RAS) are essential components of signaling networks that regulate various cellular activities, including proliferation, differentiation and survival [1, 2]. Of all human cancers, pancreatic cancer (PC) gives the lowest survival rate [3, 4], and its KRAS mutation goes as high as nearly 100 % [5], which indicates KRAS could be an attractive therapeutic target. Nevertheless, inhibiting this single critical target did not eliminate the malignancy of PC thoroughly [6, 7]. Drug resistance remains tough issue to all cancer therapy including PC [8, 9].
RNA interference (RNAi) was reported and has been used on KRAS to study the relationship between KRAS and PC [10]. However, the members of the Ras GTPase family are crucial players in many signaling networks connecting a great variety of upstream signals to an even wider set of downstream effectors pathways. Specific knockdown of activated N-RAS via RNAi in human hepatoma cells resulted in cell response to N-RAS knockdown [7]. The cell response involved in changing expression of many genes that may be a compensation of N-RAS knockdown for cell growth. Similarly, shock of KRAS may allow other genes act as salvage with compensatory mechanisms [11]. Study on such compensation may help to discover more therapeutic targets for PC.
It is noted that long-term KRAS silencing means that cancer cells probably make more compensation. Aimed at discovering novel therapeutic approach for PC through investigating the intracellular regulatory system countered the KRAS silencing; in this study, we report the result of our efforts to construct stable transfected cell lines through the lentivirus-mediated shRNA interference method, and to find out the intracellular regulatory characteristics through human genome microarray assay in this long-term suppression model. The data analysis showed that a number of genes expression were changed after KRAS-shRNA treatment. Among them, ribosomal proteins L26 and L29 (RPL26 and RPL29) were upregulated and picked for further validation because of the extraribosomal functions in many cancer. Furthermore, the effects of silencing RPL26 and RPL29 with siRNA were examined on proliferation of the pancreatic cancer PANC-1 cells.
Materials and methods
Tumor cell line and culture condition
Human pancreatic cancer cell line PANC-1 and HEK293T cell line were maintained in vitro in DMEM high-glucose medium (Gibco, CA, USA) supplemented with 10 % (v/v) fetal bovine serum (FBS) (Gibco). Cells were incubated at 37 °C in a humidified incubator with 5 % CO2. All cells were obtained from the Committee on Type Culture Collection of Chinese Academy of Sciences (Shanghai, China).
Lentivirus-mediated shRNA interference
Lentivirus vector pLKO.1-GFP was a gift from Dr Jianzhong Xi (Peking University, Department of Biomedical Engineering College of Engineering, Beijing, China). The target sequences for KRAS gene (Genbank NM_004985.3) were 5′-TAGTTGGAGCTGATGGCGTAG-3′ (KRAS-shRNA-1, targeting mutation site at codon 12) and 5′-AAGAGTGCCTTGACGATACAG-3′ (KRAS-shRNA-2). Two oligonucleotides, encoding short hairpin transcripts directed against different portion of the KRAS mRNA respectively, were synthesized (Table 1). The oligonucleotide pairs were subcloned into the vector after annealing to generate pLKO.1-KRAS shRNA vector (Fig. 1a). The empty vector served as control at the same time. Lentiviruses were produced by transfecting HEK293T packaging cells with a 3-plasmid system and lentiviral infection as described by Moffat [12]. Thereafter, cells were cultured in DMEM with 20 % (v/v) FBS for several generations to increase the survival rate of silencing KRAS. GFP-positive cells were sorted by automated cell deposition unit (ACDU) in BD FACSAriaTM (Institute of Biochemistry and Cell Biology, Shanghai, China). Cells were collected in 96-well plates at single cell/well for expansion of cultivation. Single-cell clonings were obtained for further analysis.
Lentivirus-mediated shRNA targeting KRAS of PANC-1 constructs P-M and P-W cell lines. a The structures of the pLKO.1-GFP vector and the schematic diagram of siRNA generating process. The DNA template about KRAS-shRNA was ligated in the downstream U6 promoter. Transcripts of the template will produce shRNA which has a 21-nt double-strand stem with a 6-nt loop and targets KRAS mRNA. b GFP-positive cells sorting by ACDU FCM. The R1 area (red) shows GFP-negative cells. The R2 area (green) shows the GFP-positive cells that were collected and cultured in the 96-well plates. c Photos of GFP expression in cells. P-M and P-W cell lines gave the green fluorescence (magnification: ×100). PANC-1 cells have no fluorescence under the same conditions. (Color figure online)
Stable transfected cell lines validation
Stability of single-cell clonings was verified by Flow Cytometry (FCM) and genomic DNA PCR amplification method. FCM was used to detect the GFP expression of different generation cells. Briefly, the cells were spread to the first generation (P1), the sixth generation (P6) and the 12th generation (P12), and then the GFP fluorescence intensity was detected at 10,000 events by FCM, respectively. The genomic DNA PCR primers (Table 2) were designed according to the sequence of both sides of shRNA in pLKO.1-GFP vector. PCR was implemented and the anticipated sequences were detected by Invitrogen (Shanghai, China).
