Abstract
Antimicrobial peptides (AMPs) offer a novel alternative to present day antibiotics which are becoming ineffective due to development of resistance in microbes. In this study, a short synthetic β sheet forming antimicrobial peptide-IRK (IRIKIRIK) was synthesized and we studied the effect of multimerization of peptides on its antimicrobial activity. The activity of linear, dimeric and tetrameric forms of IRK peptide was evaluated against reference strain of Staphylococcus aureus (ATCC 29,213) and Methicillin Resistant Staphylococcus aureus (MRSA) clinical isolates (n = 25). It was observed that the linear IRK peptide showed a minimum inhibitory concentration (MIC) of 16 µM, whereas the dimeric IRK showed 65% and 76% inhibition of bacterial growth at 16 µM and 32 µM respectively. However, tetrameric form of IRK peptide showed only 60% inhibition at both 16 µM and 32 µM concentration. At higher concentration, i.e., 64 µM (equivalent to 4 X MIC for linear IRK), all forms of IRK peptide inhibited the growth of more than 92% of MRSA isolates. While antibacterial activity decreased with the branching of IRK peptide, the haemolytic activity increased with the degree of branching. Structural changes arising due to multimerization may have attributed to this change in biological activity. We can conclude that linear and branched forms of IRK peptide have immense potential to inhibit the growth of MRSA and can be used as an alternative to antibiotics, especially in formulations for topical applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The World Health Organization (WHO) categorizes the emergence of antibiotic resistance as one of the biggest threats to human health. The antibiotics that once saved millions of lives have now been transformed into a menace for living beings. The increase in bacterial resistance against conventional antibiotics and saturation in development of novel antibiotics have become a major setback in treatment of bacterial infections (Schäberle and Hack 2014). The antimicrobial resistance loom up has been contributed by the excessive and imprudent use of antibiotics (Sengupta et al. 2013) further aided by reduced economic incentives and challenging regulatory requirements (Gould and Bal 2013). Presently, the menace of Methicillin Resistant Staphylococcus aureus (MRSA) is of global concern (Sengupta et al. 2013). Previously, MRSA was considered as a major pathogen responsible for nosocomial infections but now it has emerged as a community-associated infection causing agent (Lakhundi and Zhang 2018).
Mastitis is an infectious disease of the mammary glands of dairy cattle, with high worldwide prevalence at both the cow and udder quarter levels (Abrahmsén et al. 2014). Many microorganisms are responsible for causing mastitis, with Streptococcus agalactiae, Staphylococcus aureus and Mycoplasma spp. as the major contagious pathogens (Gomes and Henriques 2016). Several reports have revealed isolation of multi drug resistant S. aureus from mastitic cow milk (Salauddin et al. 2020; Ismail 2017; Mohammed et al. 2018; Kozerski et al. 2014; Wang et al. 2018; Haran et al. 2012; Ren et al. 2020). Whenever a new molecule has been introduced these smart little bugs always find their ways to survive by altering shields, thus search for alternate sources of antibiotics is of utmost importance (Lewis 2013; Gajdács and Albericio 2019). Antibiotic resistance may arise by numerous mechanisms in bacteria which involve inactivation of antimicrobials enzymatically, antimicrobial target modifications/ alterations through acquired point mutations, increased efflux of antimicrobials, decrease in interaction between the antimicrobial and bacteria through altered drug accessibility followed by acquisition of resistance through mobile genetic elements (Vestergaard et al. 2019; Garg et al. 2020). Recent studies reveal that more than 99% of S. aureus clinical isolates are resistant to penicillin through the acquisition of blaZ gene responsible for coding beta lactamase enzyme. Methicillin was introduced as a semisynthetic alternative resistant to beta lactamase in the 60’s (Boyle-Vavra and Daum 2016). MRSA strains are resistant to Methicillin by acquiring the mecA gene that encodes for PBP2a, a transpeptidase effective against most beta lactams and its derivatives. Another gene, mecC has also been acquired by MRSA strains and aids to further cases of resistance.
Having the potential to fight against the alarming situation of antimicrobial resistance, Antimicrobial peptides (AMPs) are being contemplated as an alternative to conventional antibiotics (Wang et al. 2016). AMPs are potential, viable therapeutic agents which can be used to treat the MDR infections (Gupta et al. 2021). The unique mode of action of antimicrobial peptides with relatively no resistance development have lured many researchers to this newer approach of fighting superbugs since last few years.
There is constant effort to enhance the activity of antimicrobial peptides for which several strategies are in progress like N-terminal modification, C-terminal modification, Dimerization etc. There are reports that dimerization not only increases the peptide activity by many folds but also improves stability against proteases (Tam et al. 2002; Falciani et al. 2007) over their monomeric forms. However, there are reports that show decrease in antimicrobial activities, and increase in toxicity of peptides upon dimerization (Yang et al. 2009; Lorenzon et al. 2012). Reported synthetic β-sheet forming short amphiphilic peptide IK8L (IRIKIRIK) designed by Ong et al. (2014) was selected for this study because of its potent antimicrobial activity. In the present work we studied the influence of dimerization and tetramerization on the antimicrobial activity of the peptide against S. aureus ATCC 29,213 and MRSA isolates and further elucidated their haemolytic activity.
