Abstract
BMAP-27 and melittin are two antimicrobial peptides (AMPs) that display potent antimicrobial activities against a wide range of microbes including multidrug resistant (MDR) strains of bacteria. Unfortunately, their significant toxicity against eukaryotic cells has hampered their development into clinically useful antibiotics. In this study, we have rationally designed a novel hybrid AMP aiming to retain the potent antimicrobial activities of BMAP-27 and melittin while enhancing their therapeutic index. The strategy employed in our design was based on combining the activities of individual α-helical fragments of each AMP to generate a novel hybrid AMP with improved characteristics compared to the parent peptides. Named as BMAP27-Melittin, the novel peptide displayed broad spectrum antimicrobial activity against standard representative Gram-positive and Gram-negative bacterial strains in the range of 1–7.5 µM. Moreover, the peptide managed to kill standard resistant strains of MDR bacteria with significant potency and with MIC values as low as 1 µM. BMAP27-Melittin also proved to exhibit potent antibiofilm activities while the hemolytic and antiproliferative studies against eukaryotic cells revealed that the peptide is exhibiting minimal toxicity at antimicrobial concentrations. Additionally, the molecular dynamics simulations of the peptide folding displayed that the hybrid peptide has folded into a well-defined α-helical structure; supporting the experimental findings. Overall, this work highlights the potential of rational design in generating improved AMPs with enhanced specificity that could be developed into successful therapeutics for the treatment of bacterial infections.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The wide scale overuse of antibiotics in the last decades has contributed significantly to the development of several bacterial strains resistant to traditional antibiotics (Carlet et al. 2012). The emergence of resistant bacterial strains has been accompanied by a sharp decrease in the number of antibiotics being developed for clinical use and successfully reaching the clinic (Freire-Moran et al. 2011). This has recently raised several worldwide concerns regarding the future management of bacterial infections and imposed a huge pressure on the medical community to develop novel classes of antimicrobial agents with novel mechanisms of action to control the escalating problem of microbial resistance (Carlet et al. 2014). Antimicrobial peptides (AMPs) represent an integral component of the innate host defense system of a variety of organisms including vertebrates, plants, insects and bacteria (Wiesner and Vilcinskas 2010). AMPs are small molecules and display potent wide spectrum antimicrobial activities against several organisms including bacteria, fungi, parasites and enveloped viruses (Jenssen et al. 2006). AMPs are generally short, 10–50 amino acids in length, exhibit a cationic nature and form amphipathic structures when in contact with cell membranes or membrane mimetics (Brogden 2005; Giuliani et al. 2007). Due to their attractive properties regarding the broad-spectrum antimicrobial activity and rapid killing kinetics, AMPs gained a significant interest as potential antibiotic pharmaceuticals for commercial development (Fox 2013). Unfortunately and despite the great potential of these compounds and the extensive research efforts that were carried out to move these agents to the clinic, the results have been limited. This is mainly attributed to several obstacles with the most important being related to AMPs low antibacterial target selectivity and high toxicity against mammalian cells (Marr et al. 2006). Current research is focused on overcoming the problem of AMPs induced mammalian cell toxicity, an issue that is hampering clinical development of this class of agents.
Several AMPs reported in literature have been found to exhibit potent broad-spectrum antimicrobial activities against different strains of Gram-positive and Gram-negative bacteria but suffered from strong hemolytic activities and significant cytotoxicity against mammalian cells. Examples of such peptides include the bovine derived AMP BMAP-27 and the Bee venom derived AMP melittin. Both peptides display potent antimicrobial activities against a wide spectrum of microbes including multidrug resistant (MDR) strains of bacteria (Giacometti et al. 2003; Pompilio et al. 2011). Additionally, both peptides exhibit significant toxicity profiles that limited their applicability for clinical development (Asthana et al. 2004; Skerlavaj et al. 1996). Various strategies have been employed in order to enhance the selectivity of AMPs and improve their therapeutic index including AMP congeners, AMP mimetics and hybrid antimicrobial peptides (Peters et al. 2010). Hybridization of two peptides with different physico-chemical properties is an effective method to obtain novel AMPs with enhanced antibacterial activities or decreased cytotoxicity against mammalian cells (Andreu et al. 1992). Several hybrid AMPs have been designed based on the hybridization of different segments resulting in significant improvements in the biological activities and toxicity profiles over the parental ones (Shin et al. 1998; Park et al. 2004). These hybrid AMPs have great application potential as novel therapeutic agents. In this study, we have rationally designed a novel hybrid AMP with the aim of retaining the potent antimicrobial activity of both BMAP-27 and melttin while enhancing their therapeutic index. The strategy employed in the design was based on combining the activities of individual α-helical fragments of each AMP to generate a novel hybrid AMP with improved characteristics compared to the parent peptides. Named as BMAP27-Melittin, we investigated the antimicrobial and antibiofilm activity of the hybrid peptide against a range of Gram-positive and Gram-negative bacterial strains in addition to bacterial strains displaying multi drug antibiotic resistance. Additionally, we have assessed the antibiofilm activity of BMAP27-Melittin against biofilm growing cells of Staphylococcus aureus and Pseudomonas aeruginosa and also analyzed the underlying molecular mechanisms responsible for the antimicrobial activity of the peptide. BMAP27-Melittin’s toxicity in adittion to the parental peptides was also evaluated using hemolytic assays in addition to antiproliferative studies against mammalian cells.