RNA preparation and microarray analysis
Stable transfected cells and control PANC-1 cells were lysed using TRNzol™ Reagent (TIANGEN, Beijing, China) according to standard protocol. Briefly, 107 cells were collected and rinsed twice with phosphate-buffered saline (PBS), and then 1-ml TRNzol was used to lysis cells. Total RNA were isolated and further purified with Total Prep RNA amplification kit (Illumina, CA, USA) by Genergy Co., Ltd (Shanghai, China). HumanHT-12 Expression Bead Chip Microarray (Illumina, CA, USA) was used to analyze the gene expression profile by Genergy Co., Ltd. Gene Cluster 3.0 was applied to compare the similarity among individual samples.
siRNA transfection
The gene-specific siRNAs of RPL26 (Genbank NM_000987.3) and RPL29 (Genbank NM_000992.2) were designed and synthesized by GenePharma Co., Ltd (Shanghai, China). The Mock-siRNA as negative control (NC) also was provided. siRNA sequences were shown in Table 3. About 24 h before transfection, PANC-1 cells were trypsinized and plated on 12-well plates at a density of 5 × 104 cells/well. Transfection was performed at 30–50 % cell confluence with above siRNAs (40 nM), respectively using lipofectamine2000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s guidelines. At 6 h after transfection, the media were removed and replaced with fresh growth media. Cells were assayed at 48 h after transfection.
Quantitative real-time PCR and western blot analysis
Total RNA was extracted from treated or untreated cells using the TRNzol™ Reagent (TIANGEN, Beijing, China). Reverse transcription reactions were carried out using 2 μg of total RNA and the First Strand cDNA Synthesis Kit (Fermentas, Hanover, MD, USA). Quantitative real-time PCR (qPCR) was performed on the Master cycler ep realplex Real-Time System (Eppendorf, Germany) using the SYBR Green qPCR kit (Fermentas, Hanover, MD, USA) and gene-specific primers. 2−(∆∆ct) normalization [13] was used to calculate the results. Primers designed for detecting KRAS mRNA, RPL26 mRNA, RPL29 mRNA and GAPDH mRNA were shown in Table 2.
For protein expression analysis, immunoblotting was performed as described by Jiang [14]. Briefly, cells were rinsed twice with ice-cold PBS, harvested and lysed with RIPA Lysis Buffer (Beyotime, Shanghai, China). Subsequently, the whole-cell extracts were cleared by centrifugation at 1.2 × 104×g for 20 min at 4 °C, and the protein concentration was determined using BCA Protein Assay Kit (Beyotime, Shanghai, China). Equal amounts (20 μg) of each protein sample were electrophoresed on 12 % SDS–polyacrylamide gelsand and transferred onto PVDF membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5 % nonfat milk PBS-T buffer at room temperature for 1 h and incubated for 2 h with primary antibodies. Then, the membranes were incubated for 1 h with an appropriate horseradish peroxidase-linked secondary antibody (1:5,000 dilution) at room temperature. Immunoreactive proteins were visualized using an enhanced chemiluminescence detection system (Thermo Fisher Scientific, Waltham, MA, USA).
Primary monoclonal antibodies to KRAS (sc-30) and to GAPDH (sc-3233) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Primary polyclonal antibody to RPL26 (AP15410b) was purchased from Abgent Inc. (San Diego, CA, USA). Primary polyclonal antibody to RPL29 (A01) was purchased from Abnova Inc. (Taipei, Taiwan). Secondary antibodies (Goat Anti-Mouse IgG and Goat Anti-Rabbit IgG) were purchased from ZSGB-BIO Inc. (Beijing, China).
