Abstract
Estrogens exert their biological roles mainly through estrogen receptors (ER) that function as ligand-activated transcription factors. ER content in a cell is regulated by many factors and is decisive for estrogen action. The purpose of the present study was to investigate the influence of an 8-wk endurance training program on ER expression in the liver, right atrium (RA), and left ventricle (LV) of intact and ovariectomized (Ovx) rats. We measured ERα and ERβ mRNA content by reverse transcription-polymerase chain reaction (RT-PCR). We found an important increase in ERα mRNA levels in the liver (300%; P < 0.01) and in ERβ mRNA levels in the RA (200%; P < 0.05), and a marked decrease in ERα (80%; P < 0.01) and ERβ (40%; P < 0.05) transcripts content in the LV of intact rats after endurance training. On the other hand, ERα mRNA levels were depressed by 50% (P < 0.01) in the liver, and increased by 60% (P < 0.01) in LV of Ovx rats after exercise training. These results first indicate that endurance training is associated with modifications of ER transcripts levels in the liver, LV, and RA of female rats. More specifically, these effects are tissue and isoform-specific and the direction of the response (increase or decrease) is different in intact and Ovx rats. It is suggested that some of the adaptations to endurance training in liver and heart may be mediated by an ER-dependent mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is now well recognized that the effects of estrogens are not limited to the female reproductive system [1, 2]. Almost all organs of the body in both male and female are under estrogenic influence, and especially metabolic tissues such as liver and heart [1]. The biological actions of estrogens occur mainly when they bind to one of the two known estrogen receptor (ER) isoforms—ERα and Erβ—specifically distributed among tissues [1, 2]. These nuclear receptors are ligand-activated transcription factors and can stimulate or repress the expression of genes encoding proteins with important physiological functions [1, 2]. In addition, ER can be stimulated independently of estrogen-binding, by phosphorylation of the N-terminal ligand-independent activation function (AF-1 domain), present on the ER gene [3]. Activation of the AF-1 domain allows ER to stimulate the transcription of estrogen-regulated genes [3]. Therefore, some estrogenic effects involving an ER-mechanism are mediated in the absence of estrogens [3].
In the liver, the synthesis of angiotensinogen, blood coagulation factors, LDL receptor, and lipoproteins are under the control of an ERα-dependent mechanism [4–7], the ERβ isoform being absent in liver [8]. Estrogens also act in heart through ERα, where estradiol treatment reduces myocardial injury associated with ischemia-reperfusion [9, 10], and through ERβ, where estrogens exert a protection against left ventricular hypertrophy [11]. Since the capacity of a cell to mediate an estrogenic response is correlated to the intracellular ER content, it is of interest to study the stimuli that influence ER expression [7, 12, 13].
Hepatic ERα content is principally influenced by growth hormone action, but other pituitary and glucocorticoid hormones are involved [14–16]. It is known from the ovariectomized (Ovx) rat model that endogenous estradiol negatively regulates hepatic ERα content [17–19], whereas hyperphysiological doses of 17β-estradiol after exogenous administration or during pregnancy stimulates its synthesis [20–22]. Nevertheless, the changes in circulating estradiol levels associated with the estrous cycle do not affect ERα content in liver, unlike the fluctuations in ERα levels reported in uterus [16, 20, 21]. On the other hand, a positive regulation of ER levels by endogenous estrogens was reported in female rat atria using an Ovx model [23]. However, few studies looked at the regulation of ER content in heart, making it less defined and maybe not only endogenous estrogens are implicated. Taken together, it can be concluded that the regulation of ER expression is tissue-specific and can be influenced by several factors.
In addition to the above-mentioned stimuli, an endurance-trained state has also been reported to influence ER expression [24–26]. However, the information in this regard is scarce and limited to skeletal muscle. Two studies conducted by Lemoine et al. [24, 25] indicated changes in ERα transcripts levels after a 7-wk endurance training program in skeletal muscle. Furthermore, Wiik et al. [26] found higher ERα and ERβ transcripts levels in vastus lateralis of highly endurance-trained than in moderately active men. Lemoine et al. [24, 25] put forward the concept that an ER-mediated mechanism could contribute to the training adaptations of skeletal muscle. We hypothetized that ER expression is also modulated following endurance training in liver and heart. The rationale for this hypothesis is based on the evidence that both endurance training and estrogen signaling pathway share physiological targets in liver and heart. Examples of this are repression of the expression and the activity of hepatic lipase in liver [27, 28] and improvement of cardiac recovery after ischemia/reperfusion [29, 30]. In a first step, to increase our understanding of the link between ER and exercise training in these tissues, we determined the effects of an 8-wk endurance training program on ERα and ERβ transcripts expression in liver and 2 heart chambers, left ventricle (LV) and right atrium (RA) of intact female rats.
