Abstract
Nipah virus was first appeared in Malaysian 1998, which further found be outburst in neighbor countries i.e. Bangladesh, Singapore, and India. Currently there is no effective drug or vaccine available only supportive care and prevention are way to manage it. Epitope based vaccine could be the best way to cure Nipah virus infection. In this study, the proteome of Nipah virus was retrieve from UniProt database and were subjected to check for allergenicity using Allergen FP v.1.0 tool. NetMHCII 2.3 server screened epitopes from non-allergen proteins and Vaxijen tool was used to identify the most antigenic epitopes binds with MHC class II molecules. Two potent T cell epitopes LLFVFGPNL and KYKIKSNPL were found, which binds with HLA-DRB1*01:01 and HLA-DRB1*07:01 MHC class II alleles. PepstrMod and Swiss-Model server were used to build 3D structure model of epitopes and alleles, respectively. Further identified epitopes were docked with HLA alleles using AutoDock vina tool to confirm binding ability. The epitopes LLFVFGPNL and KYKIKSNPL had binding affinity of − 8.1 kcal/mol and − 5.7 kcal/mol with HLA-DRB1*01:01 and HLA-DRB1*07:01 alleles, respectively. Solidity of predicted epitope—allele docked complex were evaluating by molecular dynamics simulation.HLA distribution analysis was performed for predicted epitopes using the population coverage tool of Immune Epitope Database (IEDB). These predicted epitopes cover maximum number of populations in India as well as worldwide. Therefore, these epitopes could be potent vaccine candidates to counter Nipah virus by testing in wet lab.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nipahvirus (NiV) originates from the Paramyxoviridae family and genus Henipavirus. In 1999 it was isolated and recognized in Malaysia and Singapore during an outbreak of encephalitis mostly amongst farmers concerned to pigs and also the people having close contact to the same. Nipah virus named after the village of its origin, Sungai Nipah in Malaysia Peninsula. In the Malaysia and Singapore Nipah virus outbreak, transmission happened via interaction between people and pigs, while in Bangladesh and India, it occurred by absorption of date palm fluid which is contaminated and also through human-to-human transmission (Ang et al. 2018). There are as of now no drugs or vaccines available for either people or animals for Nipah virus infection in spite of the fact that WHO have distinguished Nipah as a priority disease for the WHO Research and Development. Intensive supportive care is recommended to cure Nipah virus infection. Nipah virus can also be communicated to human-to-human and animals to humans or through contaminated foods (WHO 2018). Humans infected with Nipah virus had a range of clinical presentations, which may range from infection (asymptomatic) to sever diseases related to respiratory system and may finally be converted to lethal encephalitis.
Fruit bats that belong to Pteropodidae family are nominated as the natural host of this virus. Nipah virus was first outbreaks in Malaysia, Bangladesh, Singapore, and India (Chua et al. 1999; Gurley et al. 2007; Paton et al. 1999; Chadha et al. 2006; Arankalle et al. 2011). Zoonotic communication of the Nipah virus through pigs and bats to human has also been reported of (Chua et al. 2000; Luby et al. 2009). Recently in May, 2018, a Nipah virus outbreak was reported from Kozhikode district of Kerala, India, in which 17 deaths and 18 confirmed cases found as of 1 June 2018 (WHO 2018).
To solve the problem of Nipah virus infection, vital need of a vaccine that could potentially cure Nipah virus by providing immunity against it. Here, epitopes based vaccines design approach was used to find out potential vaccine candidate. This method is rather more precise, easy, economic, consumes less time, and harmless when compared to the vaccine designs conveniently available in the market (Kumar et al. 2015).In this study, six proteins from proteome of Nipah virus were checked for allergenicity. Non-allergen proteins were subjected for prediction of epitopes. Predicted epitopes will be subjected for immunogenic properties, structural modeling and the docking with corresponding MHC II alleles to investigate the strong binding interaction. Population coverage analysis of predicted epitopes was also performed.
Methodology
Retrieval of Proteome Data
Full-genome sequence (18,252 nt) of Nipah virus amplified from lung tissue of human of West Bengal, India (Arankalle et al. 2011) were taken as sample in this study. UniProt database has been used for the complete retrieval of the proteome of the Nipah virus, which has proteome id UniProt UP000128950 (http://www.uniprot.org/proteomes/), which displays the sum of 6 coding sequence of protein.
Prediction of Allergic Properties of Proteins
The sequences of protein were then tested, for the prediction of allergenicity with the help of Allergen FP v.1.0 (Dimitrov et al. 2014).