Re-expression of KRAS in stable transfected cell lines
The pcDNA3.1/myc-His(-)A plasmid was a gift of Dr Wenwei Mao (Shanghai Jiao Tong University, School of Pharmacy, Shanghai, China). Total RNA was extracted from PANC-1 cells using the TRNzol™ Reagent (TIANGEN, Beijing, China). Full-length cDNA template of PANC-1 cells was synthesized by First Strand cDNA Synthesis Kit (Fermentas, Hanover, MD, USA). The PCR primers of full-length KRAS gene were designed and synthesized (Table 2). The pcDNA3.1/myc-His(-)A plasmid containing the full-length KRAS was transfected into PANC-1 cells by lipofectamine2000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s guidelines. The empty pcDNA3.1/myc-His(-)A plasmid was the control plasmid for the re-expression of KRAS. After 48 h of transfection, the cells were lysed using TRNzol™ Reagent (TIANGEN, Beijing, China) to extract total RNA. Reverse transcription reactions were carried out using 2 μg of total RNA to obtain cDNA template of quantitative real-time PCR (qPCR). Then, the qPCR assays were performed to detect the expression of KRAS, RPL26 and RPL29 at mRNA level.
Cell proliferation assay and colony-forming assay
To observe cell proliferation, PANC-1 cells were seeded in 12-well plates (Corning, NY, USA) at a cell density of 5 × 104 cells/well and cultured in DMEM with 10 % fetal bovine serum overnight, and then were transiently transfected with Mock-siRNA, RPL26-siRNA and RPL29-siRNA (40 nM). At 24 h after transfection, the cells were trypsinized and seeded onto 96-well plates (Corning, NY, USA) at a cell density of 3,000 cells/well with 200 μl media. On each day for five consecutive days, the number of viable cells was determined by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay.
The MTT assay was performed as described by Mossmann [15]. Briefly, cells were incubated for 4 h at 37 °C with 0.8 mg/ml of MTT. Then, the reaction was stopped by lysing the cells with addition of 200 μl dimethyl sulfoxide (DMSO). Gentle shaking for 5 min so that complete dissolution was achieved. Absorbance was recorded at 490 nm with a microplate reader (Bio-Rad, Hercules, CA, USA).
To detect colony formation, cells were seeded into 35-mm dishes at a density of 1,000 cells/dish, after transfection (40 nM, 24 h). Cells were fed with new media every 4 days. After 8 days, colonies were washed three times with PBS and dyed with hematoxylin and counted.
Cell cycle and apoptosis analysis
PANC-1 cells (1.5 × 105) were seeded in six-well plates and transfected with Mock-siRNA, RPL26-siRNA and RPL29-siRNA (40 nM, 72 h). For cell cycle analysis, the cell DNA was stained with propidium iodide (PI) using Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China). Briefly, cells were harvested by trypsinization and fixed with cold 75 % ethanol at 4 °C overnight. The fixed cells were collected and suspended in PBS containing 10 μg/ml PI and 10 μg/ml RNase A, and then incubated at room temperature for 30 min. DNA content was analyzed by the BD FACSCalibur (BD Biosciences), and each histogram was constructed with the data from at 10,000 events. The data were analyzed and expressed as percentages of total gated cells using the Modfit LT™ Software (BD Biosciences).
For apoptosis analysis, the cells were washed twice with ice-cold PBS and stained with Annexin V-FITC and PI by Apoptosis Detection Kit (Keygen, Nanjing, China) in the dark at room temperature for 10 min. Then cells were analyzed with the BD FACSCalibur and FlowJo software. For analysis of changes in nuclear morphology during apoptosis, cells were stained with Hoechst 33258 using Hoechst Staining Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Fluorescence microscopy was used to examine condensed chromatin and nuclear fragment of apoptotic cells.
Determination of intracellular reactive oxygen species
Reactive oxygen species (ROS) were measured based on the intracellular peroxide-dependent oxidation of 2,7-dichlorfluorescein-diacetate (DCFH-DA), which forms the fluorescence compound 2,7-dichlorfluorescein (DCF). PANC-1 cells were seeded in 12-well plates at a density of 5 × 104 cells/well and cultured overnight. The cells were then transfected with Mock-siRNA, RPL26-siRNA and RPL29-siRNA (40 nM, 48 h). Washed twice with cold PBS and added with 10-μM DCFH-DA (Beyotime, Shanghai, China), the cells were incubated for 30 min at 37 °C in the dark. The fluorescence intensity was determined with FCM.
Statistical analysis
The statistical analysis was performed with SPSS 12.0 software (SPSS Inc., Chicago, IL, USA). Results were expressed as mean values ± standard deviation (SD). Student’s t test was used to analysis multiple variable comparisons. P < 0.05 was considered to be significant.