Materials and Methods
Peptide Design and Analysis
The network protein sequence analysis (NPS) secondary structure prediction software was used for the determination of the α-helical fragments of the parent peptides for the purpose of identifying the suitable segments that can be employed in the design of the new hybrid peptide. The software was also used for the calculation of the percentage of the α-helical content of each of the newly designed hybrid sequences (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_hnn.html). The mean hydrophobicity (〈H〉), and the hydrophobic moment (〈µH〉) were calculated using the HydroMCalc software for the parent and hybrid peptides (http://www.bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html). The Innovagen’s peptide calculator was employed for calculating the water solubility, net charge at neutral pH, molecular weight and isoelectrical point of the parent and hybrid peptides (http://www.innovagen.se/custom-peptide-synthesis/peptide-property-calculator/peptide-property-calculator.asp). Finally, The ProtParam software from the ExPASy server was employed for the evaluation of the physicochemical parameters of the generated hybrid peptide.
Peptide Synthesis and Purification
BMAP27-Melittin was synthesized by solid-phase methods and Fmoc chemistry (G.L. Biochem, China). The purity and the identity (>95 %) of the synthetic peptide were confirmed by high performance liquid chromatography (HPLC) and Electron spray Ionization (ESI-MS) mass spectrometry.
Microorganisms
The antimicrobial activity and potency of BMAP27-Melittin was evaluated and determined against nine standard model bacterial strains that were acquired from the American type tissue culture collection (ATCC) including Listeria ivanovii (ATCC 19119), Enterococcus faecalis (ATCC 19433), S. aureus (ATCC 29213), Staphylococcus epidermidis (ATCC 12228), Escherichia coli (ATCC 25922), Salmonella enterica (ATCC 10708), Salmonella typhimurium (ATCC 29629), P. aeruginosa (ATCC 27853) and Klebsiella pneumoniae (ATCC 13883). Additionally, the antimicrobial activity of BMAP27-Melittin was also evaluated against standard resistant strains of bacteria including two Gram-positive Methicillin-resistant S. aureus (ATCC 43300 & 33591) and two Gram-negative bacterial strains including MDR P. aeruginosa (ATCC 2114) and MDR Acinetobacter baumannii (ATCC 1790).
Bacterial Susceptibility Assay
The minimum inhibitory concentration (MIC) of BMAP27-Melittin against the bacterial strains mentioned previously was determined by adapting the microbroth dilution method outlined by the Clinical and Laboratory Standards Institute (CLSI) guidelines (Ouhara et al. 2008). Briefly, bacterial cells were grown overnight in Mueller-Hinton Broth (MHB) and diluted to 106 CFU/ml in the same medium prior to use. Serial dilutions of BMAP27-Melittin were prepared in culture medium at a volume of 50 μl/well in 96-well plates. BMAP27-Melittin was tested against planktonic bacteria within the concentration range of 0.5–10 μM. Different concentrations of BMAP27-Melittin were inoculated with microorganism cultures to give a final bacterial concentration of 5 × 105 CFU/ml and placed into 96-well microtiter cell culture plates. Plates were incubated for 18 h, at 37 °C, in a humidified atmosphere. Following this, the growth of bacteria was determined by means of measuring optical density (OD) at λ = 570 nm by an ELISA plate reader. For the minimum bactericidal concentration (MBC), 10 μl of the well contents was spread on agar and grown at 37 °C for 24 or 48 h. The MBC was determined as the lowest concentration that resulted in <0.1 % survival of the subculture. All MIC and MBC determinations were made in triplicate.
Time Kill Studies of Exponential Phase Growing Bacteria
The killing kinetics of BMAP27-Melittin against bacterial strains were analyzed by time-kill assay as previously described with a slight modification (Eckert et al. 2006). Time-kill experiments were performed at 37 °C with shaking at 220 rpm under aerobic conditions. Culture aliquots of Log-phase bacteria (3 × 107 CFU/ml) were incubated with different concentrations of BMAP27-Melittin (1 × MIC, 2 × MIC, 3 × MIC and 4 × MIC) in MHB. 10 µl culture aliquots were withdrawn at different time points (0, 15, 30, 60, 120, 300 and 480 min), serially diluted and plated into pre-sterilized nutrient agar plates, afterwards the plates were incubated overnight at 37 °C followed by colony counting. Time-kill curves were constructed by plotting the log10 CFU/ml over time. All assays were performed in triplicate.
Time Kill Studies of Stationary Phase Growing Bacteria
A nutrient depleted MHB was prepared to maintain the bacteria in slow growing stationary phase and according to the method described previously (Mascio et al. 2007). Briefly, all bacterial strains were grown and cultivated in MHB at 37 °C for 46 h. Cultures were later centrifuged at 8,000×g for 30 min. The resultant supernatant was discarded and the bacterial pellet was washed three times with 1× PBS buffer. Stationary-phase cells (1 × 106 CFU/mL) were separately incubated in MHB in the presence of different concentrations of BMAP27-Melittin. The time-kill assay was performed as detailed in the previous section.
Antibiofilm Activity of BMAP27-Melittin Against Standard Resistant Strains of S. aureus and P. aeruginosa
Biofilm formation was performed as previously described in (Ceri et al. 2001; Feng et al. 2013) employing the Calgary biofilm device (Innovotech, Canada). Briefly, P. aeruginosa (ATCC 2114) and S. aureus (ATCC 4330) bacterial strains were incubated in MHB for 20 h at 37 °C, and cultures were diluted in the same medium to achieve a concentration of 107 CFU/ml. 150 μl of this bacterial culture were added to the 96 pegs-lids on which biofilm cells can build up and the pegs were incubated for 20 h under a rotation of 125 rpm at 35 °C to allow biofilm formation on the purpose-designed pegs. Once the biofilms were allowed to form, the pegs were rinsed twice with phosphate buffered saline (PBS) to remove planktonic cells. Each peg-lid was then transferred into a “challenge 96-well microtiter plate” containing two hundred microlitres of each BMAP27-Melittin concentration and the peg lids containing the biofilms were incubated for 4 h at 37 °C. The Minimum biofilm eradication concentration (MBEC) is defined as the minimum concentration needed to inhibit the regrowth of biofilms after 4 h of peptide treatment using an ELISA plate reader λ = 550 nm. Additionally, the biofilms were assessed for their minimum bactericidal concentration (MBECb), as this parameter is defined as the lowest concentration able to eradicate 3log10 of the viable microorganisms in a biofilm (99.9 % killing) after 2 h of incubation using the colony count method.
