Abstract
Klebsiella pneumoniae is a hospital-acquired pathogen that leads to various infections. There is a great interest in developing an efficient Klebsiella vaccine and epitope-based vaccines may provide a hopeful choice. Outer membrane proteins (Omps) of gram-negative bacteria play an essential role in bacterial pathogenicity; therefore, they are ideal candidates for vaccine development against these pathogens. This study proposed an in silico approach by assembling consensus and high ranked epitopes of OmpW, OmpA, OmpF, OmpC and OmpX of K. pneumoniae and other virulence Enterobacteriaceae to induce multi-epitopic responses against these pathogens. Two vaccine constructs of discontinuous (conformational) B cell and linear CD4+ T cell epitopes of the Omps were designed. The three-dimensional structure of the B cell construct was successfully modeled and validated. Discontinuous B cell epitopes predictions of the B cell construct showed that the predicted epitopes are admissibly in common with total inserted discontinuous epitopes result in stimulation an efficient humoral immune response. In order to improve the immunogenicity, the size of the constructs was increased by multiplication of the epitopes and incorporation of sequence tags. Both constructs were found to be soluble, non-allergen and free of much posttranslational modification that could be effective on immunogenicity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Klebsiella pneumoniae, a member of Enterobacteriaceae family, is an important cause of nosocomial infections and is responsible for significant morbidity and mortality in patients with deficiencies in the immune system (Tsai et al. 2010). Klebsiella is an extracellular parasite and immunity against it is largely mediated by antibodies (Schulz 1996). Besides, there are several lines of evidence to support an essential role of CD4+ T cell in protection against this pathogen (Hagen et al. 1998). Several studies have shown the requirement of major histocompatibility complex (MHC) class II for protection in mice (Hagen et al. 1998; Zisman et al. 1998). Additionally, CD4+ T cell has an important role in supporting both B cell and CD8+ T cell function (Li et al. 2010). These findings suggest that an effective immunotherapeutic strategy against K. pneumoniae should include B and CD4+ T helper cell (Th) epitopes. Nowadays, most of the available vaccines against K. pneumoniae are based on native components such as capsular polysaccharides (CPSs) and lipopolysaccharides (LPSs) (Clements et al. 2008; Yadav et al. 2005). Toxic reactions, which arise from LPS-based vaccines and a high number of antigens applied in CPS-containing vaccines indicate critical disadvantages of such vaccines (Lundberg et al. 2013). An efficient modality to fight K. pneumoniae needs exploration of new antigens and making novel recombinants (Li et al. 2010).
Outer membrane proteins (Omps) are a series of channel proteins that span the outer membrane of gram-negative bacteria (Schulz 1996). Thus, they allow the permeation of a broad range of components, which are necessary for growth and function of the cell (Hong et al. 2006). In addition to transport function, Omps act as virulence factors during bacterial infection (Galdiero et al. 2003). On other hand, sequence comparisons between members of each enterobacterial Omp family have implied that their sequences are highly conserved (Braun and Cole 1984). Therefore, Omps are ideal candidates for vaccine development against Enterobacteriaceae since they have the ability to induce strong immunity response and cover a wider spectrum of pathogens. Previous research has recognized a number of Omps such as OmpA, FepA, OmpC, OmpX and OmpW from K. pneumoniae which are able to induce robust immune responses against this pathogen (Galdiero et al. 2003; Kurupati et al. 2006).
Active immunotherapy, such as an epitope-based vaccine, has recently drawn much attention in treating infectious diseases (Yang and Yu 2009). Immunization based on epitope-based vaccines is powerful in stimulation of the cellular and/or humoral arms of the immune system (Bijker et al. 2007). These types of vaccines consist of highly immunogenic T and/or B cell epitopes, which provoke cytotoxic T cells (CTL), Th or B to specific epitopes (Baloria et al. 2012; Akhoon et al. 2011). B and Th cells play an important role in induction of a protective immune response in many bacterial infections; thus, determination of peptides that induce T and B cell responses is a crucial requirement for the design of effective epitope-based vaccines (Gupta et al. 2010, 2012). The epitope-based vaccines have some potential advantages, such as ability to choose the type of immunity, cost effective production and increased safety. In contrast, they have a few limitations such as low immunogenicity of single epitope (Sbai et al. 2001). Generally, different strategies such as increasing the number of antigenic epitopes and enabling the insertion of antigenic epitopes into immunogenic adjuvants or a carrier protein are being used to prevail epitope vaccine deficiencies (Yi et al. 2004; Coban et al. 2011).
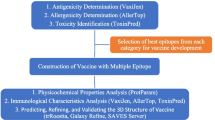
Immune-informatics or computational biology have added an unavoidable contribution to design epitope-based vaccines. In this context, identification of potential epitopes from an antigen protein by in silico methods can be considered in such vaccines reducing the lengthy process for discovery of appropriate epitopes (Srivastava et al. 2011). Nowadays, many online servers are available for predicting B and T cell epitopes. In this respect, Immune Epitope Database (IEDB) server website (Vita et al. 2010) provides tools to predict both B and T cell epitopes. Other online servers such as MetaMHCII (Hu et al. 2010), ProPred (Singh and Raghava 2001), MHCpred (Guan et al. 2003) and SVMHC (Dönnes and Elofsson 2002) have different tools for finding T cell epitopes. Besides, several web-servers such as Discotope (Kringelum et al. 2012) and CBTOPE (Ansari and Raghava 2010) provide access to prediction of conformational B cell epitopes on an antigen sequence. However, determining the 3D structure of a discontinuous B cell polytopic construct is critical since such construct representing discontinuous epitopes should mimic the structure of antigenic protein epitopes (Ponomarenko and Regenmortel 2009). In order to the technical difficulties and labor intensiveness of experimental techniques for the structural characterization of proteins, the numbers of designed epitope-based or chimeric protein vaccines were frequently modeled by computational approaches by different researchers (Nezafat et al. 2014; Nazarian et al. 2012). There exist several methodologies for the structure prediction of proteins including comparative modeling, threading and ab initio modeling (Yang et al. 2015; Roy et al. 2010). It is demonstrated that the composite approaches, which combine various techniques of protein structure prediction, have significant advantages in protein structure prediction (Srivastava et al. 2011). I-TASSER is one of such servers, which combines threading and ab initio structure prediction methods to obtain the full-length model (Yang et al. 2015; Roy et al. 2010).
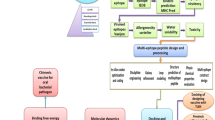
In the present study, two multi-epitope vaccinal constructs, which are based on the Omps of the K. pneumoniae have been designed. Besides, in order to minimize possible problems that occur in designing epitope-based vaccines, and the resulting low efficiency, the design process was modified to create a broad spectrum vaccine that covers virulence Enterobacteriaceae.