Materials and Methods
Synthesis and Purification of Peptide
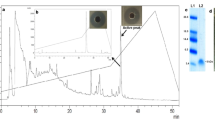
The antimicrobial peptide IRK (IRIKIRIK) was synthesized in linear, dimer and tetramer form by stepwise solid phase peptide synthesis on Rink amide resin (Novabiochem, Germany) using Fmoc chemistry (Merrifield 1963) (Table 1). For synthesis of linear IRK, Rink amide resin with loading efficiency of 0.8 mmol/gm and for synthesis of dimeric and tertrameric IRK peptides Rink amide resin with loading efficiency of 0.6 mmol/gm were used. C-terminal dimerization by incorporation of one whereas for C-terminal tetramerization three di-Fmoc protected lysines were used which enabled simultaneous synthesis of the two and four chains of dimeric and tetrameric peptide respectively (Fig. 1). After sequential synthesis of peptides, the crude peptide was cleaved from the resin by using K-reagent comprising of phenol, TFA (Trifluoroacetic Acid, Merck, Germany), EDT (1,2-Ethanedithiol, HiMedia, India) and thioanisole (HiMedia, India). The peptides were precipitated using chilled diethyl ether (SD Fine Chemicals, India) and washed several times with diethyl ether to remove salts and impurities. The peptides were then de-salted on ODS-Silica column using a gradient of acetonitrile (Merck, Germany) and finally solubilized in water. The peptides were analyzed for purity by performing reverse-phase high-performance liquid chromatography (RP-HPLC, Shimadzu LC20AR) on a C-18 column (Shim-pack GIST C18 5 μm, 250 X 14 mm) under a linear gradient of acetonitrile containing 0.05% TFA.
Screening for Antibacterial Activity
Bacterial Strains
For determining the MIC of peptides, we used the reference strain of S. aureus (ATCC 29213). Besides this the activity of peptides was also determined on clinical isolates of S. aureus isolated from mastitic milk samples collected from dairy farms.
Isolation of S. aureus from Mastitic Milk Samples
Milk samples were collected aseptically from each quarter of the mastitic cow in a sterile 15 ml centrifuge tube. At first, udder was disinfected with 5% potassium permanganate then milk was collected and transported to the laboratory in an icebox. Milk samples were first enriched in Brain Heart Infusion broth (BHI, HiMedia, India). 1 ml of sample was homogenized in 9 ml of sterile BHI broth and further incubated at 37 ℃ for 16–18 h. A loopful from this enrichment broth was spread on Mannitol Salt Agar (MSA, HiMedia, India) plates and incubated at 37 ℃ for 16–18 h. Single yellow- colored colonies were picked from MSA plates and inoculated into BHI broth and incubated at 37 ℃ for 6–8 h. These log phase cultures in BHI broth were used to prepare 25% glycerol stocks and further utilized for genotypic and phenotypic screening.
Genotypic Identification and Confirmation of MRSA
Overnight log phase cultures in BHI broth were used for DNA isolation. DNA was isolated by CTAB method described by Minas et al. (2011) (CTAB, Cetyl Trimethyl Ammonium Bromide, HiMedia, India). A multiplex polymerase chain reaction (PCR) was carried out for identification of S. aureus with nuc and 23S rRNA genes whereas another multiplex PCR was set up for antibiotic resistant genes mecA and mecC. Details of the primers used and annealing temperatures for the multiplex PCR are given in Table 2. Genomic DNA from Pseudomonas aeruginosa ATCC 27853 was used as negative control for PCR.
Phenotypic Confirmation (Antibiogram) of MRSA
The susceptibility of the isolates against antibiotics was evaluated by antibiotic sensitivity test following Kirby-Bauer disk diffusion method (Bauer et al. 1966). The antibiotic discs (Himedia, India) used for this study were Chloramphenicol (30 mcg), Tetracycline (30 mcg), Erythromycin (15 mcg), Enrofloxacin (5 mcg), Co-Trimoxazole (Sulpha/ Trimethoprim) (25 mcg), Vancomycin (5 mcg), Linezolid (30 mcg), Penicillin-G (10 units), Oxacillin (1 mcg), and Cefoxitin (30 mcg).
Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Determination
The MIC for the linear form of the peptide IRK was determined against S. aureus, ATCC 29213by using broth micro dilution method (Wiegand et al. 2008) in a 96 well flat bottom cell culture plate. Briefly, the bacteria were streaked onto a Mueller Hinton Agar (MHA, HiMedia, India) plate and kept for overnight incubation at 37 ℃ for growth. Single colony from the plate was then inoculated into Mueller Hinton Broth (MHB, HiMedia, India) and then incubated till mid-exponential phase at 37 ℃. Turbidity of the cultures was measured spectrophotometrically (600 nm) and an OD of 0.3 was equivalent to 2 × 108 CFU/ mL bacterial cells approximately. It was further diluted to 1 × 106 CFU/mL in fresh MHB for seeding in wells. A stock solution of 2000 µg/mL for AMP was prepared and serially diluted to make a two-fold serial dilution starting from 128 to 1 µM in 50 µl of sterile water. To each of these wells 50 µL of MHB containing 1 × 106 CFU/mL were added. The wells with bacteria in MHB supplemented with 50 µL of sterile water was taken as control. The wells containing erythromycin (2 μg/mL) along with bacterial culture was taken as positive control and wells with 50 µL of MHB and 50 µL of sterile water were taken as blank. Each concentration was tested in duplicate wells. The microplate was incubated for 24 h at 37 ℃ in a humidified chamber and growth inhibition was measured at optical density (OD) 600 nm in a microplate reader (Multiskan Go, Thermo Fisher Scientific, USA). For determination of minimum bactericidal concentration (MBC), 10 µL from all the wells of known MIC was taken and serially diluted 10,000 times in sterile PBS and spread onto MHA plates and incubated at 37 ℃ for 18 h. Then, plates were evaluated for bacterial inhibition. The lowest concentration at which bacterial growth were reduced to 99.9% was determined as MBC. This experiment was performed in triplicate.
Bactericidal Kinetics
Bactericidal killing kinetics was performed for all the three linear, dimeric and tetrameric forms of the peptide IRK against S. aureus, ATCC 29213 as per CLSI (Clinical and Laboratory Standards Institute 2016) guidelines. Broth microdilution assay was performed similar to MIC determination but only 16 µM for all the peptides was taken for this study. The micro plate readings were taken at OD 600 nm at 0 h, 3 h, 6 h, 9 h, 12 h and 24 h. Each concentration was taken in duplicate.
Screening of Linear, Dimeric and Tetrameric Forms of IRK Against S. aureus ATCC 29213 and MRSA Isolates
Three concentrations of all the peptides viz., 8 µM, 16 µM, 32 µM were taken and their activity was evaluated against S. aureus ATCC 29213. For screening of linear, dimeric, and tetrameric forms of the peptide on 25 MRSA clinical isolates fixed concentration of the three peptides i.e., 64 µM was taken, which is four times of the MIC determined for the linear peptide. The study was performed by broth micro dilution method as described in MIC determination section.
Haemolytic Activity
The haemotoxicity of the linear, dimeric and tetrameric forms of the peptide IRK on erythrocytes of healthy rabbit was tested following the methods described by Bhagavathula et al. (2017) and Tan et al. (2017). The blood collected was tranferred to a 5 ml micro centrifuge tube and resuspended with equal volume of Alsever’s solution (pH 6.1) and centrifuged at 1000×g for 5 min at 4 ℃. The supernatant containing haemolysed RBC cells was discarded. Further, equal volume of Alsever’s solution was added and the tubes were kept at 40C overnight for ageing. Next day the erythrocytes were washed twice with Alsever’s soluion and a 4% erythrocyte solution was made. 80 µL of this 4% erythrocyte solution was added to the 20 µL of peptide concentrations of 8 µM and 16 µM in a 0.6 mL microcentrifuge tube. This mixture was incubated at 37 ℃ for 1 h and then centrifuged at 1000×g for 5 min at 4 ℃. The supernatant (75 µL) was pipetted out in 96-well plate and readings were taken at 570 nm using microplate reader. 0.1% Triton-X 100 (Sigma Aldrich, USA) was used as the positive control (100% hemolysis) and phosphate buffer saline (PBS) was used as the negative control. This experiment was performed in duplicates. Percent haemolysis was calculated by follwing formula:
Statistical Analysis
All the results are reported as mean ± SEM and total variation, present in between groups was determined by one way and two- way analysis of variance (ANOVA). Tukey’s multiple comparison test was used for determining significance.
Results
Antimicrobial Peptides: IRK Linear, Dimeric and tetrameric Forms
The peptides were synthesized and purified using RP-HPLC where > 90% purity was achieved (Table 1). The stock solutions of each lyophilized peptide were prepared at a concentration of 2000 μg/mL in sterile water, filter sterilized by passing through 0.22 μm filter, and stored at − 20 ℃ for subsequent experiments. The dissolved peptide samples were analyzed using RP-HPLC at different time points during storage and compared with original molecule. We observed no disintegration of any of these peptides over a period of one month after reconstitution.