Erythrocyte Hemolysis Assay
In order to examine the ability of BMAP27-Melittin and the parental peptides BMAP-27 and melittin to induce hemolysis when exposed to human erythrocytes, the hemolytic activity was performed as described previously (Almaaytah et al. 2012). Briefly, Aliquots of 4 % suspension of human erythrocytes in 0.9 % NaCl were incubated with different concentrations of each peptide at 37 °C for 30 min. 100 % hemolysis was determined by addition of Triton X-100. Negative controls represent the erythrocyte suspension in 0.9 % NaCl without the peptide. The percent hemolysis was calculated using the following equation: hemolysis = (A–A0)/(AX–A0) × 100, where A is OD 570 with the peptide solution, A0 is OD 570 in NaCl, and AX is OD 570 nm with 0.1 % Triton X-100.
Cytotoxicity Assay
The cytotoxicity of the hybrid peptide BMAP27-Melittin in addition to the parental peptides BMAP-27 and melittin against cultured cells was determined by the MTT assay and using two mammalian cell lines, HEK 293 and Vero. Each cell line used in this experiment was seeded at a density of 5 × 103 cells per well on a 96-well plate. Cells were treated with various concentrations of each peptide or with medium alone. Following this, the plates were incubated for 24 h, after which, 20 µl of 5 mg/ml MTT solution was added to each well and the plates incubated again for 3 h. The growth medium was later removed using a 1 ml syringe fitted with a needle and 200 μl of DMSO were added to each well and mixed vigorously to dissolve the formazan crystals that had developed. The absorbance was measured using an absorbance microplate reader at 550 nm.
β-Galactosidase Assay
The ability of BMAP27-Melittin to damage cytoplasmic membrane of bacterial cells and the ability of the peptide to induce membrane permeabilization were assessed by measuring the activity and the release of cytoplasmic beta-galactosidase from E. coli (ML-35) cells into the culture medium using ONPG as a substrate as described previously (Ouhara et al. 2008). Escherichia coli (ML-35) were harvested, washed and centrifuged for 10 min (1.4×g) and resuspended in PBS solution with an absorbance of 0.8 at 420 nm. About 1 × 106 cells/ml were incubated with different concentrations of BMAP27-Melittin (1× to 4 × MIC). The hydrolysis of ONPG to O-nitro phenol by beta-galactosidase enzyme was monitored at 405 nm using an ELISA plate reader over different time points (10, 20, 30, 40, 50, 60 and 120 min).
Molecular Modelling of BMAP27-Melittin Peptide
Construction and preparation of the structural model of BMAP27-Melittin peptide was performed using discovery studio (DS) 3.5 from BIOVIA® (formerly Accelrys®) Software Inc. (Discovery Studio 2013). Molecular dynamics (MD) simulations were performed using the molecular dynamics software CHARMm (Chemistry at HARvard Macromolecular Mechanics) (Brooks et al. 2009) implemented in the simulation protocols within DS. Presentation quality images were either generated using DS or the PyMol molecular graphics system (Berman et al. 2002). The computer hosting DS runs 64-bit windows 6 Enterprise Intel® Core i7-3770 3.4 GHz CPUs and 4 Gb RAM.
Construction and Preparation of the Simulated BMAP27-Melittin Model
The structural model of the BMAP27-Melittin novel peptide was constructed using the Build and Edit Protein Tools within DS, where the hybrid peptide was constructed as an extended (linear) peptide chain based on its primary sequence (KFKKLFKKLSPVIGAVLKVLT). Then it was typed using the simulation tools by applying the CHARMm force field in order to add hydrogens appropriate for the selected forcefield, estimate and assign partial charges to the system, and determine forcefield atom types.
Folding Simulations of BMAP27-Melittin Peptide
Once the initial structures have been created, the simulation process was started. CHARMm allows the simulation of the energetics and dynamics of a molecular system; in this study it was used to perform energy minimization and MD simulations.
Firstly, a short minimization was performed before running MD in order to clean the structure, fix hydrogen positions, and remove any steric clashes so that subsequent MD runs will be stable. Minimization was performed using the minimization protocol within DS. Within this protocol, smart minimizer was applied for 1,500 steps were the first 1,000 was utilizing the Steepest Descent algorithm followed by 500 steps of conjugate gradient minimization with an RMS gradient of 0.1 kcal/(mol × Å). To approximate the solvent effect, the system was implicitly solvated using the Generalized Born solvent model (Dominy and Brooks 1999). Non-bonded interactions were treated using spherical cutoff of a 14 Å radius.
The next step was to slowly heat up the system. Heating was accomplished using the dynamics (heating or cooling) protocol within DS were the system was slowly heated from 0.0 to 325 K over the course of 50 ps. Since we manually built our structure, it is likely to initially be much less stable than an experimental crystal structure. Therefore, we used a very short time step of 0.5 fs for the heating stage in order to allow the initial system to relax in a controlled manner. All other parameters were left at their default values. This was followed by another 50 ps of equilibration at 325 K using dynamics (equilibration) protocol within DS with the same parameters as the heating stage except for the time step which was 1 fs.