It is well recognized that Ovx in rodents results in an increased adiposity [31, 32], and induces several metabolic disturbances including hepatic steatosis, hypercholesterolemia, hyperinsulinemia [31, 33], and decreased insulin sensitivity [34]. Several of these disturbances are prevented by exercise training [34, 35]. Furthermore, exercise training and estrogen therapy augment cardiac and endothelial function in Ovx rats [36–39]. It is not known if ER transcripts are affected after exercise training in Ovx rats. This question is of interest especially that ER mRNA levels cannot rely on endogenous ovarian hormones anymore. Therefore, a second aim of the present study was to determine the changes in ER transcripts expression following endurance training in liver and heart of ovariectomized rats. We have shown that endurance training is associated with tissue- and isoform-specific ER changes in intact female rats and that the direction of the response is modified by the removal of ovaries.
Materials and methods
Animal care
Female Sprague–Dawley rats (n = 32) weighing 180–200 g (8-wk old) were obtained from Charles River (St-Constant, PQ) and housed individually. The 12:12-h light–dark cycle started at 6:00 AM, and room temperature was maintained at 20–23°C. All animals were given free access to the usual pellet rat chow (12.5% fat; 63.2% carbohydrate; 24.3% protein; kJ, Agribrands Purina Canada, Woodstock, ON) and tap water. They were treated similarly in terms of daily manipulations. The experiments described in this report were conducted according to the guidelines of the Canadian Council on Animal Care after institutional approval.
Surgery
Two days after their arrival in our laboratory, the rats were randomly assigned to ovariectomy (Ovx) or sham-operation (Sham) groups. Ovx was performed according to the technique described by Robertson et al. [40]. The animals were injected with antibiotics (Tribrissen 24%; 0.125 ml/kg sc) for 3 days, beginning on the day before surgery. For surgery, the rats were anesthetized with a mixture of ketamine–xylazine (61.5–7.6 mg/kg, i.p.).
Groups
Two days after surgery, Ovx and Sham rats were submitted to training (Tr), or remained sedentary (Sed), which resulted in a total of 4 groups (n = 8/group): sham-operated (Sham), sham-operated + endurance training (ShamTr), ovariectomized (Ovx), and ovariectomized + endurance training (OvxTr). Body weight and food intake in g were monitored every other day. All rats were killed 8 wk after surgical manipulation in the Tr or Sed state.
Endurance training protocol
Exercise training consisted of continuous running on a motor-driven rodent treadmill (Quinton Instruments, Seattle, WA), 5 times/wk for 8 wk. The rats progressively ran from 15 min/day at 15 m/min, 0% slope, up to 60 min/day at 26 m/min, 4% slope, for the last 4 wk. At the end of this 8-wk period, animals were sacrificed 48 h after the last exercise bout.
Tissue sampling
Rats were killed between 08:00 and 11:00 AM. Food was removed from the cage 2–3 h before sacrifice. Immediately after complete anesthesia (pentobarbital sodium; 50 mg/kg i.p.), the abdominal cavity was opened along the median line of the abdomen. Several organs and tissues were removed in the following order: liver, heart, uterus, femur, mesenteric, urogenital, and retroperitoneal fat depots, along with the right triceps surae (soleus, plantaris, and gastrocnemius). The liver median lobe was freeze-clamped and the heart was divided in its 4 chambers. The liver median lobe, the right atrium (RA), and the left ventricle (LV) were processed for ERα and ERβ mRNA determination and quantification. All tissue samples were weighed (Mettler AE 100), immediately frozen in liquid nitrogen, and stored at −78°C until further analysis. Finally, the right femur wet weight was obtained following a short-boiling period in a 10% KOH solution in order to remove the surrounding tissue.