Antibiogram and PCR Screening of MRSA Isolates
A total of 80 isolates were obtained from different mastitic milk samples as confirmed by PCR for nuc and 23S rRNA genes (data given for selected resistant 25 isolates in Table 3). Out of these 80 isolates, antibiotic sensitivity assay of 25 clinical isolates of S. aureus showed high resistance against different antibiotics. All the 25 isolates were found to be resistant for oxacillin and cefoxitin (Table 3). On PCR screening, all these isolates were found positive for mecA gene and 7 isolates were found to be positive for mecC gene suggesting MRSA (Fig. 2a and b).
a Lane 1-1 Kb ladder (Gene ruler); Lane 2-23S rRNA gene (894 bp); Lane 3-nuc gene (278 bp), Lane 4-23S rRNA and nuc gene, Lane-5 Negative control (PCR performed with Pseudomonas aeruginosa ATCC 27853 genomic DNA), b Lane 1–50 bp ladder (Gene ruler); Lane 2-mecA gene (162 bp); Lane 3-mecC gene (138 bp), Lane-4 Negative control (PCR performed with Pseudomonas aeruginosa ATCC 27853 genomic DNA)
Antibacterial Screening Assay
Determination of MIC and MBC for Linear IRK Peptide
Different concentrations of linear IRK peptide were screened against S. aureus ATCC 29213 (Fig. 3). Broth microdilution assay was performed with S. aureus ATCC 29213 and different concentrations of the peptide and a range of percent growth inhibition was obtained. Incubation of S.aureus for 24 h with concentrations ranging from 1 to 4 µM showed a growth inhibition of < 35% whereas, at 8 µM the inhibition was > 75%. At a range of 16 µM to 128 µM of peptide concentrationthe inhibition was found to be > 95%. Erythromycin showed > 95% inhibition at its breakpoint of 2 µg/mL. The results elucidate the potent antimicrobial activity of the peptide at 16 µM and above concentrations. Therefore, MIC of the linear IRK can be considered as 16 µM against S. aureus. The MBC of IRK linear was found to be the same as MIC concentration i.e., 16 µM where no growth was observed on the plates.
Determination of Minimum Inhibitory Concentration (MIC) for linear form of IRK, where all values represent mean (n = 2) ± SEM. E(2) stands for Erythromycin at 2 µg/mL.One way ANOVA was applied to compare treatments with control where P value vs control: *< 0.05, **< 0.01, ***< 0.001, ns not significant
IRK Linear, Dimeric and Tetrameric Peptide Killing Kinetics
The antimicrobial activity of linear, dimeric and tetrameric forms of the peptide IRK was determined for bacterial isolates at 16 µM (concentration equivalent to MIC for linear peptide) with respect to time (Fig. 4). Linear, dimer and tetramer peptides showed 30%, 20%, and 19% inhibition of bacterial growth respectively, within 3 h of incubation. The inhibition increased for linear, dimeric and tetrameric IRK at 6 h to 68%, 45%,40% respectively whereas at 9 h it further increased to 76%, 61%, 59% respectively. After 12 h of incubation 81%, 76%, 75% inhibition of growth was obtained for IRK linear, dimer and tetramer respectively which finally after 24 h of incubation was > 99%, > 85%, > 83%, respectively. It was deduced from this study that more than 50% of bacteria were killed within 6 h by the linear form while dimeric and tetrameric forms could kill more than 50% bacteria only after 9 h of incubation.
Activity of Linear, Dimeric and Tetrameric Forms of IRK Against S. aureus ATCC 29213
The effect of different forms of the peptide IRK on its antimicrobial activity against S. aureus ATCC 29213 was assessed (Fig. 5). Three concentrations viz., half of the MIC (8 µM), MIC (16 µM) and 2xMIC (32 µM) of the linear form of IRK peptide were selected for the study. The dimeric and tetrameric forms of IRK were also evaluated at the same concentrations. It was found that at 8 µM concentration the percent growth inhibition for linear, dimeric and tetrameric forms of IRK were 82%, 59%, and 57% respectively; at 16 µM the percent growth inhibition for linear, dimeric, tetrameric forms were > 95%, 65%, and 60% respectively and at 32 µM the inhibition for linear, dimeric and tetrameric forms increased to > 95%, 76% and 61% respectively. At all the three concentrations linear form of the peptide showed more potent activity than the branched forms of the peptide. These findings demonstrate that upon branching the peptide, its activity against bacteria decreases.
Inhibition of S. aureus (ATCC 29213) by linear, dimeric and tetrameric forms of IRK. All values represent mean (n = 2) ± SEM. Two-way ANOVA was applied to compare groups and multiple group analysis was done by Tukey’s test where P value vs linear peptide treatment: a < 0.001, b < 0.01, c < 0.05, d- not significant; P value vs dimeric peptide treatment: p < 0.001, q < 0.01, r < 0.05, s- not significant
Activity of Linear, Dimeric and Tetrameric Forms of IRK Against MRSA Isolates
All the three forms of the peptides were screened against clinical MRSA isolates (Fig. 6). Growth of around 92% of isolates was inhibited by all the three forms of the peptide at a concentration of 64 µM (equivalent to 4 X MIC for linear IRK).