The final stage of the simulation was to run a long production simulation at 325 K. This was done using the dynamics (production) protocol within DS. Production stage was run in segments of 5 ns (2500000 steps and 2 fs time steps) each at constant temperature dynamics (NVT, the canonical ensemble) using Berendsenweak coupling method (Berendsen et al. 1984). Since the motion of hydrogen atoms in the structure is of little importance in MD, the SHAKE algorithm (Ryckaert et al. 1977) was applied to constrain all bonds involving hydrogen. This in turn reduces the computational complexity and allows longer time steps. Snapshots of the simulations were saved every 2 ps (every 1,000 steps) for analysis.
Results
Design of BMAP27-Melittin Hybrid Peptide
In order to design a novel hybrid AMP for this study, the peptide sequences of BMAP-27 and melittin were evaluated using the NPS HNN secondary structure prediction software for the calculation of the percentage helicity of the parent peptides (Table 1). This was followed by identifying the α-helical fragments within the primary sequence of the peptides in order to be used as platforms for the design and generation of the novel hybrid peptide. The results shown in (Table 1) display that BMAP-27 displays 70.37 % helicity with four potential α-helical suitable for rational design while melittin exhibits a 62.96 % helicity with three potential α-helical fragments suitable for hybridization. The predicted structural data obtained from the NPS software for melittin indicate that the peptide displays 63 % helicity which closely match the data obtained previously from circular dichroism studies which is around 60 % (Eisenberg et al. 1980). Additionally, the structural studies performed on melittin display that the melittin peptide chain is composed of two α-helical segments and an overall shape of a bent rod due to the presence of a large bend around Pro-14 (Vogel and Jaehnig 1986). The three helical fragments generated in this study are all devoid of this proline bend reported earlier and are with agreement with the structural data reported previously for melittin. Recently, several researchers have constructed different hybrid peptides with increased antimicrobial activity compared to the parent peptides due to improved secondary structure parameters. The approach used for the selection and screening of the generated peptides was based on the improvement of the total helicity of the generated hybrid peptides while optimizing other physico-chemical properties related to the activity of AMPs. Using this approach, three novel hybrid peptides were designed, synthesized and were evaluated for antimicrobial screening. Out of the three generated peptides only one peptide displayed antimicrobial activity in the initial screening assays (Supplementary material). This hybrid peptide was named BMAP27-Melittin, and it consists of an N-terminal fragment obtained from residues 9–20 of BMAP-27 and a C-terminal fragment from residues 2–9 of melttin (Table 2). BMAP27-Melittin is composed of 21 amino acid residues and displays a total percentage helicity of 76.2 %, a net charge of +6, a hydrophobicity of 0.531 and a hydrophobic moment of 0.64 (Table 2). Compared to the parent peptides, BMAP27-Melittin displays an enhanced helicity profile in addition to an optimized net charge and hydrophobicity index.
In Vitro Antimicrobial Activity of BMAP27-Melittin
The in vitro antimicrobial activity of BMAP27-Melittin was carried out to determine the spectrum and antimicrobial potency of the designed hybrid peptide against representative standard control and resistant strains of Gram-positive and Gram-negative bacteria. The details of the MIC values of BMAP27-Melittin are listed in Table 3. BMAP27-Melittin displayed potent antimicrobial activities against G+ bacteria with S. epidermidis and S. aureus (ATCC 29213) being the most sensitive with a MIC value of 1 µM, other G+ bacterial strains including L. ivanovii, and E. faecalis reported MIC values of 1.5 µM. The same potent antimicrobial activity of BMAP27-Melittin was also reported with the two Gram-positive methicillin-resistant S. aureus (ATCC 33591 & 4330) bacterial strains with MICs values of 1 and 1.5 µM, respectively. Additionally, BMAP27-Melittin displayed potent antimicrobial activities against G− bacteria with P. aeruginosa (ATCC 27853) being the most sensitive strain with a MIC value of 2 µM while the most resistant being the K. pneumoniae strain with a MIC value of 7.5 µM. MDR resistant strains of G− bacteria were also susceptible to the antimicrobial activity of BMAP27-Melittin as the peptide displayed potent activities against MDR P. aeruginosa (ATCC 2114) and A. baumannii with equal MIC values of 3.5 µM. The MBC values reported for BMAP27-Melittin against all bacterial strains studied were equal to the MIC values (Table 3).This observation indicates that BMAP27-Melittin is probably displaying a bactericidal effect since it is generally accepted that bactericidal drugs have MBC values that are usually within the same range as their MICs and generally not more than fourfold that of the MIC. In contrast, the MBC of bacteriostatic drugs are many-fold higher than their corresponding MICs.
Killing Kinetics
The results of the killing kinetics analysis of BMAP27-Melittin against all exponentially growing bacterial strains employed in this study are shown in (Fig. 1 Supplementary material). For Gram-negative bacteria BMAP27-Melittin managed to totally eradicate (99.9 % killing) all tested strains of bacteria within 30 min of incubation when exposed to peptide concentration equivalent to the MIC. For Gram positive-bacteria, BMAP27-Melittin managed to eradicate L. ivanovii within 30 min of exposure to peptide concentration equivalent to the MIC. At twofold MIC, E. faecalis were killed within 60 min of exposure. At threefold MIC, S. aureus (43300) were killed within 180 min of exposure while 300 min of exposure were needed to totally kill both S. aureus (33591) and S. aureus (29213). For the slow growing stationary bacteria, BMAP27-Melittin displayed bactericidal effects (99.9 % killing) against Gram-positive L. ivanovii and E. faecalis bacteria at peptide concentration equivalent to the MIC within 60 min of exposure, while 180 min of incubation were needed to totally kill S. epidermidis. At twofold MIC, 30 min of incubation were required for the eradication of both S. aeureus (29213) and S. aeureus (43300). At threefold MIC, the S. aureus (335921) bacterial strains were killed with 30 min of exposure to the peptide. For the Gram-negative bacteria, peptide concentration equivalent to the MIC and 30 min of incubation were sufficient to kill the A. baumanii, K. pneumoniae and P. aeruginosa (2114) strains, while 60 min of incubation were needed to kill S. typhi, S. enterica and P. aeuriginosa (27853) and 180 min of incubation were needed for the S. epdermidis strain. threefold MIC concentration and 60 min of incubation were needed to totally eradicate the S. epidermidis strain (Fig. 2 Supplementary material). The rapid killing kinetics observed in the time kill assays for BMAP27-Melittin is consistent with the results of several bactericidal agents and other cationic antimicrobial peptides reported in literature (Saravolatz et al. 2012; Wang et al. 2012). Additionally, the rapid killing behaviour observed with BMAP27-Melittin indicates that the peptide is displaying a mechanism of action similar to that reported for the parent peptides and that the peptide is exhibiting a membranolytic behaviour with the bacterial membrane probably being the main target responsible bacterial killing.