Results
Construction of stable transfected RNAi cell lines
Two KRAS-shRNA viruses 1 and 2 were constructed using lentivirus vector pLKO.1-GFP to target on different portion of the KRAS mRNA, respectively as described in “Materials and methods” section. Human pancreatic cell line PANC-1 cells were infected by the KRAS-shRNA viruses and control viruses with identical titer. This lentivirus vector carries the GFP marker and drives shRNA expression from a human U6 promoter. The shRNA folds back to form a hairpin loop structure after being transcribed, then it can be cleaved by endonuclease Dicer, and finally triggers degradation of the KRAS gene mRNA inside PANC-1 cells (Fig. 1a). It should be pointed out that the cells were difficult to survive after KRAS-shRNA viruses infection. We increased the serum content in culture medium up to 20 % (v/v), and the cells proliferated stably about 2 weeks later. Thereafter, the GFP-positive cells were sorted with ACDU FCM for single-cell cloning (Fig. 1b). Two stable cell lines (P-M cell line for KRAS-shRNA-1 and P-W cell line for KRAS-shRNA-2) were generated as indicated in Fig. 1c.
To confirm the stability, the FCM was used to detect the GFP expression in the cells undergoing successive passage. The results showed that when P-M and P-W were spread to different generation (P1, P6 and P12), the GFP expression was stable (Fig. 2a). Subsequently, the genomic DNA was extracted, PCR was performed and KRAS-shRNA sequences were detected (Fig. 2b). These results indicated that GFP and KRAS-shRNA had been integrated into the PANC-1 cells genome.
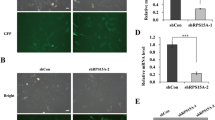
The stability and KRAS expression of P-M and P-W cell lines. a GFP fluorescence intensity of P-M and P-W were confirmed though FCM analysis. GFP expression remained unchanged at different generation (P1, P6 and P12). Three independent experiments were of similar results. b Sequencing results in P-M and P-W cell lines. Primers were designed according to the sequence of both sides of KRAS-shRNA in pLKO.1-GFP vector. PCR was implemented, and the correct sequence was detected. c KRAS expression in P-M and P-W cells. QPCR assays to show the KRAS mRNA levels was downregulated compared with PANC-1 cells (untreated) and control cells (empty viruses), and protein levels assays gave the similar results. GAPDH was used as an internal control for qPCR and western blot. Data are presented as the mean ± SD (n = 3). * indicates P < 0.05
To evaluate the efficiency of shRNA-mediated inhibition of KRAS in PANC-1 cells, qPCR and western blot were used to detect KRAS mRNA and protein levels in the P-M and P-W cell lines. As indicated in Fig. 2c, the expression of KRAS in P-M and P-W cells was specifically downregulated at both the mRNA and protein levels. Expression of the GAPDH internal control remained unchanged at the same time. All these results indicated that P-M and P-W cell lines were stable transfected with significant inhibition of KRAS expression.
KRAS-shRNA alters gene expression profile in PANC-1 cells
Total RNA were extracted from PANC-1, P-M and P-W cells for microarray experiments to show the alteration of gene expression profile. The sample diffscore values below −13 or >13 between PANC-1 and P-M or P-W were regarded as a significant change in mRNA expression. Comparing with PANC-1 cells, 133 genes were upregulated and 392 genes were downregulated in P-M cells, while 96 were upregulated and 439 were downregulated in P-W cells (data was not shown). The alternated profile between PANC-1 and P-M were different from PANC-1 and P-W. Then, we selected the overlapped genes, 43 genes were upregulated and 182 genes were downregulated in both P-M and P-W cell lines (Supplementary Table S1). Based on the roles in specific biological functions, the 225 overlapped genes were grouped by Gene Ontology (GO) (Fig. 3a). We focused our attention on those affecting the cellular process. According to GO descriptions, 22 upregulated genes were associated with the regulation of cellular process. These genes are LOC442454, HIST2H2AA3, HAPLN3, SNX5, PSAP, CRABP2, SNCA, RPL26, UGDH, MFGE8, RPL24, RPL39, CPS1, FTH1, RPL29, RPL12P6, KRT18, SQSTM1, RPL31, HMOX1, RPL21 and RPL10A.
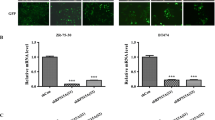
Summary of microarray results. a Functional categorization of genes from overlapped genes. 1 cellular process, 2 intracellular part, 3 intracellular organelle part, 4 organelle part, 5 intracellular non-membrane-bounded organelle, 6 non-membrane-bounded organelle, 7 anatomical structure morphogenesis, 8 chemical homeostasis and 9 alcohol metabolic process. b Validation of microarray results by qPCR with the expression of RPL26 and RPL29. c RPL26 and RPL29 downregulated by corresponding siRNAs (RPL26-siRNA and RPL29-siRNA, 40 nM, 48 h) both at mRNA and protein levels in PANC-1 cells compared with control (untransfected) and NC (Mock-siRNA) groups. GAPDH was used as an internal control. Each bar represents the mean ± SD of three separate experiments with triplicate wells per condition. * indicates P < 0.05
RPL26 and RPL29 upregulated by KRAS-shRNA and downregulated by re-expression of KRAS in P-M and P-W cells
Based on the observation from studies in many cancer [16, 17], two ribosomal protein genes RPL26 and RPL29 in 225 overlapped genes were selected for further verification by qPCR analysis which showed the same trend as microarray analysis in upregulated expression of RPL26 and 29 at mRNA level (Fig. 3b). Then, we considered whether re-expression of KRAS downregulated the expression of RPL26 and RPL29. As indicated in Fig. 4, the RPL26 and RPL29 were downregulated accompanied by upregulation of KRAS in P-M and P-W cells. These results indicated that RPL26 and RPL29 were upregulated after knockdown of KRAS, and this response was KRAS-specific.