Isolation of RNA and RT-PCR for hepatic ERα
ERα mRNA assessment in liver was conducted using RT-PCR while real time RT-PCR was used for determination of ERα and ERβ mRNA in heart because of technical problems. Total RNA was isolated from rat frozen liver with Trizol reagent (Life Technologies, Rockville, MD) according to manufacturer’s specifications. Total RNA was then treated with RNase-free DNase I under a standard protocol. The integrity and quality of the purified RNA were controlled by formaldehyde denaturing agarose gel electrophoresis and measurement of the A 260/A 280 ratio. First-strand cDNA was synthesized in a final volume of 40 μl containing first-strand buffer, 2 μg RNA, 2 μg hexanucleotide primer (Life Technologies), and avian myeloblastosis virus reverse transcriptase (12 units/μg RNA, Life Technologies). About 5 μl of first-strand cDNA were added to a PCR mixture and amplified for 27 cycles by incubation at 95°C for 1 min, at 55°C for 50 s and at 72°C for 1 min 10 s, with final incubation at 72°C for 3 min, all in a Robocycler gradient 40 thermocycler (Stratagene, La Jolla, CA). The ERα primer sequences are enumerated in Table 1. Amplification of 18S RNA by oligonucleotides was achieved according to the protocol provided by the manufacturer (Ambion, Austin, TX). Control RT-PCR omitted reverse transcriptase or RNA from the reaction mixture. After amplification, the samples were loaded and electrophoresed on 1.5% agarose gel. Bands stained by ethidium bromide were counted and analyzed with the Storm 840 imaging system and ImageQuant software (Version 4.2, Molecular Dynamics, Sunnyvale, CA). To validate RT-PCR as a tool for the semi-quantitative measurement of mRNA, dose–response curves were charted for different amounts of total RNA extracted from the rat liver, and the samples were quantified in the linear phase of PCR amplification. These data were normalized to the corresponding values of 18S RNA. All samples were run in duplicate. The intra- and inter-assay coefficients of variation were 6 and 8%, respectively.
Isolation of RNA and semi-quantitative real-time PCR for ERα and ERβ in heart
Total RNA was extracted from frozen hearts with Trizol (Invitrogen Life Technologies, Inc., Burlington, ON) according to manufacturer’s protocol. To remove genomic DNA, RNA samples were incubated with 2 U deoxyribonuclease I (DNase I; Invitrogen Life Technologies, Inc., Burlingon, ON)/μg RNA for 30 min at 37°C. The cDNA synthesis was performed as previously described. PCR was carried out in the iCycler IQ Real time PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA), using SYBR® green chemistry. Samples were analyzed in duplicate or triplicate. For amplification, 2 μl of diluted cDNA was added to a 20 μl reaction mixture containing 1× iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.) and 200 nM forward and reverse primers. The thermal cycling program was 95°C for 2 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Primers were purchased from Invitrogen Life Technologies, Inc. Primer pairs were chosen to minimize primer dimerization and to generate an amplicon between 150 and 350 bp. The primers used are enumerated in Table 1. Optical data were collected during the annealing step of each cycle. Following PCR, reaction products were melted for 1 min at 95°C, and then the temperature was lowered to 55°C, and then gradually increased to 95°C in 1.0°C increments, 10 s per increment. Optical data were collected over the duration of the temperature increase with a dramatic drop in fluorescence occurring. This was done to ensure that only one PCR product was amplified per reaction.
The relative expression of the RT-PCR products was determined using the ΔΔCt method. This method calculates relative expression using the equation: Fold induction = 2−[ΔΔCt], where Ct = the threshold cycle, i.e. the cycle number at which the sample’s relative fluorescence rises above the background fluorescence and ΔΔCt = [Ct gene of interest (unknown sample) − Ct GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (unknown sample)] − [Ct gene of interest (calibrator sample) − Ct GAPDH (calibrator sample)]. One of the control samples was chosen as the calibrator sample and used in each PCR. Each sample was run in duplicate and the mean Ct was used in the ΔΔCt equation. GAPDH was chosen for normalization instead of 18S RNA because the latter is rapidly amplified and taking into consideration the high sensitivity of real time RT-PCR, it is difficult to determine Ct for 18S RNA. GAPDH showed consistent expression relative to other housekeeping genes among the treatment groups in our array experiments.
Statistical analysis
Values are expressed as means ± SE. Statistical analyses were performed by a two-way ANOVA for non-repeated measures. Fisher’s PLSD post hoc test was used in the event of a significant (P < 0.05) ratio.
Results
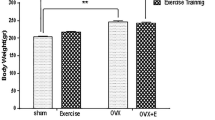
Comparison of body composition parameters in Sham and Ovx rats confirms that ovariectomies were well performed (Table 2) [31]. Ovx rats showed higher (P < 0.01) body weight, energy intake, intra-abdominal fat depot, and right triceps surae weights than Sham rats. Moreover, uterus weight was significantly lower in Ovx rats (P < 0.01), indicating total ovariectomy.