Materials and Methods
Retrieving Reference Sequences of OmpA, FepA, OmpW, OmpX and OmpC of K. pneumoniae
Complete putative OmpA (Accession number [AN]: NC_012731.1), FepA (AN: NZ_JQSE01000018.1), OmpW (AN: NZ_AJVY01000178.1), OmpX (AN: NZ_JQSE01000029.1) and OmpC (AN: NZ_JQSE01000032.1) of K. pneumoniae mentioned as reference sequences in the National Center for Biotechnol-ogy Information (NCBI) Databases (http://ncbi.nlm.nih.gov/) were retrieved, separately. The sequences were saved in FASTA format and performed for subsequent analysis.
Entropy Plot and Alignment for Finding the Mutational/Conserved Regions
Thirteen sequences of OmpA of K. pneumoniae and other Enterobacteriaceae were retrieved from NCBI by direct searching. Eleven sequences of FepA, 14 sequences of OmpW, 22 sequences of OmpX and 12 sequences of OmpC were also obtained by the mentioned strategy. Selected sequences and their accession numbers are given in Online Resource 1. Retrieved sequences of each Omp were aligned, analyzed and trimmed using Bioedit software version 7.7.9, separately. Partial sequences and areas with ambigu-ous alignment were omitted and Shannon entropy values (Shannon 1948) were measured for retrieved sequences of the five mentioned proteins, separately. Shannon entropy analysis measures the variable and conserved regions in the set of aligned sequences. The Shannon entropy score (Hx) ranges from 0 to 4.322 for every position in an alignment. Typically, positions with Hx ≤ 1.0 are considered highly conserved positions (Litwin and Jores 1992). The epitopes from highly conserved regions are likely to evoke more immune responses (Sánchez-Burgos et al. 2010; Gupta et al. 2011).
Prediction of Topology of the Omps
While designating B cell epitopes of transmembrane proteins, it is important to determine the amino acid positions, with respect to the lipid bilayer. In order to determine the topology of sequences of OmpA (AN: NC_012731.1), FepA (AN: NZ_JQSE01000018.1), OmpW (AN: NZ_AJVY01000178.1), OmpX (AN: NZ_JQSE01000029.1) and OmpC (AN: NZ_JQSE01000032.1) of K. pneumoniae, PRED-TMBB server (http://bioinformatics.biol.uoa.gr/PRED-TMBB) was employed. PRED-TMBB predicts the transmembrane strands and the topology of β-barrel Omps of gram-negative bacteria based on a Hidden Markov Model.
3D Structure Prediction and Validation of the Omps
Since 3D structures of FepA, OmpX and OmpW of K. pneumoniae were not available in RCSB Protein Data Bank; therefore, modeling of these proteins were done by using I-Tasser server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). I-TASSER is an integrated platform based on multiple threading alignment for automated protein structure prediction (Yang et al. 2015; Roy et al. 2010). The tool PyMOL was used to visualize the modeled 3D structures.
To recognize the errors in the generated FepA, OmpX and OmpW models, coordinates were supplied by uploading 3D structures in PDB format into ProSAweb, separately. ProSAweb, which is frequently exploited in protein structure validation, analyzes the energy distribution in protein structure to determine a structure as native- like or fault (Wiederstein and Sippl 2007). ProSAweb z-score indicates overall model quality, and its value is displayed in a plot that contains the z-scores of all experimentally determined protein chains in current PDB.
The modeled structures of the FepA, OmpX and OmpW were evaluated to see the quality of the resulting stereochemistry of structure by using Ramachandran plot in PROCHECK (http://swissmodel.expasy.org/workspace) (Laskowski et al. 1993), separately. Ramachandran plot, a way to visualize backbone dihedral angles psi against phi of amino acid residues in protein structure, is a regular tool utilized in determining protein structure (Lovell et al. 2003). All residues in an adequate packed protein have fractional volumes near to 1.0 ± 0.1.
Gromos96 forcefield (Gunsteren et al. 1996), implemented in Swiss-PdbViewer v.4.2 (Kaplan and Littlejohn 2001) was performed for energy minimization of the modeled proteins. Energy minimization procedure helps to correct the stereochemistry of the model via eliminating bad contacts between protein atoms and structural water molecules (Laskowski et al. 1993). Minimum energy arrays of atoms correspond to stable states of the system (Srivastava et al. 2011).
Representation of Discontinuous B Cell Epitopes of Omps
Discontinuous B cell epitopes in 3D structure of OmpA (PDB: 2K0L), OmpC (PDB: 1OSM), and modeled FepA, OmpW and OmpX were predicted by using Discotope Server (http://www.cbs.dtu.dk/services/DiscoTope/) (Kringelum et al. 2012). DiscoTope predicts discontinuous epitopes from 3D structure of proteins by applying a linear combination of the normalized values of the hydrophilicity, amino acid statistics, number of contacts, and area of relative solvent accessibility for each residue. In the current study, in order to insert in the B cell construct, discontinuous epitopes from each Omp, which were located in the extracellular surface not in the highly variable regions of the original Omp, were selected.
Prediction of CD4+ T Cells Epitopes
MetaMHCII online tool at http://www.biokdd.fudan.edu.cn/Service/MetaMHCII/server.html (Hu et al. 2010) and Propred at http://www.imtech.res.in/raghava/propred/ (Singh and Raghava 2001) were performed to predict 9mer linear CD4+ T cell epitopes of OmpA of K. pneumoniae. This strategy was also used for identification of 9mer CD4+ T epitopes from FepA, OmpW, OmpC and OmpX of the mentioned pathogen, separately. MetaMHCII implements consensus, probabilistic meta-predictor (PM), AvgTanh and MetaSVMp approaches for combining the results of different servers and, therefore, it has a better performance than discrete predictors (Hu et al. 2010). ProPred implements matrix-based prediction algorithm employing amino-acid/position coefficient table deduced from the literature (Singh and Raghava 2001). The maximum accuracy of ProPred is 75 at a 4 % threshold (default threshold). HLA-DRB1*0101 is one of the most frequent alleles in Caucasians (Pedron et al. 2005). Hence, predictions of epitopes were checked for this allele.