Re-expression of KRAS in P-M and P-W cells resulted in downregulation of RPL26 and RPL29. a, c Re-expression of KRAS in P-M and P-W cells. Compared with P-M/P-W cells (untransfected) and control cells (empty vector), the KRAS was upregulated significantly. b, d Accompanied with upregulation of KRAS, the RPL26 and RPL29 were downregulated in P-M and P-W cells. Each bar represents the mean ± SD of three separate experiments with triplicate wells per condition. * indicates P < 0.05
RPL26 and RPL29 downregulated by corresponding siRNAs at both the mRNA and protein levels
To selectively inhibit the expression of RPL26 and RPL29, we synthesized the gene-specific siRNAs (RPL26-siRNA and RPL29-siRNA, 40 nM, 48 h) for the post-transcriptional gene silencing against RPL26 and RPL29 in PANC-1 cells. QPCR and western blot results revealed that RPL26 and RPL29 were significantly decreased by RPL26-siRNA and RPL29-siRNA at both the mRNA and protein levels compared with control (untransfected) and NC (Mock-siRNA) (Fig. 3c) groups, indicating that RPL26 and RPL29 were expressed in PANC-1 cells and selectively downregulated by the corresponding siRNA.
Silencing of RPL26 or RPL29 reduces PANC-1 cells proliferation and colony formation
To examine the effect of knockdown of RPL26 or RPL29 in PANC-1 cells, the proliferation and colony formation were determined. MTT analysis showed that the silencing of RPL26 or RPL29 genes markedly inhibited cell proliferation of PANC-1 cells compared with the control (untransfected) and NC (Mock-siRNA) groups (Fig. 5a). Consistent with MTT results, the ability of colony formation of PANC-1 cells was also dramatically decreased (Fig. 5b). These findings indicated that ribosomal protein genes RPL26 and RPL29 were closely related to PANC-1 cells proliferation.
Knockdown of RPL26 and RPL29 suppress PANC-1 cells proliferation in vitro. a The effect of transfection with RPL26-siRNA and RPL29-siRNA on cell proliferation. Cells were detected by MTT assay on each day for five consecutive days. Viable cell numbers were significantly different compared with control (untransfected) and NC (Mock-siRNA) groups. b For colony formation, PANC-1 cells were treated as described in “Materials and methods” section. The cells transfected with RPL26-siRNA and RPL29-siRNA showed much less colonies than control (untransfected) and NC (Mock-siRNA) groups. Each bar represents the mean ± SD of three separate experiments with triplicate wells per condition. * indicates P < 0.05
G1 arrest and apoptosis induced by RPL26 and RPL29 silencing in PANC-1 cells
Next, we got interested in whether cell cycles would be affected by knockdown of RPL26 and RPL29 in PANC-1 cells. FCM analysis revealed that the treatment of RPL26-siRNA or RPL29-siRNA (40 nM, 72 h) induced an accumulation in G1 phase and reduction in S phase (Fig. 6a). The ratio of G0/G1 phase cells was increased significantly (Fig. 6b) compared with the control (untransfected) and NC (Mock-siRNA) groups. Then, apoptosis of cells was evaluated by Annexin V-FITC/PI double-staining assay with FCM. The results suggested that knockdown of RPL26 or RPL29 enhanced apoptosis of PANC-1 cells (Fig. 6c) compared with the control (untransfected) and NC (Mock-siRNA) cells. The nuclear morphology for apoptotic cells was observed after transfection (40 nM, 72 h) with Hoechst 33258 by fluorescence microscopy (Fig. 6d). In apoptotic cells, the cell membrane increased the uptake of Hoechst 33258, and highly enriched chromosomes enhanced binding to Hoechst 33258. Therefore, the apoptotic cells showed strong blue fluorescence, while normal cells showed weak fluorescence. Hoechst 33258 staining assay indicated that a small proportion of cells was characterized with nuclear fragmentation and chromatin condensation, which were the typical hallmarks of apoptosis (Fig. 6d). No apoptotic nuclei or diffusely stained ones were observed in the control and NC cells (Fig. 6d). These changes led to the logic elucidation that suppression of cell proliferation might be caused by apoptotic cell death resulting from silencing of RPL26 or RPL29.