Endurance training led to increased (P < 0.05) body weight and energy intake in Sham rats, suggesting that intact animals did not experience negative energetic balance during the training period (Table 2). Exercise training resulted in a significant (P < 0.01) decrease in intra-abdominal fat depot weight when expressed per unit of body weight in both Sham and Ovx rats reflecting the effectiveness of the running program. In addition, femur weight was significantly increased (P < 0.01) with training in both Sham and Ovx groups.
The RT-PCR technique was used to demonstrate ER transcripts changes in response to endurance training in the liver and heart chambers (LV and RA) of intact and Ovx female rats. Our results confirmed that ovariectomy induced tissue-specific alterations of ER mRNA levels [41], and showed for the first time that endurance training affects ER expression in liver and heart.
A summary of the results on ER transcripts is presented in Table 3. ERα mRNA levels were increased by 200% in the liver and RA and decreased by 80% in LV of sedentary Ovx compared to Sham rats (P < 0.05; Fig. 1). Different effects of endurance training on ERα expression were observed in the liver and heart chambers, and between intact and Ovx rats. Hepatic ERα transcripts content was significantly higher (300%, P < 0.01) in Sham, but 50% lower (P < 0.01) in Ovx rats after endurance training (Fig. 1A). On the other hand, ERα mRNA levels in LV were decreased by 80% (P < 0.05) in Sham, and increased by 60% (P < 0.01) in Ovx rats after endurance training (Fig. 1B). There were no significant effects of training on ERα transcripts content in RA (Fig. 1C).
ERα mRNA levels in the liver, left ventricule (LV), and right atrium (RA) of sham-operated (Sham), sham-operated + endurance training (ShamTr), ovariectomized (Ovx), and ovariectomized + endurance training (OvxTr) rats. Values are means ± SE, n = 8 rats/group. *P < 0.05 and **P < 0.01: significantly different from corresponding Sham group. + P < 0.05 and ++ P < 0.01: significantly different from corresponding sedentary rats. Values were normalized to values of Sham-sed groups in (B) and (C)
ERβ transcripts levels were completely depressed by Ovx in LV (100%; P < 0.01; Fig. 2A), but remained unchanged in RA of sedentary rats (Fig. 2B). ERβ mRNA levels were 40% lower (P < 0.05; Fig. 2A) in LV, but 200% higher (P < 0.05; Fig. 2B) in RA of Sham rats after endurance training. However, endurance training did not change ERβ mRNA content in either LV or RA of Ovx rats (Fig. 2A and B).
ERβ mRNA levels in left ventricule (LV) and right atrium (RA) of sham-operated (Sham), sham-operated + endurance training (ShamTr), ovariectomized (Ovx), and ovariectomized + endurance training (OvxTr) rats. Values are means ± SE, n = 8 rats/group. *P < 0.05 and **P < 0.01: significantly different from corresponding Sham group. + P < 0.05: significantly different from corresponding sedentary rats. Values were normalized to values of Sham-sed groups. Values for Ovx and OvxTr rats were 0.001 ± 0.0003 and 0.001 ± 0.001, respectively
Discussion
The purpose of the present study was to investigate the effects of an 8-wk endurance training regimen on ER transcripts levels in liver and heart of female rats. We found significant alterations of ER expression in the liver, left ventricle, and right atrium of endurance-trained animals. More specifically, we showed that these effects are tissue and isoform-specific and that the direction of the changes in transcripts levels is different in Ovx than in intact rats.
Training in Sham rats
The most novel finding of the present study is the clear effect of endurance training on ER transcripts content in liver and heart. We found an important increase (300%) in hepatic ERα mRNA levels, accompanied by a decreased expression of both ERα and ERβ genes in LV, and an increase (200%) in ERβ transcripts content in RA following endurance training in intact rats. Our results are in line with the tissue-specific regulation of ER, which has been shown to be under the control of several factors including pituitary and gonadal hormones [14, 15, 23]. The present results extend these findings by indicating that an endurance-trained state also influence ER gene expression in liver and heart.