Construct Design, Fusion of Epitopes and Improving Immunogenicity
In order to acquire maximum yield in immunization, it is valuable to consider placing patterns of epitopes in the right positions near each other. Tandem fusion of epitopes to each other and/or adjuvant without proper linkers can result in generation of a new protein with novel properties (Livingston et al. 2002; Yano et al. 2005). To overcome these challenges, a linker sequence of NH2-EAAAK-COOH was inserted within the B cell construct. In the case of CD4+ T epitopes, to provide target-specific cleavage in lysosomal degradation machineries, linker sequences of GPGPG or NH2-AAY-COOH or KK were incorporated between epitopes within the T cell construct sequence. Epitopes were arranged randomly in each construct. In the case of B cell construct, two repetitions of each epitope were inserted in the construct. The frequency of each linker and their connection order with epitopes in T cell construct were optimized by predicting physiochemical characteristics.
The addition of an immunogenic protein sequence can also enhance epitope-based vaccine potency and efficacy (Capone et al. 2006). Hence, for improving immunogenicity of the B cell construct, the complete sequence (548 AA) of the GroEL protein (HSP60) of Salmonella typhi (AN: NP_458769.1) was retrieved from NCBI in FASTA format and added to N-terminal of the polytope via the helical linker (NH2-EAAAK-COOH). The role of GroEL in priming of the humoral and cellular immune responses is being exploited in vaccine development in infectious diseases (Panchanathan et al. 1998; Chitradevi et al. 2013). For T cell construct, the complete sequence of the Heparin-binding hemagglutinin (HBHA) of Mycobacterium tuberculosis (AN: ZP_07011362.1) was obtained from NCBI, and its functional amino acid residues were added to the N-terminal of the construct as an adjuvant. HBHA is an immune adjuvant that has the functional role in binding to toll like receptor 4 (TLR4) (Jung et al. 2011, Adams 2009). TLR ligands agonists, such as TLR4, have strong immunostimulatory effects and can be employed as adjuvant in immunotherapy (van der Burg et al. 2006). Since HBHA is a functional protein, the helical EAAAK linker was placed at both the -NH2 and -COOH termini of the partial selected sequence of this protein to reduce the interaction with other construct regions and cause more efficient separation (Arai et al. 2001).
Evaluation of the Physicochemical Parameters
Protein sequence statistics for B and T cell constructs including amino acid composition, theoretical pI, instability index, in vitro and in vivo half-life, aliphatic index, grand average of hydropathicity (GRAVY) and molecular weight were computed applying ProtParam tool (http://web.expasy.org/protparam/). ProtParam results present the physicochemical parameters of uncharacterized proteins. SOLpro server at http://scratch.proteomics.ics.uci.edu/ was used to predict the propensity of protein solubility upon over-expression in Escherichia coli. SOLpro performs a two-stage SVM architecture method based on multiple representations of the primary sequence (Magnan et al. 2009). The overall accuracy of this server is evaluated over 74 % using multiple runs of tenfold cross-validation.
Posttranslational Modification Analysis
For posttranslational modification analysis of B and T cell constructs, the NetNGlyc1.0 and NetOGlyc4.0 online tools available at http://www.cbs.dtu.dk/services/ were applied. To predict N-glycosylation sites in human proteins, the NetNGlyc server performs artificial neural networks (ANNs) that examine the sequence context of asparagine- any amino acid- serine/threonine (Asn-Xaa-Ser/Thr) sequence (Cai et al. 2003). The NetOGlyc predicts mucin-type GalNAc O-glycosylation sites in mammalian proteins based on neural network (Steentoft et al. 2013).
Calculation of Hydrophobic Regions
In order to evaluate hydrophobic behavior of amino acid sequences of each construct, different methods were exploited in BioEdit software. To evaluate the hydrophobic and hydrophilic regions of the constructs, the algorithm of Kyte and Doolittle (1982) was employed. Peak and trough regions in the profile diagram exhibit hydrophobicity and hydrophilicity, respectively.
Reverse Translation and Codon Optimization
Reverse translation of the B and T cell constructs into DNA sequence and adaptation of the DNA sequences to E. coli codon usage (codon optimization) were performed by JCAT (http://www.jcat.de) and OPTIMIZER (Puigbo et al. 2007), separately.
Allergenicity Evaluation
To analyze the allergenicity of the B and T cell constructs, AlgPred web server at http://www.imtech.res.in/raghava/algpred/ was employed. AlgPred projects the allergenicity based on similarity of known epitope with any region of the protein. Hybrid prediction approach (SVMc+IgEepitope+ARPsBLAST+MAST), exploited at AlgPred, predicts protein allergenicity with a high accuracy (85 % at a threshold _0.4) (Saha and Raghava 2006).
B Cell Construct Modeling and Evaluation
For tertiary structure prediction of the B cell construct, I-TASSER server was used. The tool PyMOL software was used to visualize the modeled 3D structure. ProSAweb was used to recognize the errors in the generated models. In order to see the quality of the resulting stereochemistry of the best model, Ramachandran plot in PROCHECK software was performed (Laskowski et al. 1993). The energy minimization of the structure was done by GROMOS96 implemented in Swiss-PdbViewer v.4.2 (Kaplan and Littlejohn 2001).
Prediction of Immunogenic Epitopes of the B Cell Construct
A construct including discontinuous B cell epitopes should produce B cell mediated immunity to be a good vaccine candidate. For prediction of discontinuous epitopes of the B cell construct, modeled construct was subjected to Discotope server.
Results
Entropy Plot for Finding the Conserved Sites
Based on the entropy plot, a high conservation (Hx ≤ 1) was observed along the OmpA sequence (Fig. 1a). Results of the entropy plot also reflected that the conservation (Hx ≤ 1) is very high along the FepA, OmpW, OmpX and OmpC sequences, separately (Fig. 1b–e). More specifically, according to the entropy plot (Fig. 1a), four highly variable regions (regions above threshold 1) were observed along OmpA that is located in residues 47–49, 143, 182 and 303. Besides, three highly variable regions were detected along FepA that is located in residues 146, 514–516 and 569–570 (Fig. 1b). No highly variable regions were observed along the amino acid sequence of OmpW (Fig. 1c). In the case of OmpX, six highly variable regions were found along this protein that is located in residues 41, 46, 76–77, 115, 120, 116–117 (Fig. 1d). Finally, 11 highly variable regions were found along OmpC that is located in residues 57, 59, 88–92, 102, 164, 178–186, 191–200, 232–244, 277–285, 320–328 and 363–364 (Fig. 1e).
Prediction of the Topology of the Omps
The topology of each Omp of K. pneumoniae was predicted by using PRED-TMBB. Graphical representation of the position of the transmembrane β strands with respect to the lipid bilayer and the location of the loops (periplasmic/extracellular) for OmpA, OmpC, FepA, OmpX and OmpW are represented in Online Resource 2a–e, respectively.