Silencing of RPL26 and RPL29 induces cell arrest at G0/G1 phase and enhances cell apoptosis in PANC-1 cells. a PANC-1 cells were transfected with RPL26-siRNA or RPL29-siRNA (40 nM) for 72 h, and then cell cycle measured using FCM analysis. b Percentage of cell cycle distribution. After treated with RPL26-siRNA or RPL29-siRNA, the proportion of G1 phase cells increased, and the percentage of S phase cells decreased. c PANC-1 cells were treated with RPL26-siRNA or RPL29-siRNA analyzed by Annexin V-FITC/PI staining using FCM analysis. The lower right area shows early apoptotic cells. Cell apoptosis was increased compared with control (untransfected) and NC (Mock-siRNA) groups. d Cell morphological changes with Hoechst 33258 staining. Nuclear shrinkage and chromatin condensation were examined by fluorescence microscopy (magnification: ×100). Red arrows indicate apoptotic cells. Three independent experiments were of similar results. Each bar represents the mean ± SD of triplicate analysis. * indicates P < 0.05. (Color figure online)
Silencing of RPL26 and RPL29 leading to ROS reduction in PANC-1 cells
The cancer cells have the lower Reactive oxygen species (ROS) clearance compared with normal cells. Normally, ROS play a positive role in the anti-inflammatory and antibacterial to protect cells. Abnormal high ROS level promotes cell DNA damage, genetic instability and drug resistance [18]. Therefore, through the oxidation of DNA and subsequent mutation of genes, ROS promote carcinogenesis [19]. Reducing intracellular ROS level, such as the use of antioxidants, represents a therapeutic strategies in cancer [20]. Intracellular ROS generation was evaluated in this study using intracellular peroxide-dependent oxidation of DCFH-DA, which forms fluorescence DCF. DCF fluorescence was detected following cell treatment with 40-nM siRNA for 48 h. As indicated in Fig. 7a and b, ROS production was decreased in the PANC-1 cells on treatment with RPL26-siRNA or RPL29-siRNA compared with control (untransfected) or treatment with Mock-siRNA. The results showed that silencing of RPL26 or RPL29 reduced intracellular ROS production, and further indicated that silencing of RPL26 and RPL29 is of potential value for intervention of pancreatic cancer.
Silencing of RPL26 and RPL29 decreases the intracellular reactive oxygen species (ROS) generation. a PANC-1 cells were treated by RPL26-siRNA or RPL29-siRNA (40 nM, 48 h), and the level of intracellular peroxide was detected by DCHF-DA assay using FCM analysis. First, the cells without adding DCHF-DA were detected by FCM, and then the M1 region was set. Next, each treated cells with DCHF-DA were detected by FCM at the same condition. Therefore, the M2 regions were cells with fluorescence DCF. b Decrease in intracellular fluorescence intensity detected in PANC-1 cells after treatment of RPL26-siRNA or RPL29-siRNA. The “M2-Geo Mean” represents the total intracellular fluorescence intensity of the M2 cells. Each bar represents the mean ± SD of three separate experiments, and each experiment was carried out in triplicate. * indicates P < 0.05
Discussion
Discovering novel target(s) for treatment of pancreatic cancer (PC) is a tough challenge. Despite that oncogenes such as KRAS have been characterized as therapeutic targets and various strategies have been used for clinic treatment to improve survival rate [21, 22], the current strategies may still not bring the survival rate to a satisfactory level. It is realized that better understanding of the molecular characterization of PC and exploration of more effective therapeutic targets are quite desirable.
RNA interference (RNAi) is a powerful tool for silencing the function of specific genes either by small interfering RNAs (siRNAs) or by short hairpin RNAs (shRNAs) [23, 24]. Lentivirus-mediated shRNA interference [12, 25] is useful for long-term gene silencing in target cells. Thus, the human immunodeficiency virus-based VSV-G-pseudotyped lentiviral vector was used to construct two stable transfected cell lines (P-M and P-W) in our study. However, as RNAi directed toward mutant KRAS disrupts the malignant phenotype of PC cells [26], it is difficult to get stable cell lines with low KRAS expression. Therefore, we increased the serum content in culture medium to promote cell survival. With serial passage, the cells were able to proliferate stably, indicating that the cells got adapted to decrease of KRAS expression. Study on such mechanism of intracellular systematic regulation can help to discover some new therapeutic targets [7].