Our results raise the question as to why ER expression changes following endurance training in liver and heart. Endurance training is known to stimulate the expression of transcription factors, leading to the synthesis of proteins involved in the adaptations of skeletal muscle to training. For instance, the increased expression of the transcription factor NRF-1 takes part in the mitochondrial biogenesis processes in muscle in response to endurance training [42]. ERs are themselves transcription factors and variations in their transcripts levels may lead to stimulation or repression of target genes [1, 2]. Lemoine et al. [24, 25] were first to report an effect of endurance training on ERα gene expression in skeletal muscle. They put forward the concept that an ER-mediated mechanism could contribute to the training adaptations of skeletal muscle [24, 25]. The stimulation of mitochondrial biogenesis is an adaptation to training that may also be mediated by an ER-dependent mechanism in skeletal muscle, as a positive correlation was reported between ERs levels and citrate synthase activity [26]. Our data suggest that this concept could be extended to liver and heart. It seems likely that endurance training and estrogens share target genes in the liver. For instance, both endurance training and estrogens, through ERα, repressed the expression and activity of hepatic lipase, leading to increased levels of circulating HDL [22, 27, 28, 43]. We found important increases in hepatic ERα mRNA levels in endurance-trained rats. It is thus possible that the stimulation of the estrogen signaling pathway through endurance training participates in the repression of hepatic lipase by training.
Aside from the liver, the physiological role of estrogens on the myocardium is poorly understood, making the interpretation of our training effects on ER transcript levels more difficult. However, several associations exist for the ER content and myocardial functions and dysfunctions [44, 45]. For instance, ER are involved in the stability of cardiac intercalated discs and an up-regulation of ER content in myocardium is related to myocardial pressure load as a compensatory mechanism [44, 45]. The decreased levels of ERα mRNA content found in LV of our endurance-trained rats may, therefore, be interpreted as an indicator of an improvement in heart function. Nevertheless, it is difficult to reconcile the important decrease in both ERα and ERβ genes in LV following endurance training with the known cardioprotective effect associated with training and with the administration of estrogens [46, 47]. It is possible that both of these actions occur through a different mechanism. Recent evidence suggests that ERβ attenuation of cardiac dysfunction following trauma-hemorrhage is mediated via a nuclear as well as a mitochondrial action [48]. This reemphasizes our limited understanding of the effects and the mechanism of action of ERα and ERβ on the cardiovascular system [49]. The 80% decrease in LV ERα in Sham rats subjected to exercise can be related to observed increase of LV mass in these animals. It is well known that physiological cardiac hypertrophy, caused by chronic exercise training and defined as athlete’s heart is part of beneficial adaptive responses of the cardiovascular system to exercise training. Recently it was reported that estrogen treatment attenuates the increase in relative heart weight in the ovariectomized rats in association with ERα [50]. Therefore, training-related lowering of ERα in Sham can serve to promote development of physiological LV hypertrophy.
Effects of Ovx
It has been proposed that endogenous estrogens may be repressive on hepatic ERα gene expression of intact rats, as an elevation of ERα mRNA levels is observed after Ovx [17, 18]. Accordingly, we found an increase in hepatic ERα mRNA content after Ovx. On the other hand, the consequences of ovaries removal on ER transcripts levels in heart are poorly defined. A positive regulation of ERα levels by endogenous estrogens was reported in female rat atria following Ovx [23], while other reports showed no significant changes [41]. Our study is the first, to our knowledge, to report effects of Ovx on both ER isoforms transcripts levels in two different heart chambers of the female rat. We observed decreased ERα and ERβ mRNA levels in LV, and increased content of ERα transcripts in RA by Ovx. These results indicate the positive and negative regulation of ER mRNA levels in LV and RA, respectively, by sex steroids. Therefore, our findings in liver and heart chambers of Ovx rats further highlight the reported tissue-specific regulation of ERs by endogenous ovarian hormones.
Training in Ovx rats
Taking into account the fact that ER transcripts were largely modified by the Ovx, the second major finding of the present study is the different effect of endurance training found on ER transcripts levels according to the presence or the absence of ovaries. The training adaptations of ERα mRNA content in the liver and LV of Ovx animals were opposite to the training response observed in Sham rats. In addition, while exercise training altered ERβ mRNA content in LV and RA of Sham rats, it had no effect in Ovx rats. These findings suggest that the presence or absence of ovaries influence the response of ER gene expression to the stimulus of exercise. This is in line with the action of estradiol in liver that has been reported to depend on the presence of the pituitary hormones [12, 14, 15]. Thus, it seems that the responsiveness of the ER gene in liver and heart to the stimulus of exercise training is different according to the presence of ovarian steroids. The response of ER transcripts to exercise training in the present Ovx rats also suggest that exercise training compensated for the ovarian steroid deprivation in liver and heart. It is important to recall that the transcriptional activity of ER can be modified by factors other than estrogens [3, 51]. Exercise-induced activation of MAPK can lead to phosphorylation of the AF-1 domain of the ER gene, thus resulting in the stimulation or the repression of ER transcriptional activity [52, 53].