Antibiofilm Activity of BMAP27-Melittin
The antibiofilm activity of BMAP27-Melittin was investigated against pre-formed biofilms of resistant standard strains of S. aureus (43330) and P. aeruginosa (2114). The Calgary device was employed for the evaluation of the antibiofilm activity of the peptide and the identification of the minimal biofilm eradication concentration (MBEC) that is required to eradicate biofilm formation after 4 h of exposure to BMAP27-Melittin. For both bacterial strains the MBEC values reported were equal to 10 µM (Table 4), a clear indication that the peptide is exhibiting significant antibiofilm activity at relatively low concentrations. Additionally, the minimum BMAP27-Melittin concentration needed to reduce the number of viable biofilm cells to almost zero (99.9 % killing) (MBECb) against biofilm cells of P. aeruginosa was found to be 100 µM which was tenfold higher than the MBEC value, while the MBECb value observed with the S. aureus biofilms was 60 µM which represents a sixfold increase over the MBEC value (Table 4). These results clearly display the antibiofilm potential of BMAP27-Melittin and the innate antimicrobial resistance of biofilms towards antimicrobial peptides.
Hemolytic Assay
When tested against human erythrocytes, at a concentrations equivalent to its MICs (1–7.5 µM) against both G+ and G− bacteria, BMAP27-Melittin caused 0 % haemolysis after 60 min of incubation with human erythrocytes (Table 5). Additionally, when BMAP27-Melittin was incubated with human erythrocytes at concentrations equal to the MBEC values (5 and 10 µM) needed to inhibit biofilm growth of P. aeruginosa and S. aureus respectively, the peptide as with the MIC values induced 0 % haemolysis. The parental peptides BMAP-27 and melittin were also evaluated for their haemolytic activity. Both peptides induced significant haemolysis at the antimicrobial concentrations reported earlier with the hybrid peptide BMAP27-Melittin, when incubated with human RBCs for 1 h, the hemolytic activity of melittin at the same concentrations were significantly higher as the percentage hemolysis was in the range of (17–32.8 %) (Table 5). Regarding BMAP-27, the same hemolytic behavior against human RBCs was observed as the percentage hemolysis was in the range of (7–38 %) (Table 5). The maximal haemolytic activity reported for BMAP27-Melittin was 2.9 % at a concentration 100 µM which is tenfold higher than the concentrations required to eliminate bacterial growth while BMAP-27 and melittin induced a percentage haemolysis of 51 and 48 % respectively at the same concentration of 100 µM. The results obtained from the hemolytic assay indicate that BMAP27-Melittin is exerting minimal toxicity against eukaryotic cells at the antimicrobial concentrations and is displaying significant selectivity against microbial cells when compared with the parental peptides BMAP-27 and Melittin.
Cell Cytotoxicity
When assessed for its antiproliferative activity against mammalian cells, BMAP27-Melittin managed to inhibit the proliferation of both mammalian cell lines HEK 293 and Vero with average IC50 values of 21.72 and 19.8 µM, respectively (Table 6). A fivefold increase in the values of the geometric MIC was needed to inhibit 50 % proliferation of mammalian cells when treated with BMAP27-Melittin. The cytotoxicity of the parental peptides BMAP-27 and melittin were also evaluated against mammalian cells. BMAP-27 managed to inhibit the proliferation of both mammalian cell lines HEK 293 and Vero with average IC50 values of 7.8 and 12.9 µM, respectively (Table 6). Melittin on the other hand displayed even higher toxicity against mammalian cells as it managed to inhibit the proliferation of both mammalian cell lines HEK 293 and Vero with average IC50 values of 3 and 7.8 µM, respectively (Table 6). The results of the cytotoxicity studies confirm the results from the previous haemolytic assays that BMAP27-Melittin displays significant selectivity against microbial cells and exhibits minimal toxicity against mammalian cells when compared with the parental peptides BMAP-27 and Melittin.
Cytoplasmic Membrane Permeability
In order to determine BMAP27-Melittin’s ability to inflict damage to the bacterial membranes and cause an increase in the leakage of bacterial intracellular components, we carried out the cytoplasmic membrane permeability assay that depends on the ability of the bacterial intracellular enzyme β-galactosidase to convert ONPG to galactose and ortho-nitrophenol which can be monitored spectrophotometrically at 405 nm. As shown in Fig. 1, BMAP27-Melittin at different concentrations induced significant changes in the membrane permeability of E. coli cells as indicated by the rapid increase of OD values over time which corresponds to o-nitrophenol formation. The absorbance increased in a dose dependent manner and reaches a maximum after 60 min of incubation for all four concentrations employed. The results of the membrane permeability assay clearly demonstrate that BMAP27-Melittin is inducing significant bacterial membrane damage and confirms our previous observations that the peptide is behaving in a similar manner to the parent peptides and that the main mechanism of action responsible for bacterial cell death is probably membrane related.