Intracellular compensation regulation in tumor cells could develop resistance to various treatments such as chemotherapy and radiotherapy on cancer patients [27, 28]. The microarray data in this study showed that expression profile of a series of genes in P-M and P-W cell lines was altered in responding to inhibitory effect of KRAS by KRAS-shRNA. These genes are likely to be a kind of compensation for KRAS block. Interestingly, ribosomal proteins genes within 225 overlapped genes in P-M and P-W cells were all upregulated (Supplementary Table S1). The consistent trend indicated that these ribosomal proteins might be critical for PANC-1 cells’ development and survival. Therefore, P-M and P-W cell lines may be useful to study the compensation mechanism and to discover more effective therapeutic targets for PC.
Reports have demonstrated that upregulation of transcripts for several ribosomal proteins have been shown in several malignancies, including carcinomas of the colorectum, prostate and esophagus [29]. RPL26 or RPL29 encode a ribosomal protein that is a component of the 60S subunit. RPL26 and nucleolin may bind to 5′untranslated region (UTR) of p53 mRNA, which could control the translation and induction of p53 after DNA damage [30]. RPL29 (also known as Heparin/heparan sulfate interacting protein, HIP) may play a role in carcinogenesis [31]. In addition, it is an anti-apoptotic peptide, involved in regulation of apoptosis induced by anticancer drugs and in maintaining cancer phenotype [32, 33]. The extraribosomal function of RPL26 and RPL29 in pancreatic cancer is not yet clear. Our data showed that knockdown of KRAS in PANC-1 cells with two different shRNA (KRAS-shRNA-1 and KRAS-shRNA-2) upregulated the expression of RPL26 and RPL29. This fact suggested that RPL26 and RPL29 might be crucial for PC in cell proliferation. Further data showed that silencing of RPL26 or RPL29 resulted in significant affecting to the physiological activities of PANC-1 cells, such as cell proliferation, cell cycle, cell apoptosis and ROS generation. ROS, generated in mitochondria and as the byproducts of normal cellular oxidative processes, were mutagenic and promote cancer [19]. By detecting and comparing the changes of the intracellular ROS levels after knockdown of RPL26 or RPL29, which confirmed that RPL26/RPL29-specific siRNAs reduced intracellular ROS production. Knockdown of RPL26 and RPL29 is of potential value for treatment of PC.
As the ribosomal proteins, RPL26 and RPL29 are composed of the ribosome and control the protein translation. Our results showed that knockdown of RPL26 and RPL29 blocked cell cycle and promoted apoptosis in parental PANC-1 cells (Fig. 6). These effects may be KRAS-dependent or a general consequence of disturbing ribosomal function. Therefore, we deleted the RPL26 and RPL29 in KRAS knockdown cells (P-M and P-W cells) to observe the cell cycle and apoptosis. The results showed that knockdown of RPL26 and RPL29 in P-M and P-W cells induced cell arrest at G0/G1 phase and promoted apoptosis (Fig. S1), which was similar to PANC-1 cells. Our results indicated that although knockdown of KRAS caused upregulation of RPL26 and RPL29 specifically, the RPL26 and RPL29 played a role in cell cycle and apoptosis did not via KRAS. However, whether knockdown of RPL26 and RPL29 blocked cell cycle and promoted apoptosis was because of disturbing ribosomal function needed further research.
In summary, we constructed two stable transfected cell lines (P-M and P-W) through lentivirus vector-mediated silencing of KRAS in PANC-1 cells. By investigating the cell response to knockdown of KRAS, we identified that RPL26 and RPL29 are crucial for PANC-1 cells proliferation. To the best of our knowledge, this is the first report about the anti-pancreatic cancer activity of RPL26-specific siRNA and RPL29-specific siRNA in PANC-1 cells in vitro. Stable silencing of significant oncogenes might be a novel approach to discover the potential therapeutic targets in PC and in other malignant tumors.