Haemolytic Assay
Haemolytic activity of the linear, dimeric and tetrameric forms of the peptide IRK was studied against rabbit erythrocytes. At 8 µM concentration linear, dimeric and tetrameric forms showed 8%, 14%, and 16% haemolysis, respectively whereas at 16 µM linear, dimeric and tetrameric forms showed 12%, 16%, and 20% haemolysis, respectively (Fig. 7). This demonstrates that the haemolytic activity increases with the branching of the peptide.
Haemolytic activity of linear, dimeric and tetrameric forms of IRK against 4% Rabbit RBC. All values represent mean (n = 2) ± SEM. Two-way ANOVA was applied to compare groups and multiple group analysis was done by Tukey’s test where P value vs linear peptide treatment: a < 0.001, b < 0.01, c < 0.05, d-not significant; P value vs dimeric peptide treatment: p < 0.001, q < 0.01, r < 0.05, s-not significant
Discussion
Currently, an alternative search for new antimicrobials to combat multidrug resistance bacteria is a pressing priority among the researchers. Various efforts are being made either to develop new antimicrobials or improve the present antimicrobials in terms of their efficacy against MDR bacteria. Antimicrobial peptides (AMPs) are small proteins having potent antimicrobial and immunomodulatory activities against gram-negative and gram-positive bacteria, fungi and viruses. AMPs based on their mode of action include both membrane acting and non-membrane acting peptides (Boparai and Sharma, 2019). Most membrane active peptides are cationic with hydrophobic residues that interact with the bacterial membrane and lead to membrane disruptions. AMPs can also target the formation of structural components, such as the cell wall. Non-membrane active peptides cross the membrane without causing any damage but interfere with intracellular functions such as transcription and translation. Several others, modulate the host immunity by either recruiting or activating of immunocytes or by modulating recognition of microbial products and nucleic acids released upon tissue damage by Toll like receptors (TLRs) (Zhang and Gallo 2016). Several databases have been developed for the collection of antimicrobial peptide sequences of both natural and synthetic origin. With advancement in artificial intelligence and other computer assisted technologies, identification and prediction of new AMP molecules have become quicker and different AI based methods have been employed to search for new molecules. Recently Deep ABPpred classifier has been proposed to identify new AMP sequences in proteins (Sharma et al. 2021). The model predicts AMP sequences with high accuracy and few of the predicted molecule have been experimentally validated in laboratory.
Recent advances in bioinformatics have helped redesign and discover several other antimicrobial agents apart from AMPs. Gupta et al. (2019) have established that statin drugs used for hypercholesteremia can also have antibacterial potential after analyzing them in silico and validating them in vitro against S. aureus, B. pumilus, P. aeruginosa and S. enterica. In a similar study, Fluvastatin has been reestablished as a potent antifungal agent against Candida albicans by in silico docking and analysis, potentiated by in vitro studies (Rana et al. 2019). Newer modifications in existing molecules such as thiophenes have been done by computer aided drug design and validated in silico by ADME prediction. In vitro studies against S. aureus, B. subtilis, P. aeruginosa and E. coli showed the potent antimicrobial activity of 1,3,4-Oxadiazole/1,2,4-Triazole-Substituted Thiophenes (Singla et al. 2020).
AMPs offer a viable and effective alternative to present day antibiotics to overcome the ever-increasing menace of antimicrobial resistance as observed in MRSA. Selective, rational chemical modification of AMPs offers great prospective for development of novel next generation antimicrobials. These modifications may include glycosylation, lipidation and multimerization (Li et al. 2021). In the present work the effect of multimerization by C terminal dimerization and tetramerization on the antimicrobial activity of IRK peptide was studied. The short peptide IRK in its linear, dimeric and tetrameric forms was synthesized and their efficacy against MRSA strains was demonstrated. IRK is a short synthetic β sheet folding amphiphilic peptide which strongly interacts with the bacterial cell membrane. It was initially designed by Ong et al. (2014) based on the sequence (X1Y1X2Y2)n-NH2 where X stands for hydrophobic amino acids, Y stands for cationic amino acids and n is the number of repetitions. MIC of the linear IRK was determined to be 16 µM against antibiotic sensitive standard S. aureus ATCC 29213 and at this concentration inhibition pattern of IRK dimer and tetramer was also evaluated. As per the previous report killing kinetics of IK8 L was dose dependent; killing time was halved from 2 to 1 h with doubling of MIC from 2 × MIC to 4 × MIC (Zhong et al. 2017). In our present study also with increase in concentration a higher activity of this peptide was observed for all the forms.