Structure Prediction and Folding Simulations of BMAP27-Melittin Peptide
The starting structure in this simulation (Fig. 2) corresponds to the linear hybrid peptide whose design was based on combining the well formed α-helical fragments from the parent peptides. Figure 5 displays that the N-terminal half of the structure is highly positively charged compared to the C-terminal part which might have an effect on the folding of the structure since it will exhibit a very strong polar interactions with the surrounding aqueous environment. In this study we ran a 50 ns simulation time. Our system managed to fold into a stable conformation in the first 5 ns. However, the results of the 50 ns will be presented.
System Stability and Conformational Flexibility
The first thing to check in any simulation is the stability of the simulated system which can be assessed by tracking changes in its thermodynamic properties such as temperature and energy that, in a healthy simulation, are supposed to be stable throughout the simulation time. Figure 3 shows that the temperature is well equilibrated around 325 K which indicates that the thermostat used to control the temperature was successful. Also, all energies were stable during the simulation time indicating that the total energy was very well conserved and the system was stable (note that we were saving results every 2 ps (1,000 steps) that is why the time axis in the plots was multiplied by two). In order to investigate the conformational changes our system has experienced throughout the simulation time we calculated the root-mean-square deviation (RMSD) of all atoms from the corresponding starting structure versus time (Fig. 4).
Folding of the Hybrid Peptide
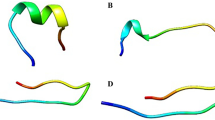
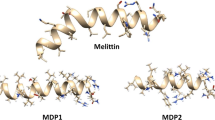
Since we are simulating the folding process of the hybrid peptide, we are most interested in the potential energy plot because we want to locate the most stable conformers (the lowest energy structures) which are supposed to include the folded state of our peptide. Plotting only the potential energy versus time can clearly reveal the lowest energy conformers (Fig. 5). The lowest energy conformers were extracted and presented in Fig. 6. Besides the lowest energy structures in Fig. 6, we got a structure (frame 4356) that is similar to the one in box 2 of Fig. 6 but has a better defined α-helical parts. Comparing the conformation of this structure with that of the parent peptides (Mellitin and BMAP-27) shows that the two segments of the hybrid peptide are adopting α-helical structure similar to their corresponding parts within the parent ones (Fig. 7).
Comparison of the folded conformation of the BMAP27-Melittin hybrid peptide with that of the parent ones. In the lower panel the red part of BMAP27-Melittin corresponds to the segment taken from Melittin, and the green part corresponds to the segment taken from BMAP-27. Both segments of BMAP27-Melittin are α-helical similar to that of the parent peptide from which they have been taken (Color figure online)
Discussion
As the number of antibiotic resistant pathogenic bacteria continues to rise worldwide, the need to develop novel classes of antimicrobial agents to tackle this serious problem is urgently needed. AMPs represent a novel class of potent antimicrobial agents with a great potential of being developed clinically for the treatment of several infectious diseases. Despite the clear potential of these compounds and the great efforts to move them into clinical setting, successful results have been limited. One of the most significant obstacles for the development of AMPs as future therapeutics is related to their lack of microbial target selectivity and their toxicity against mammalian cells. Both are considered fundamental requirements for any successful antimicrobial drug. Several AMPs reported in literature display potent broad-spectrum antimicrobial activity but suffer from high hemolytic activity and cytotoxicity against mammalian cells. The employment of rational design in the development of highly selective AMPs could accelerate the rate at which this class of agents could successfully reach the clinic.
In this study, we have designed a hybrid peptide that is structurally based on the amino acid sequence of two potent naturally occurring AMPs that suffer from high hemolytic activity and a low selectivity index. The rationale behind choosing these peptides was to employ our design method to generate hybrid peptides that retain the potent antimicrobial activity of the parent peptides while enhancing their therapeutic index. The design of novel synthetic AMP analogues requires careful consideration of several physico-chemical parameters that are known to influence AMPs activity and consequently its differential selectivity. The most important parameters that govern AMPs activity include charge, hydrophobicity, and secondary structure (Huang et al. 2010). The positively charged amino acid residues found in AMPs are responsible for the initial binding step of the peptides to the negatively charged phospholipids found on bacterial membranes (Pasupuleti et al. 2012). As AMPs bind bacterial membranes through electrostatic attractions, their non-polar faces insert themselves into the bacterial membranes forming transient pore like structures that destabilize the membranes and consequently cause cell lysis and death (Fjell et al. 2012). The secondary structure of AMPs is known to be a crucial factor in this membrane permeation step and since both the parent peptides employed in this study are reported to display α-helical secondary structures, the design strategy employed depended on combining the activities of the two parent peptides by merging the active α-helical fragments identified in each peptide with the aim of generating novel peptides that capture and combine the potential benefits of each individual fragment and consequently would have improved characteristics over the parent peptides (Huang et al. 2014). Computer aided helical analysis of the parent AMPs display that melittin exhibits three potential α-helical fragments suitable for rational design while BMAP-27 contains four potential α-helical fragments suitable for hybridization. The design strategy generated three different peptides that were screened for their antimicrobial activity with only one hybrid peptide showing potent antimicrobial activity. The resultant peptide named BMAP27-Melittin displayed potent broad spectrum antimicrobial activity against standard representative bacterial strains of Gram positive and Gram-negative bacteria in the range of 1–7.5 µM. Additionally, the peptide managed to kill