Abbreviations
- RNAi:
-
RNA interference
- shRNA:
-
Short hairpin RNA
- siRNA:
-
Small interfering RNA
- ROS:
-
Reactive oxygen species
- PC:
-
Pancreatic cancer
- FBS:
-
Fetal bovine serum
- ACDU:
-
Automated cell deposition unit
- FCM:
-
Flow cytometry
- NC:
-
Negative control
- qPCR:
-
Quantitative real-time PCR
- FITC:
-
Fluorescein isothiocyanate
- PI:
-
Propidium iodide
- PBS:
-
Phosphate-buffered saline
- DCFH-DA:
-
2,7-Dichlorfluorescein-diacetate
- DCF:
-
2,7-Dichlorfluorescein
- MTT:
-
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- DMSO:
-
Dimethyl sulfoxide
- GO:
-
Gene Ontology
- PVDF:
-
Polyvinylidene fluoride
References
Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ (1998) Increasing complexity of Ras signaling. Oncogene 17:1395–1413
McCormick F (1989) ras GTPase activating protein: signal transmitter and signal terminator. Cell 56:5–8
Jaffee EM, Hruban RH, Canto M, Kern SE (2002) Focus on pancreas cancer. Cancer Cell 2:25–28
Jemal A, Murray T, Ward E et al (2005) Cancer statistics, 2005. CA Cancer J Clin 55:10–30
Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M (1988) Most human carcinomas of the exocrine pancreas contain mutant cK-ras genes. Cell 53:549–554
Shi X, Liang Z, Ren X, Liu T (2008) Combined silencing of K-ras and Akt2 oncogenes achieves synergistic effects in inhibiting pancreatic cancer cell growth in vitro and in vivo. Cancer Gene Ther 16:227–236
Zhao M, He H, Sun H, Ren K, Shao R (2009) Dual knockdown of N-ras and epiregulin synergistically suppressed the growth of human hepatoma cells. Biochem Biophys Res Commun 387:239–244
Dean M, Fojo T, Bates S (2005) Tumour stem cells and drug resistance. Nat Rev Cancer 5:275–284
Gottesman MM (2002) Mechanisms of cancer drug resistance. Annu Rev Med 53:615–627
Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA (2005) Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-C directed therapy. Mol Cancer Res 3:413–423
Hopfner M, Schuppan D, Scherubl H (2008) Growth factor receptors and related signalling pathways as targets for novel treatment strategies of hepatocellular cancer. World J Gastroenterol 14:1–14
Moffat J, Grueneberg DA, Yang X et al (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298
Kenneth JL, Thomas DS (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆ Ct method. Methods 25:402–408
Jiang R, Cabras G, Sheng W, Zeng Y, Ooka T (2009) Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia 11:964–973
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Ruggero D, Pandolfi PP (2003) Does the ribosome translate cancer? Nat Rev Cancer 3:179–192
Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N (2004) Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2:690–698
Trachootham D, Alexandre J, Huang P (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8:579–591
DeNicola GM, Karreth FA, Humpton TJ et al (2011) Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475:106–109
Fang J, Seki T, Maeda H (2009) Therapeutic strategies by modulating oxygen stress in cancer and inflammation. Adv Drug Deliv Rev 61:290–302
Friday BB, Adjei AA (2005) K-ras as a target for cancer therapy. Biochim Biophys Acta 1756:127–144
Ji Z, Mei FC, Lory PL, Gilbertson SR, Chen Y, Cheng X (2009) Chemical genetic screening of KRAS-based synthetic lethal inhibitors for pancreatic cancer. Front Biosci 14:2904–2910
Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553
Lee NS, Dohjima T, Bauer G et al (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol 20:500–505
Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P (2006) A versatile tool for conditional gene expression and knockdown. Nat Methods 3:109–116
Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243–247
Links M, Brown R (1999) Clinical relevance of the molecular mechanisms of resistance to anti-cancer drugs. Expert Rev Mol Med 1:1–21
Naito S, Yokomizo A, Koga H (1999) Mechanisms of drug resistance in chemotherapy for urogenital carcinoma. Int J Urol 6:427–439
Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34:3–11
Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123:49–63
Wang Y, Cheong D, Chan S, Hooi SC (1999) Heparin/heparan sulfate interacting protein gene expression is up-regulated in human colorectal carcinoma and correlated with differentiation status and metastasis. Cancer Res 59:2989–2994
Liu JJ, Huang BH, Zhang J, Carson DD, Hooi SC (2006) Repression of HIP/RPL29 expression induces differentiation in colon cancer cells. J Cell Physiol 207:287–292
Liu JJ, Zhang J, Ramanan S, Julian JA, Carson DD, Hooi SC (2004) Heparin/heparan sulfate interacting protein plays a role in apoptosis induced by anticancer drugs. Carcinogenesis 25:873–879
Acknowledgments
We are grateful to Dr Jianzhong Xi (Peking University, Department of Biomedical Engineering College of Engineering, Beijing, China) for lentiviruses production and critical reading of the manuscript. National New Drug Research and Development Project (Grant 2010ZX09401-403) supported this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, C., Ge, M., Yin, Y. et al. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol Cell Biochem 370, 127–139 (2012). https://doi.org/10.1007/s11010-012-1404-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-012-1404-x