Killing kinetics of linear, dimer, and tetramer forms of IRK demonstrated that more than 95% of S. aureus was inhibited at 16 µM concentration of linear IRK with IC50 (Inhibitory Concentration 50%) achieved within 6 h of incubation. On the other hand, IC50 was achieved after 9 h of incubation in case of dimer and tetramer. Thus, it is inferred that all the forms of IRK are highly effective against S. aureus (ATCC 29,213) with linear form having the highest antibacterial potency. When, the inhibition pattern of linear, dimer and tetramer forms of IRK was compared against S. aureus ATCC 29,213 at different concentrations it was found that efficacy and ability to inhibit the strain reduces with the branching of the peptide. This was in agreement with Lorenzón et al. (2012, 2014), who reported that dimerization of the peptides Ctx-Ha and Aurein 1.2, reduced their ability to inhibit bacterial growth. In another study it was found that antimicrobial activity decreased by 2- to 4-fold against Gram-positive and Gram-negative bacteria upon dimerization of PST13-RK by Yang et al. (2009). Similar reduction in antimicrobial activity was observed for Mellitin after its dimerization (Lorenzón et al. 2019). This reduction in activity can be attributed to the fact that dimerization and tetramerization leads to change in the two-dimensional structure of the peptide and in turn may affect its interaction with the bacterial cell membrane and thus changes its biological activity. When MRSA isolates were screened with 64 µM concentration (equivalent to 4XMIC of linear IRK) for all the three forms, they could inhibit 92% of the isolates with > 50% inhibition. This elucidates that the application of all the three forms of the peptide IRK showed potent activity against MRSA isolates but with higher MIC for dimer and tetramer forms.
Present study showed higher haemolytic activity with increase in branching of the peptide. This was in agreement with the finding that hybrid peptides have slightly enhanced hemolytic activity compared with the parent linear peptides (Liu et al. 2017; Vega et al. 2018; Lorenzón et al. 2012). This is also in compliance with the reports that hemolytic activity increases with increasing hydrophobicity of peptides leading to non selective binding to any cell membrane. Red blood cells have only lipids on their surface and lack the other bacterial membrane components like cell wall, made of peptidoglycan. Dimerization and tetramerization of the peptide lead to change of peptide structure in solution and thus lead to differential interaction with different membranes based on their composition (Lorenzón et al. 2019).
Present study demonstrated the influence of multimerization on the activity of peptide IRK and offers a better understanding on how changes in structure can influence the biological activity of the peptide. Synthetic peptides based on rational design solely focused on structural interaction with the bacterial membrane also offer newer aspects of study and research for development of novel antimicrobials. More detailed and in-depth studies are required to completely unravel the mode of action of such branched AMPs. The branched peptides have shown enhanced inhibition in the growth of MDR strains of bacteria, which could be advantageous in the long run, however, one must think of studying the detailed mechanism of action of these AMPs and also devising methods to reduce the haemolytic activity.
Conclusion
The synthesized linear, dimeric and tetrameric forms of the peptide IRK were tested against S. aureus (ATCC 29,213) and MRSA clinical isolates (n = 25). All the forms of the peptide were active against MRSA isolates at higher concentrations than the MIC determined against S. aureus (ATCC 29,213). In the standard strain the antimicrobial activity was in the following order linear > dimeric > tetrameric whereas haemolytic activity was the highest for tetrameric followed by dimeric and lastly by linear form of the peptide. Structural changes arising due to multimerization may have attributed to this change in biological activity. Thus, it can be concluded that peptide IRK is highly effective against MRSA bacteria and the antimicrobial activity of the peptide IRK decreases with increase in branching.
Data Availability
All data related to this work are presented in the MS and as such there is no supplementary data associated with this MS.
References
Abrahmsén M, Persson Y, Kanyima BM, Båge R (2014) Prevalence of subclinical mastitis in dairy farms in urban and peri-urban areas of Kampala, Uganda. Trop Anim Health Prod 46:99–105. https://doi.org/10.1007/s11250-013-0455-7
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Boparai J, Sharma P (2019) Mini review on antimicrobial peptides, sources, mechanism and recent applications. Protein Pep Lett. https://doi.org/10.2174/0929866526666190822165812
Boyle-Vavra S, Daum RS (2016) Molecular strategies of Staphylococcus aureus for resisting antibiotics. In: Somerville GA (ed) Staphylococcus: genetics and pshysiology. Caister Academic Press, UK, pp 249–300