standard resistant strains of MDR Gram-positive and Gram-negative bacteria with significant potency and with MIC values as low as 1 µM. The MICs of BMAP27-Melittin against standard wild-type and multidrug resistant bacteria were almost the same, suggesting that the antimicrobial mechanism of action of BMAP27-Melittin against bacteria is the same with no relationship to their resistance spectrum. The MBC values for BMAP27-Melittin were equal to the MIC values suggesting that the peptide is bactericidal rather than bacteriostatic. The results of the antimicrobial susceptibility assays proved that the design strategy employed for the generation of the hybrid peptide was successful in generating a highly potent bactericidal agent. The time kill assays revealed that BMAP27-Melittin is exhibiting fast killing kinetics as most microbial strains were eradicated within a short period of time and thus confirming the bactericidal nature of the peptide. BMAP27-Melittin also proved to exhibit potent antibiofilm activities, traditional antibiotics often require 10–1,000 fold higher concentrations to be effective against biofilms when compared with their planktonic counterparts due to the structural features exhibited by cells living within the biofilm environment and these include the extracellular matrix and the low growth rates experienced by biofilm cells (Hoiby et al. 2011; Olson et al. 2002). However, the MBEC reported with BMAP27-Melittin was 10 µM which clearly displays the significant ability of this hybrid peptide to eradicate biofilm formation. The mechanism of action responsible for BMAP27-Melittin’s antibiofilm activity remains to be elucidated but the preliminary antibiofilm results reported in this study indicate that BMAP27-Melittin could be a potent potential antibiofilm agent. The aim of this study was to generate hybrid peptides with significant microbial selectivity and low mammalian cell toxicity. The hemolytic assays of BMAP27-Melittin against human erythrocytes showed that the peptide did not cause any hemolysis at the concentrations required to kill planktonic and biofilm-growing bacteria. Additionally, at concentrations equal to tenfold the average MIC concentrations of BMAP27-Melittin against all microbial strains; the percentage hemolysis never exceeded 3 %. The hemolytic assays with the parental peptides BMAP-27 and melittin clearly display that both peptides induce strong haemolysis when exposed to human RBCs and confirm the cytotoxic nature of the peptides. The results of the hemolytic assays prove that the design strategy and hybridization of the active helical fragments of BMAP-27 and melittin have diminished the hemolytic activity associated with the parent peptides and generate a highly selective hybrid peptide. The cytotoxicity assays on two mammalian cell lines confirmed the selectivity of BMAP27-Melittin as the IC50 values of BMAP27-Melittin were 3–20 fold higher than the bactericidal concentrations. Additionally, the cytotoxicity assays with the parental peptides against mammalian cells display that the peptides exhibit significant cytotoxicity when compared with the hybrid peptide. The cytotoxicity profile generated for BMAP27-Melittin through hemolytic and cytotoxicity assays suggests that the design strategy may be promising to keep cytotoxicity at an acceptable level. However, full animal model toxicity experiments will be needed in future studies in order to reveal the full toxicity profile of BMAP27-Melittin, but the preliminary in vitro data generated in this study are encouraging. Finally, the bacterial susceptibility assay in addition to the time kill kinetics reveal that BMAP27-Melittin is exhibiting a bactericidal mode of action which suggests that the peptide unlike conventional antibiotics is not targeting intracellular targets and appears to act mainly via pore formation or other mechanisms involving the disruption of the integrity of bacterial membranes. The membrane permeabilization assays using beta-galactosidase confirm the membrane disruption mode of action as the release of intracellular beta-galactosidase and the formation of o-nitrophenol from E. coli cells increased dramatically when the bacterial cells were exposed to the peptide and thus confirming the membrane damage hypothesis proposed earlier.
Since the helicity and the secondary structure were considered as important factors in determining the potency and activity of the hybrid peptide, simulating the folding process of this peptide will clarify whether it will fold into an α-helical structure or not. The RMSD plot showed considerable fluctuations, however there is a semi-plateau around 3,000–5,000 ps where the system became more stable. These fluctuations reflect the behavior of the system where it keeps unfolding and refolding as the system tries to locate a more stable structure. Since folded peptides are energetically more stable than the unfolded ones, we were most interested in the potential energy plot that can locate the most stable conformers (the lowest energy structures) which are supposed to include the folded state of our peptide. Interestingly, the folding simulations show that the lowest energy conformers have well-defined α-helical parts, particularly; the frames in boxes number 2 and 4 which have two α-helical parts. Also, they emphasize the behavior we expected from the RMSD plots where we can see that part of the peptide (the positively charged N-terminal part) was continuously changing conformation. Comparison of frame 4356 with those of the parent peptides showed that the helicity of the two segments of the hybrid peptide was conserved in relation to their corresponding parts within the parent ones. In accordance with the simulations findings we can support the after mentioned correlation between the percentage helicity of the hybrid peptide and its antimicrobial activity; and that the design strategy implemented in this study was successful in generating α-helical hybrid peptide with an enhanced antimicrobial properties.
In conclusion, we report the design and functional characterization of the antimicrobial and antibiofilm activities of a novel hybrid peptide named BMAP27-Melittin. The antimicrobial, haemolytic and antiproliferative studies performed in this study indicate that the peptide displays potent selective activities against a wide range of microbes including MDR bacteria. The potency of BMAP27-Melittin combined with its mild cytotoxic profile indicate that the peptide has a great potential as an effective antimicrobial agent and is worthy of being subjected to further investigation in order to optimize its pharmacokinetic and pharmacodynamic profiles.