3D Structure Prediction of the Omps
Comparative modeling of the FepA, OmpX and OmpW sequences was exploited by using I-TASSER, separately, to generate 3D models. The quality and potential errors in 3D models of each protein were checked by ProSAweb. The z-scores of starting input structures of FepA, OmpW and OmpX were −3.21, −3.68 and −3.36, respectively, which appear within the range of scores determined in native proteins of similar size. However, in order to obtain the best model of each protein with minimal bad contacts, energy minimization was done. After energy minimization process, the z-scores of the modeled FepA, OmpW and OmpX were improved to −3.49, −3.94 and −3.54, respectively. The predicted 3D structures of the FepA, OmpW and OmpX after energy minimization, visualized by PyMOL software, are shown in Online Resource 3a–c, respectively. Besides, according to energy minimization, the energy minimized FepA, OmpW and OmpX models have admissible stability (−40141.125, −8960.531 and −7712.953 kcal/mol, respectively) compared to that of the initial models (−49541.617, −10828.097, −9683.152 kcal/mol, respectively). These data show that for each protein, the minimized energy structure has more stability in proportion to the initial model.
Evaluation of Models Stability
Before and after energy minimization, the predicted structures of the FepA, OmpW and OmpX were separately validated for their reliability and structural quality based on the Ramachandran plot quality assessment analysis. Obtained results after energy minimization for FepA, OmpW and OmpX are given in Online Resource 4a–c, respectively. Data show that most residues of each afore-mentioned Omps after energy minimization are within >90 % (allowed) regions, separately.
Prediction of B Cell Epitopes in the 3D Structures of the Omps
The predicted discontinuous B cell epitopes in 3D structures of OmpA, OmpC, FepA and OmpX by Discotope server are demonstrated in Online Resource 5. No discontinuous epitope was determined in 3D structure of OmpW. The selected discontinuous epitopes for performing in the B cell construct are shown in Table 1. All of the picked epitopes are located in the extracellular surface (outside) of their original Omps (Online Resource 2a–e). Besides, selected epitopes are not located in the highly variable regions of the parental Omps (Fig. 1).
Defining CD4+ T Cell Epitopes
9mer CD4+ T epitopes of OmpA sequence of K. pneumoniae were determined using MetaMHC and ProPred servers. This strategy was also used for identification of 9mer CD4+ T epitopes from FepA, OmpW, OmpC and OmpX sequences of K. pneumoniae, separately. In order to achieve the high ranked epitopes of OmpA of K. pneumoniae, the peptides with low MetaSVMp values in MetaMHC server and high scores in ProPred server were selected. Then, the high ranked epitopes which were designated from the MetaMHC server were utilized in the final selection with the ProPred server. This strategy was also used for final prediction of CD4+ T epitopes from FepA, OmpW, OmpC and OmpX, separately. The final selected epitopes from the five above mentioned proteins are shown in Table 2. The position of the epitopes on the original protein sequences are also shown in this table. Moreover, all of the predicted epitopes are located in conserved regions (Fig. 1).
Primary Analysis of HBHA Sequence
It is reported that the important functional region of HBHA is placed between amino acid residues 1–158, and the low complexity region (LCR) of protein is located between residues 159 and 199 (Nezafat et al. 2014). Accordingly, we omitted the LCR from HBHA. Moreover, in order to obtain a shorter sequence suitable to insert in the construct, amino acid residues of signal peptide from HBHA was predicted using SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/) and excluded from HBHA sequence. The results showed that these residues are located between 1 and 27.
Construct Design
A schematic diagram of the designed B and T cell constructs are shown in Fig. 2a, b, respectively. For B cell construct, Ep1–Ep4 (two repeat for each epitope) are the epitopes fused to each other by a linker. NH2 terminus of B cell construct was fused to GroEL of S. typhi using EAAAK linker (Fig. 2a). In the case of T cell construct, E1–E25 (one repeat for each epitope) are the epitopes that were linked to each other by a linker. HBHA (residues 28–158) works as the adjuvant sequence to improve immunogenicity (Fig. 2b).
Evaluation of the Physicochemical Parameters
According to PratParam server results, molecular weights were calculated as 72.99 and 55.06 kDa for B and C cell construct, respectively. The theoretical isoelectric point value (pI) is defined as the pH at which the surface of the protein is covered with charge but the net charge of the protein is zero. pI is valuable for assessment of mobility in an electric field. The calculated pIs of the B and T cell constructs were computed to be 5.59 and 6.03, indicating that both B and T cell constructs are acidic in nature. Instability index (Ii) provides the evaluation of the stability of a protein in vitro. On the basis of Ii, Expasy’s ProtParam classified the B cell construct (Ii = 27.30) and T cell construct (Ii = 38.42) as stable. The GRAVY values of the constructs were defined −0.285 and −0.369 for B and C cell construct, respectively. The negative GRAVY value indicates hydrophilisity of the construct that results in better interaction with the surrounding water molecules. The aliphatic indexes were defined as 90.09 and 76.18 for B and C cell constructs, respectively. The high aliphatic indexes show that the constructs are stable for a wide range of temperatures. The chance of constructs solubility upon overexpression in E. coli was computed to be 0.65 (B construct) and 0.97 % (T construct) by SolPro.
Hydrophobicity is a crucial challenge in cloning and expression of constructs. Besides, it helps to estimate the efficiency of vaccines (Kyte and Doolittle 1982). BioEdit software version 7.7.9 was employed to explore the hydrophobic behavior of the B and T cell constructs (Fig. 3a, b).
Posttranslational Modification Analysis
Posttranslational modification analysis was done to check the existence of probable significant modifications of both constructs after their administration in mammal cells. For B cell construct sequence, three N-glycosylation and four O-glycosylation were predicted to exist. Two N-glycosylation and four O-glycosylation were predicted to exist within the T cell construct sequence. These results show that both constructs are free of much posttranslational modification that could be effective on immunogenicity.
Codon Optimization
From different ways to enhance the efficiency of gene expression, one of the most effective ones is codon optimization. It helps to achieve optimum expression of a cloned gene in the recombinant host cells (Sandhu et al. 2008). Reverse translation and codon optimization of the nucleotide sequences of B and T cell constructs were done by JCat and Optimizer. Codon adaptation index (CAI) >0.8 is favorable for high-level expression in different expression hosts. In this study, in the case of B cell construct, CAI of the optimized gene sequence is 0.83 (Fig. 4a). The ideal range of GC content, a measure of transcriptional and translational efficiency, is 30–70 %. The overall GC content of the B cell construct is 49.04 % (Fig. 4b). Moreover, codon with a frequency distribution of 91–100 in the gene construct is 55 % (Fig. 4c); codons with values lower than 30 may decrease the expression efficiency. For T cell construct, CAI of the optimized gene sequence is 0.82 (Fig. 4d), the overall GC content of the construct is 51.59 % (Fig. 4e) and codon with a frequency distribution of 91–100 in the gene construct is 54 % (Fig. 4f). Two negative cis-acting elements, which were found after optimization in the nucleotide sequence of both construct, were removed, separately.