Bhagavathula N, Meedidoddi V, Bourque S, Vimaladevi R, Kesavakurup S, Selvadurai D, Shrivastava S, Krishnappa C (2017) Characterization of two novel antimicrobial peptides from the cuticular extracts of the ant Trichomyrmex criniceps (Mayr), (Hymenoptera: Formicidae). Arch Insect Biochem Physiol. https://doi.org/10.1002/arch.21381
Clinical and Laboratory Standards Institute (2016) Performance standards for antimicrobial susceptibility testing. sixteenth informational supplement. CLSI document M100-S16 CLSI, Wayne, PA
Falciani C, Lozzi L, Pini A, Corti F, Fabbrini M, Bernini A, Lelli B, Niccolai N, Bracci L (2007) Molecular basis of branched peptides resistance to enzyme proteolysis. Chem Biol Drug Des 69(3):216–221. https://doi.org/10.1111/j.1747-0285.2007.00487.x
Gajdács M, Albericio F (2019) Antibiotic resistance: from the bench to patients. Antibiotics 8(3):129. https://doi.org/10.3390/antibiotics8030129
Garg A, Singh B, Sharma R, Singh A, Kumar A (2020) Selective estrogen receptor modulators (SERMs): mechanistic insights against microbial infections. Curr Mol Med 20(2):102–115
Gomes F, Henriques M (2016) Control of bovine mastitis: old and recent therapeutic approaches. Curr Microbiol 72(4):377–382. https://doi.org/10.1007/s00284-015-0958-8
Gould IM, Bal AM (2013) New antibiotic agents in the pipeline and how hey can help overcome microbial resistance. Virulence 4(2):185–191. https://doi.org/10.4161/viru.22507
Gupta M, Sharma R, Kumar A (2019) Comparative potential of Simvastatin, Rosuvastatin and Fluvastatin against bacterial infection: an in silico and in vitro study. Orient Pharm Exp Med. https://doi.org/10.1007/s13596-019-00359-z
Gupta S, Abhishek SS, Singh RJ, Gogoi P, Kumar B (2021) Evaluation of antibacterial activity of magainin and mastoparan and its novel hybrid against MDR E. coli isolates of neonatal calves. Int J Pept Res Ther 27(8):1–9. https://doi.org/10.1007/s10989-020-10154-z
Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S (2012) Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J Clin Microbiol 50(3):688–695. https://doi.org/10.1128/JCM.05214-11
Ismail ZB (2017) Molecular characteristics, antibiogram and prevalence of multi-drug resistant Staphylococcus aureus (MDRSA) isolated from milk obtained from culled dairy cows and from cows with acute clinical mastitis. Asian Pac J Trop Biomed 7(8):694–697. https://doi.org/10.1016/j.apjtb.2017.07.005
Kozerski ND, Oliveira JLP, Moura RA, Silva DR, Oliveira AF, Mello PL, Agostinis RO, Goncalves DD, Otutumi LK, Martins LA (2014) Antimicrobial profile of multidrug-resistant Staphylococcus spp. isolated from bovine mastitis cases in the northwest region of Parana State, Brazil. Afr J Microbiol Res 8(37):3392–3397
Lakhundi S, Zhang K (2018) Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31(4):e00020-e118. https://doi.org/10.1128/CMR.00020-18
Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12(5):371–387. https://doi.org/10.1038/nrd3975
Li W, Separovic F, O’Brien-Simpson NM, Wade JD (2021) Chemically modified and conugated antimicrobial peptides against superbugs. Chem Soc Rev 50(8):4932–4973. https://doi.org/10.1039/D0CS01026J
Liu B, Huang H, Yang Z, Liu B, Gou S, Zhong C, Han X, Zhang Y, Ni J, Wang R (2017) Design of novel antimicrobial peptide dimer analogues with enhanced antimicrobial activity in vitro and in vivo by intermolecular triazole bridge strategy. Peptides 88:115–125. https://doi.org/10.1016/j.peptides.2016.12.016
Lorenzón EN, Cespedes GF, Vicente EF, Nogueira LG, Bauab TM, Castro MS, Cilli EM (2012) Effects of dimerization on the structure and biological activity of antimicrobial peptide Ctx-Ha. Antimicrob Agents Chemother 56(6):3004–3010. https://doi.org/10.1128/AAC.06262-11
Lorenzón EN, Piccoli JP, Cilli EM (2014) Interaction between the antimicrobial peptide Aurein 1.2 dimer and mannans. Amino Acids 46(11):2627–2631
Lorenzon EN, Piccoli JP, Santos-Filho NA, Cilli EM (2019) Dimerization of antimicrobial peptides: a promising strategy to enhance antimicrobial peptide activity. Protein Pept Lett 26(2):98–107. https://doi.org/10.2174/0929866526666190102125304
Merrifield RB (1963) Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc 85(14):2149–2154. https://doi.org/10.1021/ja00897a025
Minas K, McEwan NR, Newbold CJ, Scott KP (2011) Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett 325(2):162–169. https://doi.org/10.1111/j.1574-6968.2011.02424.x
Mohammed J, Ziwa MH, Hounmanou YMG, Kisanga A, Tuntufye HN (2018) Molecular typing and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from Bovine milk in Tanzania. Int J Microbiol 2018:4287431. https://doi.org/10.1155/2018/4287431
Ong ZY, Cheng J, Huang Y, Xu K, Ji Z, Fan W, Yang YY (2014) Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic β-sheet forming peptide amphiphiles. Biomaterials 35(4):1315–1325. https://doi.org/10.1016/j.biomaterials.2013.10.053