Although ERα is the main receptor known to mediate the estrogenic effects, both isoforms mediate specific actions, and the relative importance of the two isoforms is not well understood [2]. Our results also suggest that the ER gene expression in RA is less sensitive to different stimuli, e.g. ovariectomy (ovarian steroids deprivation) and exercise training, than the ER gene expression in the liver and LV.
There are limitations to the present study that need to be addressed. It could be argued that our training program disrupted the estrus cycle of intact rats and altered ER expression, since circulating estrogens can regulate the levels of their own receptors [23, 54, 55]. However, training-induced disruption of the estrus cycle is often associated with body weight loss and low energy intake [56, 57]. This was hardly the case in our study since intact female rats gained body weight and increased their energy intake with training. Moreover, changes in ERmRNA levels following endurance training were also observed in Ovx rats. Although plasma estradiol levels were not measured in the present study, it seems likely that endurance training was indeed responsible for the changes that we observed on ER expression in both intact and Ovx rats. On the other hand, the physiological interpretation of the present data is limited by the sole measurement of transcripts levels. However, the demonstration that these ER gene transcripts are modified with endurance training in liver and heart constitutes an interesting opening in a new avenue of research linking ER as a possible contributor to the training adaptations in these tissues.
In conclusion, we showed, using the RT-PCR technique that endurance training is associated with modifications of ER gene expression in liver, right atrium, and left ventricle of the heart. We demonstrated that these training effects are tissue and isoform specifics and that the responsiveness of the ER gene to the stimulus of endurance training is different in intact and Ovx animals. It is suggested that ER gene expression may mediate some of the adaptative effects of endurance training in liver and heart.
References
Ciocca DR, Vargas Roig LM (1995) Estrogen receptors in human nontarget tissues: biological and clinical implications. Endocr Rev 16:35–62
Matthews J, Gustafsson JA (2003) Estrogen signaling: a subtle balance between ERα and ERβ. Mol Interv 3:281–292
Kato S, Endoh H, Masuhiro Y et al (1995) Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491–1494
Brindley DN, Salter AM (1991) Hormonal regulation of the hepatic low density lipoprotein receptor and the catabolism of low density lipoproteins: relationship with the secretion of very low density lipoproteins. Prog Lipid Res 30:349–360
Farsetti A, Misiti S, Citarella F et al (1995) Molecular basis of estrogen regulation of Hageman factor XII gene expression. Endocrinology 136:5076–5083
Klett C, Ganten D, Hellmann W (1992) Regulation of hepatic angiotensinogen synthesis and secretion by steroid hormones. Endocrinology 130:3660–3668
Stavreus-Evers A, Parini P, Freyschuss B (2001) Estrogenic influence on the regulation of hepatic estrogen receptor-α and serum level of angiotensinogen in female rats. J Steroid Biochem Mol Biol 78:83–88
Kuiper GGJM, Carlsson B, Grandien K (1997) Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138:863–870
Booth EA, Obeid NR, Lucchesi BR (2005) Activation of estrogen receptor-α protects the in vivo rabbit heart from ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol 289:H2039–H2047
Jovanovic S, Jovanovic A, Shen WK et al (2000) Low concentrations of 17β-estradiol protect single cardiac cells against metabolic stress-induced Ca2+ loading. J Am Coll Cardiol 36:948–952
Babiker FA, Lips D, Meyer R et al (2006) Estrogen receptor β protects the murine heart against left ventricular hypertrophy. Arterioscler Thromb Vasc Biol 26:1524–1530
Rudling M, Norstedt G, Olivecrona H et al (1992) Importance of growth hormone for the induction of hepatic low density lipoprotein receptors. Proc Natl Acad Sci 89:6983–6987