T Cell Epitopes’ Prediction
Non-allergen proteins of Nipah virus were subjected to NetMHCII 2.3 (Jensen et al. 2018) server to screen T cell epitopes which has the capability of binding having MHC class II molecules. To confirm higher confidence level of epitopes, Vaxijen (Doytchinova et al. 2007) tool was used in identification of epitopes having higher confidence level of binding with MHC class II alleles. High scorer epitopes having Vaxijen (Antigenicity) Score ≥ 1.1 were selected for analysis.
Epitopes’ Toxicity Prediction
Toxicity of designated peptides were predicted through using Toxin Pred (Gupta et al. 2013) tool. Non-toxic peptides were selected for study.
Molecular Modeling of Epitopes and HLA Alleles
PEPstrMOD server was used to build 3D structures of the predicted epitopes (Kaur et al. 2007). SWISS-MODEL server was utilized to build 3D structures of predicted corresponding HLA alleles (Waterhouse et al. 2018).
Epitopes’ Molecular Docking and HLA Alleles
AutoDock Vina was used to perform the docking of selected epitopes with HLA alleles (Trott and Olson 2010). Among nine conformation generated by Vina tool, best one has been nominated based on lowest binding energy. Ligand and protein preparation were done by using AutoDock tool.
Molecular Dynamic Simulation Studies
Epitope-allele complex were subjected to simulation by utilizing NAMD (Phillips et al. 2005) implanted with program VMD (Visual Molecular Dynamics) (Humphrey et al. 1996).
Population Coverage Analysis
MHCPred was used to predict epitope quantitatively to major histocompatibility complexes (Guan et al. 2003), whereas the IED tool of population coverage was used to predict population coverage of the predicted epitopes which are MHC-II restriction based (Bui et al. 2006).
Results and Discussion
Allergenicity Prediction
Total six proteins were identified as having Non-allergen property as displayed in Table 1.
T Cell Epitope Prediction
NetMHCII 2.3 server predicted T-cell epitopes with binding affinity from protein sequence against HLA class II alleles. VaxiJen server further assessed these epitopes were annotated on the basis scores ≥ 1.1, to predict antigenicity to elicit potent immune responses as shown in Table 2. Binding affinity of epitopes LLFVFGPNL and KYKIKSNPL with alleles HLA-DRB1*01:01 and HLA-DRB1*07:01 were 532.1 nm and 129.6 nm were predicted by NetMHCII 2.3 server.
Molecular Modeling of Epitopes and HLA Alleles
PEPstrMOD web server built 3D structures of the predicted epitopes. HLA-DRB1*07:01 and HLA-DRB1*13:01 homology model having template PDB Id 3C5J and 6CQL as shown in Table 3 built 3D structures of the HLA alleles using Swiss-Model server. Alleles HLA-DRB1*01:01 had crystal structure with PDB Id 4AH2 respectively. The quality of the homology model of alleles was verified with the help of Ramachandran plot. This plot was drawn by using Swiss-Model server which validates the model having > 90%, of the whole residues covering mostly the favored regions. Similarly, Kamthania and Sharma performed modeling of T-cell epitopes of Nipah virus’ antigenic proteins nulceocapsid, phosphoprotein, matrix, fusion, glycoprotein, L protein, W protein, V protein and C protein along with their corresponding MHC class I alleles (Kamthania and Sharma 2015).
Molecular Docking of Epitopes and HLA Alleles
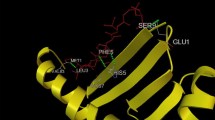
Using AutodockVina, the molecular docking between epitopes and alleles was performed. The binding affinities of epitopes with alleles were shown in Table 4.
Docked complex of epitopes and allele were shown in Figs. 1 and 2. As Similar, Ali et al. (Ali et al. 2015) docked epitopes VPATNSPEL, NPTAVPFTL and LLFVFGPNL of Nulceocapsid, V protein and Fusion protein of Nipah virus with their corresponding alleles HLA-B7, HLA-B*2705 and HLA-A2 MHC class I allele and found considerable binding energy.
Molecular Dynamics Simulation
NAMD was used for the molecular dynamics’ simulation of the docked complex, which shows the firmness of the complex structure. RMSD’s maximum value for the peptide LLFVFGPNL—HLA-DRB1*01:01 allele complex was at 1.34 Å (Fig. 3), whereas peptide KYKIKSNPL—HLA-DRB1*07:01 allele complex was at 1. 46 Å (Fig. 4). As similar, Ravichandran et al. (2018) performed molecular dynamics simulation of docked complex of predicted epitope ELRSELIGY of phosphoprotein and YPLLWSFAM of nulceocapsid protein of Nipah virus with HLA-C*12:03 alleles.