Rana R, Sharma R, Kumar A (2019) Repurposing of fluvastatin against Candida albicans CYP450 Lanosterol 14 α-demethylase, a target enzyme for antifungal therapy: an in silico and in vitro study. Curr Mol Med 19(7):506–524. https://doi.org/10.2174/1566524019666190520094644
Ren Q, Liao G, Wu Z, Lv J, Chen W (2020) Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J Dairy Sci 103(4):3368–3380. https://doi.org/10.3168/jds.2019-17420
Salauddin M, Akter MR, Hossain MK, Nazir KHMNH, Noreddin A, El Zowalaty ME (2020) Molecular detection of multidrug resistant Staphylococcus aureus isolated from bovine mastitis milk in Bangladesh. Vet Sci 7(2):36. https://doi.org/10.3390/vetsci7020036
Schäberle TF, Hack IM (2014) Overcoming the current deadlock in antibiotic research. Trends Microbiol 22(4):165–167. https://doi.org/10.1016/j.tim.2013.12.007
Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47. https://doi.org/10.3389/fmicb.2013.00047
Sharma R, Shrivastava S, Singh SK, Kumar A, Saxena S, Singh RK (2021) Deep-ABPpred: identifying antibacterial peptides in protein sequences using bidirectional LSTM with word2vec. Breif Bioinformatics bbap065. https://doi.org/10.1093/bib/bbab065
Singla N, Singh G, Bhatia R, Kumar A, Kaur R, Kaur S (2020) Design, synthesis and antimicrobial evaluation of 1,3,4-oxadiazole/1,2,4-triazole-substituted thiophenes. Chem Select 5(13):3835–3842. https://doi.org/10.1002/slct.202000191
Tam JP, Lu YA, Yang JL (2002) Antimicrobial dendrimeric peptides. Eur J Biochem 269(3):923–932
Tan T, Wu D, Li W, Zheng X, Li W, Shan A (2017) High specific selectivity and membrane-active mechanism of synthetic cationic hybrid antimicrobial peptides based on the peptide FV7. Int J Mol Sci 18(2):339. https://doi.org/10.3390/ijms18020339
Vega SC, Martínez DA, ChaláVargas HA, Rosas JE MS (2018) Design, synthesis and evaluation of branched RRWQWR-based peptides as antibacterial agents against clinically relevant gram-positive and gram-negative pathogens. Front Microbiol 9:329. https://doi.org/10.3389/fmicb.2018.00329
Vestergaard M, Frees D, Ingmer H (2019) Antibiotic resistance and the MRSA problem. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.GPP3-0057-2018
Wang S, Zeng X, Yang Q, Qiao S (2016) Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci 17(5):603–615. https://doi.org/10.3390/ijms17050603
Wang W, Lin X, Jiang T, Peng Z, Xu J, Yi L, Li F, Fanning S, Baloch Z (2018) Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front Microbiol 9:1123. https://doi.org/10.3389/fmicb.2018.01123
Wiegand I, Hilpert K, Hancock REW (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. https://doi.org/10.1038/nprot.2007.521
Yang ST, Il KJ, Shin SY (2009) Effect of dimerization of a β-turn antimicrobial peptide, PST13-RK, on antimicrobial activity and mammalian cell toxicity. Biotechnol Lett 31(2):233–237. https://doi.org/10.1007/s10529-008-9848-5
Zhang LJ, Gallo RL (2016) Antimicrobial peptides. Curr Biol 26(1):R14–R19
Zhong G, Cheng J, Liang ZC, Xu L, Lou W, Bao C, Ong ZY, Dong H, Yang YY, Fan W (2017) Short synthetic β-sheet antimicrobial peptides for the treatment of multidrug-resistant pseudomonas aeruginosa burn wound infections. Adv Healthc Mater. https://doi.org/10.1002/adhm.201601134
Acknowledgements
The work presented here was carried out under the National Agricultural Science Fund (NASF) project entitled “Detection of peptide biomarkers and development of synthetic antimicrobial peptide hydrogels for bovine mastitis (Project grant No. NASF/ABA-6014/2016-17/367). The peptide synthesis and other infrastructure available at Facility for Research & Training on Bioassays and Biosensors created under ICAR-Niche Area Excellence Programme on Biosensors at Division of Veterinary Biotechnology, ICAR-IVRI were also used for this work. The senior research fellowship received by the first author from Council for Scientific and Industrial Research (CSIR), is thankfully acknowledged.
Funding
National Agricultural Science Fund (NASF) project entitled “Detection of peptide biomarkers and development of synthetic antimicrobial peptide hydrogels for bovine mastitis (Project Grant No. NASF/ABA-6014/2016-17/367).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declare that they have no conflict of interest.
Ethical Approval
In this study, no invasive procedure was performed on human or any other animal species, except collection of whole blood from rabbit for performing the haemolytic assay, for which the permission from the Institute Animal Ethics Committee was obtained vide Reference No: F.26-1/2015-16/JD(R) dated 28th Feb, 2019.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gogoi, P., Shrivastava, S., Shah, P. et al. Linear and Branched Forms of Short Antimicrobial Peptide-IRK Inhibit Growth of Multi Drug Resistant Staphylococcus aureus Isolates from Mastitic Cow Milk. Int J Pept Res Ther 27, 2149–2159 (2021). https://doi.org/10.1007/s10989-021-10243-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-021-10243-7