References
Almaaytah A, Zhou M, Wang L, Chen T, Walker B, Shaw C (2012) Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides 35:291–299
Andreu D, Ubach J, Boman A, Wåhlin B, Wade D, Merrifield RB, Boman HG (1992) Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett 296(2):190–194
Asthana N, Yadav SP, Ghosh JK (2004) Dissection of antibacterial and toxic activity of melittin: a leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J Biol Chem 279:55042–55050
Berendsen HJC, Postma JPM, Gunsteren WFV, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K et al (2002) The protein data bank. Acta Crystallogr Sect D 58:899–907
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Micro 3:238–250
Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B et al (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614
Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D et al (2012) Ready for a world without antibiotics? The pensieres antibiotic resistance call to action. Antimicrob Resist Infect Control 1:11
Carlet J, Pulcini C, Piddock LJV (2014) Antibiotic resistance: a geo-political issue. Clin Microbiol Infect. doi:10.1111/1469-0691.12767
Ceri H, Olson M, Morck D, Storey D, Read R, Buret A et al (2001) The MBEC assay system: multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol 337:377–385
Discovery Studio (2013) version 3.5 ed. Accelrys Inc., San Diego
Dominy BN, Brooks CL (1999) Development of a generalized born model parametrization for proteins and nucleic acids. J Phys Chem B 103:3765–3773
Eckert R, Qi F, Yarbrough DK, He J, Anderson MH, Shi W (2006) Adding selectivity to antimicrobial peptides: rational design of a multidomain peptide against Pseudomonas spp. Antimicrob Agents Chemother 50:1480–1488
Eisenberg D, Terwilliger TC, Tsui F (1980) Structural studies of bee melittin. Biophys J 32(1):252–254
Feng X, Sambanthamoorthy K, Palys T, Paranavitana C (2013) The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant Acinetobacter baumannii. Peptides 49:131–137
Fjell CD, Hiss JA, Hancock REW, Schneider G (2012) Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11:37–51
Fox JL (2013) Antimicrobial peptides stage a comeback. Nat Biotechnol 31(5):379–382
Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL et al (2011) Critical shortage of new antibiotics in development against multidrug-resistant bacteria—Time to react is now. Drug Resist Updat 14:118–124
Giacometti A, Cirioni O, Kamysz W, D’Amato G, Silvestri C, Del Prete MS et al (2003) Comparative activities of cecropin A, melittin, and cecropin A—melittin peptide CA(1–7)M(2–9)NH2 against multidrug-resistant nosocomial isolates of Acinetobacter baumannii. Peptides 24:1315–1318
Giuliani A, Pirri G, Nicoletto S (2007) Antimicrobial peptides: an overview of a promising class of therapeutics. Central Eur J Biol 2:1–33
Hoiby N, Ciofu O, Johansen HK, Z-J Song, Moser C, Jensen PO et al (2011) The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65
Huang Y, Huang J, Chen Y (2010) Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1:143–152
Huang Y, He L, Li G, Zhai N, Jiang H, Chen Y (2014) Role of helicity of α-helical antimicrobial peptides to improve specificity. Protein Cell 5:631–642
Jenssen H, Hamill P, Hancock REW (2006) Peptide antimicrobial agents. Clin Microbiol Rev 19:491–511
Marr AK, Gooderham WJ, Hancock REW (2006) Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol 6:468–472
Mascio CTM, Alder JD, Silverman JA (2007) Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 51:4255–4260
Olson ME, Ceri H, Morck DW, Buret AG, Read RR (2002) Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res 66:86–92
Ouhara K, Komatsuzawa H, Kawai T, Nishi H, Fujiwara T, Fujiue Y et al (2008) Increased resistance to cationic antimicrobial peptide LL-37 in methicillin-resistant strains of Staphylococcus aureus. J Antimicrob Chemother 61:1266–1269
Park Y, Lee DG, Hahm KS (2004) HP(2–9)-magainin 2(1–12), a synthetic hybrid peptide, exerts its antifungal effect on Candida albicans by damaging the plasma membrane. J Pept Sci 10(4):204–209
Pasupuleti M, Schmidtchen A, Malmsten M (2012) Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol 32:143–171
Peters BM, Shirtliff ME, Jabra-Rizk MA (2010) Antimicrobial peptides: primeval molecules or future drugs? PLoS Pathog 6:e1001067
Pompilio A, Scocchi M, Pomponio S, Guida F, Di Primio A, Fiscarelli E et al (2011) Antibacterial and anti-biofilm effects of cathelicidin peptides against pathogens isolated from cystic fibrosis patients. Peptides 32:1807–1814
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Saravolatz LD, Pawlak J, Johnson L, Bonilla H, Fakih MG, Fugelli A et al (2012) In vitro activities of LTX-109, a synthetic antimicrobial peptide, against methicillin-resistant, vancomycin-intermediate, vancomycin-resistant, daptomycin-nonsusceptible, and linezolid-nonsusceptible Staphylococcus aureus. Antimicrob Agents Chemother 56:4478–4482
Shin SY, Kang JH, Lee MK, Kim SY, Kim Y, Hahm KS (1998) Cecropin A-magainin 2 hybrid peptides having potent antimicrobial activity with low hemolytic effect. Biochem Mol Biol Int 44(6):1119–1126
Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M (1996) Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem 271:28375–28381
Vogel H, Jaehnig F (1986) The structure of melittin in membranes. Biophys J 50(4):573
Wang W, Tao R, Tong Z, Ding Y, Kuang R, Zhai S et al (2012) Effect of a novel antimicrobial peptide chrysophsin-1 on oral pathogens and Streptococcus mutans biofilms. Peptides 33:212–219
Wiesner J, Vilcinskas A (2010) Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464
Conflict of interest
The authors of this manuscript certify that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements) in the subject matter or materials discussed in this manuscript.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Almaaytah, A., Tarazi, S., Al-Fandi, M. et al. The Design and Functional Characterization of the Antimicrobial and Antibiofilm Activities of BMAP27-Melittin, a Rationally Designed Hybrid Peptide. Int J Pept Res Ther 21, 165–177 (2015). https://doi.org/10.1007/s10989-014-9444-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-014-9444-6