Toxicity Prediction of Predicted Epitopes
Epitopes LLFVFGPNL and KYKIKSNPL having VaxiJen (Antigenicity) Score 2.3879 and 1.3649, respectively having lowest binding energy with their respective alleles, were chosen to predict toxicity and also conservancy analysis. Prediction of toxicity of epitopes as shown in the Table 5 was done using Toxin Pred.
Analysis of Population Coverage
IEDB tool was used to predict population coverage analysis of the epitopes and their corresponding MHC-II alleles. Epitopes and MHCPred were provided to IEDB population coverage tool as input. Epitopes LLFVFGPNL and KYKIKSNPL have shown the elicitation of the immune response of 28.63–22.06% total world population as shown in the Figs. 5 and 6. Epitopes LLFVFGPNL and KYKIKSNPL have population coverage 35.82 and 11.72 in India. The results of the population coverage analysis show the epitopes distribution in the different regions.
Conclusion
Two potent T cell epitopes LLFVFGPNL and KYKIKSNPL were identified having antigenicity Score 2.3879 and 1.3649 and had strong binding affinity with MHC class II alleles. These epitopes had lowest binding − 8.1 kcal/mol and − 5.7 kcal/mol with HLA-DRB1*01:01 and HLA-DRB1*07:01 alleles. These predicted epitopes cover maximum number of populations in India as well as worldwide. Therefore, these epitopes can be used in designing vaccines against Nipah virus after wet lab verification.
References
Ali MT, Morshed MM, HassanF (2015) A computational approach for designing a universal epitope-based peptide vaccine against Nipah virus. Interdiscip Sci 7(2):177–185. https://doi.org/10.1007/s12539-015-0023-0
Ang BSP, Lim TCC, Wang L (2018) Nipah virus infection. J Clin Microbiol 56:e01875-17. https://doi.org/10.1128/JCM.01875-17
Arankalle VA, Bandyopadhyay BT, Ramdasi AY, Jadi R, Patil DR, Rahman M, Majumdar M, Banerjee PS, Hati AK, Goswami RP, Neogi DK, Mishra AC (2011) Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis 17(5):907–909. https://doi.org/10.3201/eid1705.100968
Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A (2006) Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7(1):153
Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ (2006) Nipah virus–associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 12:235–240
Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PSK, Ksiazek TG (1999) Fatal encephalitis due to Nipah virus among pig farmers in Malaysia. Lancet 354:1257–1259
Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK (2000) Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2014) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30:846–851. https://doi.org/10.1093/bioinformatics/btt619
Doytchinova IA, Darren R, Flower (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8:4
Guan P, Doytchinova IA, Zygouri C, Flower DR (2003) MHCPred: a server for quantitative prediction of peptide—MHC binding. Nucleic Acids Res 31:3621–3624
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8:e73957
Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR (2007) Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 13:1031–1037
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, Nielsen M (2018) Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154(3):394–406
Kamthania M, Sharma DK (2015) Screening and structure-based modeling of T-cell epitopes of Nipah virus proteome: an immunoinformatic approach for designing peptide-based vaccine. 3 Biotech 5(6):877–882. https://doi.org/10.1007/s13205-015-0303-8
Kaur H, Garg A, Raghava GPS (2007) PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett 14:626–630
Kumar A, Hays M, Lim F, Foster LJ, Zhou M, Zhu G, Miesner T (2015) Protective enterotoxigenic Escherichia coli antigens in a murine intranasal challenge model. PLoS. https://doi.org/10.1371/journal.pntd.0003924
Luby SP, Hossain MJ, Gurley ES, Ahmed B, Banu S, Khan SU (2009) Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis 15:1229–1235
Paton NI, Leo YS, Zaki SR, Auchus AP, Lee KE, Ling AE (1999) Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354:1253–1256
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot Christophe, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
Ravichandran L, Venkatesan A, Febin J, Dass P (2018) Epitope-based immunoinformatics approach on RNA-dependent RNA polymerase (RdRp) protein complex of Nipah virus (NiV). J Cell Biochem. https://doi.org/10.1002/jcb.27979
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(1):W296–W303. https://doi.org/10.1093/nar/gky427
WHO (2018) Outbreak of Nipah virus encephalitis in Kerala state of India. http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus/en/ Accessed May 19 2018
Acknowledgements
The author thankfully recognize the needed computational services and steady supervision provided by the Department of Bioengineering and Biosciences, Lovely Professional University, Jalandhar, Punjab., India for their kind support throughout the research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author hereby declares that they have “no conflict of interest”.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaushik, V. In Silico Identification of Epitope-Based Peptide Vaccine for Nipah Virus. Int J Pept Res Ther 26, 1147–1153 (2020). https://doi.org/10.1007/s10989-019-09917-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-019-09917-0