Webb P, Lopez GN, Greene GL et al (1992) The limits of the cellular capacity to mediate an estrogen response. Mol Endocrinol 6:157–167
Freyschuss B, Sahlin L, Masironi B et al (1994) The hormonal regulation of the oestrogen receptor in rat liver: an interplay involving growth hormone, thyroid hormones and glucocorticoids. J Endocrinol 142:285–298
Norstedt G, Wrange O, Gustafsson JA (1981) Multihormonal regulation of the estrogen receptor in rat liver. Endocrinology 108:1190–1196
Stavreus-Evers AC, Freyschuss B, Eriksson HA (1997) Hormonal regulation of the estrogen receptor in primary cultures of hepatocytes from female rats. Steroids 62:647–654
Dickson RB, Eisenfeld AJ (1979) Estrogen receptor in liver of male and female rats: endocrine regulation and molecular properties. Biol Reprod 21:1105–1114
Eriksson HA (1982) Different regulation of the concentration of estrogen receptors in the rat liver and uterus following ovariectomy. FEBS Lett 149:91–95
Thompson C, Lucier GW (1983) Hepatic estrogen responsiveness. Mol Pharmacol 24:69–76
Lax ER, Tamulevicius P, Muller A et al (1983) Hepatic nuclear estrogen receptor concentrations in the rat – influence of age, sex, gestation, lactation and estrous cycle. J Steroid Biochem 19:1083–1088
Marr W, White JO, Elder MG et al (1980) Nucleo-cytoplasmic relationships of oestrogen receptors in rat liver during the oestrous cycle and in response to administered natural and synthetic oestrogen. Biochem J 190:17–25
Staels B, Jansen H, Van Tol A et al (1990) Development, food intake, and ethinylestradiol influence hepatic triglyceride lipase and LDL-receptor mRNA levels in rats. J Lipid Res 31:1211–1218
Jankowski M, Rachelska G, Donghao W et al (2001) Estrogen receptors activate atrial natriuretic peptide in the rat heart. Proc Natl Acad Sci 98:11765–11770
Lemoine S, Granier P, Tiffoche C et al (2002) Effect of endurance training on oestrogen receptor alpha transcripts in rat skeletal muscle. Acta Physiol Scand 174:283–289
Lemoine S, Granier P, Tiffoche C et al (2002) Effect of endurance training on oestrogen receptor alpha expression in different rat skeletal muscle type. Acta Physiol Scand 175:211–217
Wiik A, Gustafsson T, Esbjornsson M et al (2005) Expression of oestrogen receptor α and β is higher in skeletal muscle of highly endurance-trained than of moderately active men. Acta Physiol Scand 184:105–112
Bergeron J, Couillard C, Després JP et al (2001) Race differences in the response of postheparin plasma lipoprotein lipase and hepatic lipase activities to endurance exercise training in men. Results from the HERITAGE family study. Atherosclerosis 159:399–406
Jones DR, Schmidt RJ, Pickart RT et al (2002) Estrogen receptor-mediated repression of human hepatic lipase gene transcription. J Lipid Res 43:383–391
Quindry J, French J, Hamilton K et al (2005) Exercise training provides cardioprotection against ischemia-reperfusion induced apoptosis in young and old animals. Exp Gerontol 40:416–425
Xu Y, Arenas IA, Armstrong SJ et al (2006) Estrogen improves cardiac recovery after ischemia/reperfusion by decreasing tumor necrosis factor-α. Cardiovasc Res 69:836–844
Picard F, Deshaies Y, Lalonde J et al (2000) Effects of the estrogen antagonist EM-652.HCL on energy balance and lipid metabolism in ovariectomized rats. Int J Obes 24:830–840
Torto R, Boghossian S, Dube MG et al (2006) Central leptin gene therapy blocks ovariectomy-induced adiposity. Obesity 14:1312–1319
Lemieux C, Phaneuf D, Labrie F et al (2005) Estrogen receptor α-mediated adiposity-lowering and hypocholesterolemic actions of the selective estrogen receptor modulator acolbifen. Int J Obes 29:1236–1244
Latour MG, Shinoda M, Lavoie JM (2001) Metabolic effects of physical training in ovariectomized and hyperestrogenic rats. J Appl Physiol 90:235–241
Shinoda M, Latour MG, Lavoie JM (2002) Effects of physical training on body composition and organ weights in ovariectomized and hyperestrogenic rats. Int J Obes 26:335–343
Bupha-Intr T, Wattanapermpool J (2004) Cardioprotective effects of exercise training on myofilament calcium activation in ovariectomized rats. J Appl Physiol 96:1755–1760