Results of codon optimization of the B and T cell constructs. In the case of B construct, the CAI value of gene sequence is 0.83 (a), average GC content of gene sequence is 49.04 (b) and codon with a frequency distribution of 91–100, 81–90, 71–80, 61–70 and 51–60 are respectively 55, 9, 7, 6 and 21 % in gene sequence (c). For T cell construct, the CAI value of gene sequence is 0.82 (d), average GC content of gene sequence is 51.59 (e) and codon with a frequency distribution of 91–100, 81–90, 71–80, 61–70 and 51–60 are respectively 54, 5, 6, 13 and 20 % in gene sequence (f)
Six Histidine (His) codons were also located in the 3′ end of each construct for purification purpose. Aiming to clone the genes in prokaryotic expression vectors, the NcoI and XhoI restriction sites were added to 3′ and 5′ ends of each construct, respectively.
Allergenicity Evaluation
The allergenicity analysis was performed using the AlgPred server. Based on the hybrid approach in AlgPred, the B and T cell constructs were not detected as potential allergens.
B Cell Construct Modeling and Evaluation
Modeling of the B cell construct sequence was done by using I-TASSER to produce 3D models of the construct. The ProSAweb z-score of starting input structure of the best predicted model of the B cell construct was −11.24 and after energy minimization procedure, z-score improved to −11.45. ProSAweb result indicated that the construct is within the range of scores determined in native proteins of similar size. According to energy minimization, the energy minimized model has more acceptable stability (−31556.324 kcal/mol) compared to that of the initial model (−27960.523 kcal/mol). These data show that the minimized energy structure has more stability in proportion to the initial model. The predicted model of the B cell construct after energy minimization was visualized by PyMOL (Fig. 5). Ramachandran plot quality assessment analysis of the modeled B construct, after energy minimization, showed that most residues of the model are within >90 % (allowed) regions (Fig. 6).
Prediction of Antigenic B Cell Epitopes of the B Cell Construct
Full length B cell construct was subjected to conformational B cell epitope prediction using Discotope server. Out of 696 total residues, 72 conformational B cell epitopes were identified (Table 3).
Discussion
Immunotherapy is a prominent and effective strategy for reducing morbidity and mortality caused by infectious diseases. In recent years, using a rational step-by-step approach to multi-epitope vaccine design has attracted more global attention (Chiarella et al. 2009). By evolution of bioinformatics approaches and its associated branch, immunoinformatics, rapid advance occurred in the field of vaccinology enabling rational design methods of polytopic vaccines (Yang and Yu 2009; Tomar and De 2010). Several approaches have been performed for development of a vaccine against K. pneumoniae (Yadav et al. 2005; Edelman et al. 1994; Ahmad et al. 2012). Candidate proteins for vaccines against this pathogen are immunogenic surface antigens (Kurupati et al. 2006; Ahmad et al. 2012; Florea et al. 2003). Omps family members have been recognized to own these properties and are consequently ideal candidates for vaccine preparation (Kurupati et al. 2006).
In K. pneumoniae infection, B cells are able to induce robust humoral immunity and CD4+ T cells have a fundamental role in priming and maintenance of pathogen specific humoral and cellular immune responses (Hagen et al. 1998). Hence, identification of the discontinuous B cell epitopes and linear CD4+ T cell epitopes of bacterial antigenic Omps could contribute to better understanding of the protective immunity to K. pneumoniae and, also facilitate the preparation of effective anti Klebsiella vaccines. A discontinuous epitope consists of atoms from distant residues joined on the antigenic protein surface in the 3D space and determines antigenicity (Baloria et al. 2012). Discontinuous epitope may be bound to either a B cell receptor or an immunoglobulin and triggers humoral immune response (Ponomarenko and Regenmortel 2009). In this research, it has been attempted to engineer complex efficient polytopic vaccine based on discontinuous B cell epitopes and linear CD4+ T cell epitopes of five Omps from K. pneumoniae, using various bioinformatics approaches.
Prediction of discontinuous epitopes needs knowledge of protein structures. In case the 3D structure of the protein or its homologue is known, a discontinuous epitope can be mapped on the protein structure. Thus, by using the in silico methods, it is possible to model the structure of the pathogenic antigens and map conformational epitopes on the models (Ponomarenko and Regenmortel 2009). Accordingly, in this work, 3D structures of OmpA and OmpC of K. pneumoniae were obtained from RCSB Protein Data Bank. Besides, since 3D structures of FepA, OmpX and OmpW of K. pneumoniae were not available in Protein Data Bank; therefore, their 3D structures were predicted and evaluated using in silico approach. Discontinuous epitopes were designated from five afore-mentioned Omps using online server. The selected epitopes of each Omp, which were not located in the highly variable regions of the original protein (Fig. 1) were selected for further analysis.
B cell epitope region of the bacterial membrane proteins, including Omps, is surface-exposed which reacts with infected host antibodies (Stathopoulos 1996). For this reason, while predicting B cell epitopes from Omps, it is often very important to predict the location of membrane spanning segments along the sequence. In the current study, topology map of five Omps was predicted to find extracellular regions. It should be noted that since the input sequences were trimmed using BioEdit to find conserved regions of each Omp, we would say that this topology might not be a true representation for all the Omp homologs. In the next step, the conserved B cell epitopes of each Omp, were checked whether they were present in outer transmembrane regions of the original protein, using PRED-TMBB results. Epitopes exposed on the surface of the membrane of the Omps were performed in the B cell construct. In a part of the current research, the highly immunogenic CD4+ T cell epitopes were selected based on physicochemical properties and different prediction algorithms. A combination of MetaMHCII and ProPred servers with different algorithms was used to obtain more accurate binder peptides, which are bound to DRB1*0101 allele.
A critical issue in vaccination is the improvement of immune responses by enhancing immunogenicity. There are several methods to increase the immunogenicity of polytopic vaccines (Ingolotti et al. 2010). In this research, some of these approaches were used to enhance the immunogenicity of the constructs at different levels of designing. At first, the size of the constructs was increased by multiplication of the epitopes and incorporation of immunogenic sequence tags (adjuvants) because of the fact that construct which has small size may be rapidly cleared from the body (García-Briones et al. 2004; Gerner et al. 2006). Besides, appropriate linkers were performed to both constructs. Linkers are the non-immunogenic motifs which have an essential role in structural and functional features of a polytopic construct (Livingston et al. 2002). EAAAK linker improves the structural flexibility of a protein via prevention of non-native interactions between different domains that may interfere with the correct folding (George and Heringa 2002). KK spacer, the target sequence of lysosomal cathepsin B, is one of the linkers performed in the CD4+ multi-epitope vaccines (Livingston et al. 2002; Yano et al. 2005). Besides, GPGPG linkers induce Th responses and keep conformational dependent immunogenicity of helpers as well as antibody epitopes (Livingston et al. 2002). The AAY spacer eliminates junctional epitopes successfully and enhances epitope presentation. AAY extensions greatly decrease the binding affinity, and thus AAY-containing epitopes do not bind efficiently (Zhang et al. 2014).
To attain a high-level expression of each recombinant construct in the E. coli host, codon optimization was performed to improve the transcription efficiency and transcript stability. This was accomplished by developing CAI, the total GC content of DNA sequence and codon frequency distribution and removing negative elements that may form unfavorable secondary structures on mRNA. Solubility of overexpressed recombinant proteins in the E. coli host is one of the important requirements of many functional and biochemical assessments. The solubility chance of the B and T cell constructs (65 and 97 %) manifests that they show an acceptable percentage of solubility in an overexpressed mode.
However, in the field of peptide-based vaccine designing, it is important that discontinuous polytopic construct representing discontinuous epitopes mimics the structure of protein epitopes (Ponomarenko and Regenmortel 2009). For this reason, 3D structure of the B cell construct was determined, refined and validated. Then, discontinuous B cell epitopes on the construct were mapped. The result showed that the predicted discontinuous epitopes of the B cell construct (72 amino acid residue) (Table 3) are admissibly in common with total inserted discontinuous epitopes (108 amino acid residue) in the B construct (Fig. 2).
Existence of the high conservation between members of each Omp family indicates that these proteins may be suitable candidates for vaccine development against a wide range of gram-negative bacteria [Kurupati et al. 2006; Koebnik et al. 2000]. Consensus immunogen is a term utilized for immunogens that overcome the limitation of serotypes or amino acid variation of a pathogen that causes immune responses only to be efficient against some or only one serotype/s (Laddy et al. 2007). In this study, consensus highly immunogenic epitopes of Omps from K. pneumoniae and other pathogenic Enterobacteriaceae were predicted and inserted into the polytopic constructs. Therefore, the constructed complex vaccine based on Omps of K. pneumoniae in this work, could cover the genus Escherichia, genus Citrobacter, genus Enterobacter and other pathogenic Enterobacteriaceae. These conclusions may be useful in evaluation of the range of efficiency of the designed constructs against different bacteria.
Conclusion
The current study demonstrates an in silico approach to design efficient complex multi-epitope vaccine against K. pneumoniae. All constructs features are in line with this purpose. Epitopes of the designed constructs could potentially induce effective immune responses. Adjuvant sequences also could play a pivotal role in enhancing the immunogenicity of the constructs. This study could be useful in gaining insight towards the potential of epitope-based construct as an important protective and therapeutic approach for bacterial immunization. Ongoing studies will evaluate whether the polytopic vaccinal constructs could induce immune responses and protection against K. pneumoniae as well as other Enterobacteriaceae.
References
Adams S (2009) Toll-like receptor agonists in cancer therapy. Immunotherapy 1(6):949–964. doi:10.2217/imt.09.70
Ahmad TA, El-Sayed LH, Haroun M, Hussein AA, Ashry SH (2012) Development of immunization trials against Klebsiella pneumoniae. Vaccine 30(14):2411–2420. doi:10.1016/j.vaccine.2011.11.027
Akhoon BA, Slathia PS, Sharma P, Gupta SK, Verma V (2011) In silico identification of novel protective VSG antigens expressed by Trypanosoma brucei and an effort for designing a highly immunogenic DNA vaccine using IL-12 as adjuvant. Microb Pathog 51(1–2):77–87. doi:10.1016/j.micpath.2011.01.011
Ansari HR, Raghava GPS (2010) Identification of conformational B-cell epitopes in an antigen from its primary sequence. Immun Res 6:1–9
Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng 14(8):529–532. doi:10.1093/protein/14.8.529
Baloria U, Akhoon BA, Gupta SK, Sharma S, Verma V (2012) In silico proteomic characterization of human epidermal growth factor receptor 2 (HER-2) for the mapping of high affinity antigenic determinants against breast cancer. Amino Acids 42(4):1349–1360. doi:10.1007/s00726-00010-00830-x
Bijker, Melief CJ, Offringa R, van der Burg SH (2007) Design and development of synthetic peptide vaccines: past, present and future. Expert Rev Vaccines 6(4):591–603. doi:10.1586/14760584.6.4.591
Braun G, Cole ST (1984) DNA sequence analysis of the Serratia marcescens ompA gene: implications for the organisation of an enterobacterial outer membrane protein. Mol Gen Genet 195(1-2):321–328. doi:10.1007/BF00332766
Cai C, Han L, Ji Z, Chen X, Chen Y (2003) SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucl Acids Res 31(13):3692–3697. doi:10.1093/nar/gkg600
Capone S, Zampaglione I, Vitelli A, Pezzaner M, Kierstead L, Burns J, Ruggeri L, Arcuri M, Cappelletti M, Meola A, Ercole BB, Tafi R, Santini C, Luzzago A, Fu TM, Colloca S, Ciliberto G, Cortese R, Nicosia A, Fattori E, Folgori A (2006) Modulation of the immune response induced by gene electrotransfer of a Hepatitis C virus DNA vaccine in nonhuman primates. J Immunol 177(10):7462–7471. doi:10.4049/jimmunol.177.10.7462
Chiarella P, Massi E, De Robertis M, Fazio VM, Signori E (2009) Recent advances in epitope design for immunotherapy of cancer. Recent Pat Anti-Cancer Drug Discov 4(3):227–240. doi:10.2174/157489209789206922
Chitradevi STS, Kaur G, Singh K, Sugadev R, Bansal A (2013) Recombinant heat shock protein 60 (Hsp60/GroEL) of Salmonella enterica serovar Typhi elicits cross-protection against multiple bacterial pathogens in mice. Vaccine 31(16):2035–2041. doi:10.1016/j.vaccine.2013.02.045
Clements A, Jenney AW, Farn JL, LE Brown, Deliyannis G, Hartland EL, Pearseb MJ, Maloney MB, Wesselinghd SL, Wijburga OL, Strugnella RA (2008) Targeting subcapsular antigens for prevention of Klebsiella pneumonia infections. Vaccine 26(44):5649–5653. doi:10.1016/j.vaccine.2008.07.100