Moien-Afshari F, Kenyon E, Choy JC et al (2003) Long-term effects of ovariectomy and estrogen replacement treatment on endothelial function in mature rats. Maturitas 45:213–223
Patten RD, Pourati I, Aronovitz MJ et al (2004) 17β-estradiol reduces cardiomyocyte apoptosis in vivo and in vitro via activation of phospho-inositide-3 kinase/Akt signaling. Circ Res 95:692–699
Rogers J, Sheriff DD (2004) Role of estrogen in nitric oxide- and prostaglandin-dependant modulation of vascular conductance during treadmill locomotion in rats. J Appl Physiol 97:756–763
Robertson MC, Owens RE, Klindt J et al (1984) Ovariectomy leads to a rapid increase in rat placental lactogen secretion. Endocrinology 114:1805–1811
Mohamed MK, Abdel-Rahman AA (2000) Effect of long-term ovariectomy and estrogen replacement on the expression of estrogen receptor gene in female rat. Eur J Endocrinol 142:307–314
Hood DA, Irrcher I, Ljubicic V et al (2006) Coordination of metabolic plasticity in skeletal muscle. J Exp Biol 209:2265–2275
Thompson PD, Cullinane EM, Sady SP et al (1991) High density lipoprotein metabolism in endurance athletes and sedentary men. Circulation 84:140–152
Mahmoodzadeh S, Eder S, Nordmeyer J et al (2006) Estrogen receptor alpha up-regulation and redistribution in human heart failure. FASEB J 20:929–934
Nordmeyer J, Eder S, Mahmoodzadeh S et al (2004) Upregulation of myocardial estrogen receptors in human aortic stenosis. Circulation 110:3270–3275
Brown DA, Moore RL (2007) Cardioprotection acquired through exercise. J Appl Physiol (Epub ahead of print). DOI: 10.1152/japplphysiol.00464.2007
Yu HP, Shimizu T, Choudhry MA et al (2006) Mechanism of cardioprotection following trauma-hemorrhagic shock by a selective estrogen receptor-beta agonist: up-regulation of cardiac heat shock factor-1 and heat shock proteins. J Mol Cell Cardiol 40:185–194
Hsieh YC, Yu HP, Suzuki T et al (2006) Upregulation of mitochondrial respiratory complex IV by estrogen receptor-beta is critical for inhibiting mitochondrial apoptotic signaling and restoring cardiac functions following trauma-hemorrhage. J Mol Cell Cardiol 41:511–521
Gabel SA, Walker VR, London RE et al (2005) Estrogen receptor beta mediates gender differences in ischemia/reperfusion injury. J Mol Cell Cardiol 38:289–297
Jazbutyte V, Hu K, Kruchten P et al (2006) Aging reduces the efficacy of estrogen substitution to attenuate cardiac hypertrophy in female spontaneously hypertensive rats. Hypertension 48:579–586
Bunone G, Briand PA, Miksicek RJ et al (1996) Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J 15:2174–2183
Iemitsu M, Maeda S, Jesmin S et al (2006) Activation pattern of MAPK signaling in the hearts of trained and untrained rats following a single bout of exercise. J Appl Physiol 101:151–163
Widegren U, Jiang XJ, Krook A et al (1998) Divergent effects of exercise on metabolic and mitogenic signaling pathways in human skeletal muscle. FASEB J 12:1379–1389
Carlberg KA, Fregly MJ (1985) Disruption of estrous cycles in exercise-trained rats. Proc Soc Exp Biol Med 179:21–24
Shupnik MA, Gordon MS, Chin WW (1989) Tissue-specific regulation of rat estrogen receptor mRNAs. Mol Endocrinol 3:660–665
Harber VJ (2000) Menstrual dysfunction in athletes: an energetic challenge. Exerc Sport Sci Rev 28:19–23
Williams NI, Helmreich DL, Parfitt DB et al (2001) Evidence for a causal role of low energy availability in the induction of menstrual cycle disturbances during strenuous exercise training. J Clin Endocrinol Metab 86:5184–5193
Acknowledgements
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC; JML) and the Canadian Institutes of Health Research (CIHR; JML and DP), and from CIHR MOP 53217 and CIHR NET SRD-63193, Heart and Stroke Foundation of Canada (JG and MJ).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Paquette, A., Wang, D., Gauthier, MS. et al. Specific adaptations of estrogen receptor α and β transcripts in liver and heart after endurance training in rats. Mol Cell Biochem 306, 179–187 (2007). https://doi.org/10.1007/s11010-007-9568-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-007-9568-5