Coban C, Kobiyama K, Aoshi T, Takeshita F, Horii T, Akira S, J Ishii K (2011) Novel strategies to improve DNA vaccine immunogenicity. Curr Gene Ther 11(6):479–484. doi:10.2174/156652311798192815
Dönnes P, Elofsson A (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinform 3:25
Edelman R, Taylor DN, Wasserman SS, McClain JB, Cross AS, Sadoff JC, Que JU, Cryz SJ (1994) Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine 12(14):1288–1294. doi:10.1016/S0264-410X(94)80054-4
Florea L, Halldórsson B, Kohlbacher O, Schwartz R, Hoffman S, Istrail S (2003) Epitope prediction algorithms for peptide-based vaccine design. Proc Comput Soc Bioinform 2:17–26. doi:10.1109/CSB.2003.1227293
Galdiero M, Vitiello M, Galdiero S (2003) Eukaryotic cell signaling and transcriptional activation induced by bacterial porins. FEMS Microbiol Lett 226:57–64. doi:10.1016/S0378-1097(03)00562-7
García-Briones MM, Blanco E, Chiva C, Andreu D, Ley V, Sobrino F (2004) Immunogenicity and T cell recognition in swine of foot-and-mouth disease virus polymerase 3D. Virology 322(2):264–275. doi:10.1016/j.virol.2004.01.027
George RA, Heringa J (2002) An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng 15(11):871–879. doi:10.1093/protein/15.11.871
Gerner W, Denyer MS, Takamatsu HH, Wileman TE, Wiesmüller KH, Pfaff E, Saalmüller A (2006) Identification of novel foot-and-mouth disease virus specific T-cell epitopes in c/c and d/d haplotype miniature swine. Virus Res 121(2):223–228. doi:10.1016/j.virusres.2006.05.006
Guan P, Doytchinova IA, Zygouri C, Flower DR (2003) MHCPred: a server for quantitative prediction of peptide—MHC binding. Nucleic Acids Res 31:3621–3624
Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG (1996) Biomolecular Simulations: The GROMOS96 manual and user guide: vdf Hochschulverlag AG an der ETHZ Zürich
Gupta SK, Smita S, Sarangi AN, Srivastava M, Akhoon BA, Rahman Q, Gupta SK (2010) In silico CD4+ T-cell epitope prediction and HLA distribution analysis for the potential proteins of Neisseria meningitidis serogroup B: a clue for vaccine development. Vaccine 28(43):7092–7097. doi:10.1016/j.vaccine.2010.08.005
Gupta SK, Srivastava M, Akhoon BA, Smita S, Schmitz U, Wolkenhauer O, Vera J, Gupta SK (2011) Identification of immunogenic consensus T-cell epitopes in globally distributed influenza-A H1N1 neuraminidase. Infect Genet Evol 11(2):308–319. doi:10.1016/j.meegid.2010.10.013
Gupta SK, Srivastava M, Akhoon BA, Gupta SK, Grabe N (2012) In silico accelerated identification of structurally conserved CD8+ and CD4+ T-cell epitopes in high-risk HPV types. Infect Genet Evol 12(7):1513–1518. doi:10.1016/j.meegid.2012.02.022
Hagen TL, Vianen W, Savelkoul HF, Heremans H, Burman WA, Woudenberg IA (1998) Involvement of T cells in enhanced resistance to Klebsiella pneumoniae septicemia in mice treated with liposome encapsulated muramyl tripeptide phosphatidyl ethanolamine or gamma interferon. Infect Immun 66(5):1962–1967
Hong HR, Patel D, Tamm LK, van den Berg B (2006) The outer membrane protein OmpW forms an eight stranded β-barrel with a hydrophobic channel. J Biol Chem 281:7568–7577. doi:10.1074/jbc.M512365200
Hu X, Zhou W, Udaka K, Mamitsuka H, Zhu S (2010) MetaMHC: a meta approach to predict peptides binding to MHC molecules. Nucl Acids Res 38(2):W474–W479. doi:10.1093/nar/gkq407
Ingolotti M, Kawalekar O, Shedlock DJ, Muthumani K, Weiner DB (2010) DNA vaccines for targeting bacterial infections. Expert Rev Vaccines 9(7):747–763. doi:10.1586/erv.10.57
Jung ID, Jeong SK, Lee CM, Noh KT, Heo DR, Shin YK, Yun CH, Koh WJ, Akira S, Whang J (2011) Enhanced efficacy of therapeutic cancer vaccines produced bdoiy co-treatment with Mycobacterium tuberculosis heparin-binding hemagglutinin, a novel TLR4 agonist. Cancer Res 71:2858–2870. doi:10.1158/0008-5472.CAN-10-3487
Kaplan W, Littlejohn TG (2001) Swiss-PDB viewer [deep view]. Brief Bioinform 2(2):195–197. doi:10.1093/bib/2.2.195
Koebnik R, Locher KP, Gelder PV (2000) Structure and function of bacterial outer membrane proteins: barrels in a nut shell. Mol Microbiol 37(2):239–253. doi:10.1046/j.1365-2958.2000.01983.x
Kringelum JV, Lundegaard C, Lund O, Nielsen M (2012) Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 8(12):e1002829. doi:10.1371/journal.pcbi.1002829
Kurupati PBK, Kumarasinghe G, Poh CL (2006) Identification of vaccine candidate antigens of an ESBL producing Klebsiella pneumonia clinical strain by immunoproteome analysis. Proteomics 6(3):836–844. doi:10.1002/pmic.200500214
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157(1):105–132. doi:10.1016/0022-2836(82)90515-0
Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB (2007) Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 25(16):2984–2989
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi:10.1107/S0021889892009944
Li Y, Li ZJ, Han WY, Lei LC, Sun CJ, Feng X, Du CT, Du TF, Gu JM (2010) Identification and characterization of Th cell epitopes in MrkD adhesion of Klebsiella pneumonia. Microb Pathog 49(1–2):8–13. doi:10.1016/j.micpath.2010.03.009
Litwin S, Jores R (1992) In theoretical and experimental insights into immunology. Springer, Berlin
Livingston B, Crimi C, Newman M, Higashimoto Y, Appela E, Sidney J, Sette A (2002) A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J Immunol 168(11):5499–5506. doi:10.4049/jimmunol.168.11.5499
Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50(3):437–450. doi:10.1002/prot.10286
Lundberg U, Senn BM, Schüler W, Meinke A, Hanner M (2013) Identification and characterization of antigens as vaccine candidates against Klebsiella pneumonia. Hum Vaccin Immunother 9(3):497–505. doi:10.4161/hv.23225
Magnan CN, Randall A, Baldi P (2009) SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 25(17):2200–2207. doi:10.1093/bioinformatics/btp386
Nazarian S, Mousavi Gargari SL, Rasooli I, Amani J, Bagheri S, Alerasool M (2012) An in silico chimeric multi subunit vaccine targeting virulence factors of enterotoxigenic Escherichia coli (ETEC) with its bacterial inbuilt adjuvant. J Microbiol Met 90:36–45. doi:10.1016/j.mimet.2012.04.001
Nezafat N, Ghasemi Y, Javadi G, Khoshnoud MJ, Omidinia E (2014) A novel multi-epitope peptide vaccine against cancer: an in silico approach. J Theor Biol 349(21):121–134. doi:10.1016/j.jtbi.2014.01.018
Panchanathan V, Naidu BR, Devi S, Pasquale AD, Mason T, Pang T (1998) Immunogenic epitopes of Salmonella typhi GroEL heat shock protein reactive with both monoclonal antibody and patients sera. Immunol Lett 62(2):105–109. doi:10.1016/S0165-2478(98)00028-5
Pedron B, Yakouben K, Adjaoud D, Auvrignon A, Landman J, Guerin V, Leverger G, Vilmer E, Sterkers G (2005) Listing of common HLA alleles and haplotypes based on the study of 356 families residing in the Paris, France, area: implications for unrelated hematopoietic stem cell donor selection. Hum Immunol 66(6):721–731. doi:10.1016/j.humimm.2005.02.007
Ponomarenko JV, Regenmortel MHV (2009) B-cell epitope prediction. In: Gu J, Bourne PE (eds) Structural bioinformatics, 2nd edn. Wiley, New York, pp 849–879
Puigbo P, Guzman E, Romeu A, Garcia-Vallvé S (2007) OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucl Acids Res 35(2):W126–W131. doi:10.1093/nar/gkm219
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi:10.1038/nprot.2010.5
Saha S, Raghava G (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucl Acids Res 34(2):W202–W209. doi:10.1093/nar/gkl343
Sánchez-Burgos G, Ramos-Castañeda J, Cedillo-Rivera R, Dumonteil E (2010) Immunogenicity of novel dengue virus epitopes identified by bioinformatic analysis. Virus Res 153(1):113–120. doi:10.1016/j.virusres.2010.07.014
Sandhu KS, Pandey S, Maiti S, Pillai B (2008) GASCO: genetic algorithm simulation for codon optimization. Silico Biol 8(2):187–192
Sbai H, Mehta A, DeGroot AS (2001) Use of T cell epitopes for vaccine development. Curr Drug Targets Infect Disord 1(3):303–313. doi:10.2174/1568005014605955
Schulz GE (1996) Porins: general to specific, native to engineered passive pores. Curr Opin Struct Biol 6(4):485–490. doi:10.1016/S0959-440X(96)80113-8
Shannon CE (1948) The mathematical theory of communication. Bell Syst Tech J 27:379–423 623-656
Singh H, Raghava GPS (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17(12):1236–1237. doi:10.1093/bioinformatics/17.12.1236
Srivastava M, Gupta SK, Abhilash PC, Singh N (2011) Structure prediction and binding sites analysis of curcin protein of Jatropha curcas using computational approaches. J Mol Model 18(7):2971–2979. doi:10.1007/s00894-011-1320-0
Stathopoulos C (1996) An alternative topological model for Escherichia coli OmpA. Protein Sci 5(1):170–173. doi:10.1002/pro.5560050122
Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mande U, Brunak S, Wandall H, Levery SB, Clausen H (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32(10):1478–1488. doi:10.1038/emboj.2013.79
Tomar N, De RK (2010) Immunoinformatics: an integrated scenario. Immunology 131(2):153–168. doi:10.1111/j.1365-2567.2010.03330.x
Tsai SS, Huang JC, Chen ST, Sun JH, Wang CC, Lin SF, Hsu BRS, Lin JD, Huang SY, Huang YY (2010) Characteristics of Klebsiella pneumoniae bacteremia in community-acquired and nosocomial infections in diabetic patients. Chang Gung Med J 33:532–539. doi:10.1111/j.1365-2141.2011.08878.x
van der Burg SH, Bijker MS, Welters MJ, Offringa R, Melief CJ (2006) Improved peptide vaccine strategies, creating synthetic artificial infections to maximize immune efficacy. Adv Drug Deliv Rev 58(8):916–930. doi:10.1016/j.addr.2005.11.003
Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38:D854–D862
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(2):W407–W410. doi:10.1093/nar/gkm290
Yadav V, Sharma S, Harjai K, Mohan H, Chhibber S (2005) Lipopolysaccharide mediated protection against Klebsiella pneumoniae induced lobar pneumonia: intranasal versus intramuscular route of immunization. Folia Microbiol 50(1):83–86
Yang X, Yu X (2009) An introduction to epitope prediction methods and software. Rev Med Virol 19(2):77–96. doi:10.1002/rmv.602
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Net Methods 12(1):7–8. doi:10.1038/nmeth.3213
Yano A, Onozuka A, Asahi-Ozaki Y, Imai S, Hanada N, Miwa Y et al (2005) An ingenious design for peptide vaccines. Vaccine 23(17–18):2322–2326. doi:10.1016/j.vaccine.2005.01.031
Yi JZ, Liu MQ, Zhu CZ, Zhang Q, Sheng ZT, Du QY, Yan WY, Zheng ZX (2004) Recombinant bivalent vaccine against foot-and-mouth disease virus serotype O/A infection in guinea pig. Acta Biochim Biophys Sin 36(9):589–596. doi:10.1093/abbs/36.9.589
Zhang Yu-qing, Zhang Ting-ting, Li Jin-nian, Liu Xue-lan, Li Lin (2014) Design and evaluation of a tandemly arranged outer membrane protein U (OmpU) multi-epitope as a potential vaccine antigen against Vibrio mimicus in grass carps (Ctenopharyngodon idella). Vet Immunol Immunop 160(1–2):61–69. doi:10.1016/j.vetimm.2014.03.016
Zisman DA, Strieter RM, Kunkel SL, Tsai WC, Wilkowski JM, Bucknell KA, Standiford TJ (1998) Ethanol feeding impairs innate immunity and alters the expression of Th1-and Th2-phenotype cytokines in murine Klebsiella pneumonia. Alcool ClinExp Res 22:621e7. doi:10.1111/j.1530-0277.1998.tb04303.x
Acknowledgments
This work was supported by a grant from the Research Council of Shiraz University of Medical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran.
Conflict of interest
There is no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Farhadi, T., Nezafat, N., Ghasemi, Y. et al. Designing of Complex Multi-epitope Peptide Vaccine Based on Omps of Klebsiella pneumoniae: An In Silico Approach. Int J Pept Res Ther 21, 325–341 (2015). https://doi.org/10.1007/s10989-015-9461-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-015-9461-0