Abstract
In the first instance, mononuclear Cu(II) complexes are prepared with bis-N,N′(salicylidene)-1.3-propanediamine and derivatives. After that, these mononuclear complexes are combined with μ-bridges, by the help of azide ions, to obtain the tetranuclear complexes. Prepared complexes are characterised using IR spectroscopy, elemental analysis, and X-Ray techniques. In addition, the complexes are further analysed via TG and DSC. Molecular models of two of the nine prepared complexes are determined using X-Ray diffraction methods. The two terminal copper ions are observed to be in square pyramide coordination sphere between two oxygens of the organic ligand, two iminic nitrogens and an oxygen of the solvent while the other two cupper ions are observed to be in square pyramide coordination sphere between the fenolic oxygens of the organic ligand and the nitrogen donors of the three azide ions. It is found that the fenolic oxygens form μ-bridge and two azide ions are monodentate coordinated. In the TG analyses, the complexes are observed to decompose in a highly exothermic manner at about 200 °C. This thermal reaction is partially similar to that of explosive molecules and the data from DSC proved that the liberated heat is at explosive material levels.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The fact that azide ions form two types of μ-bridges and their tendency combine metal ions within complexes has been known since 1986 [1–10]. Also bis N,N′(salicylidene)-1,3-propanediamine type ligands are known to form polynuclear complexes with transition metals and ions like nitrate and acetate [11–17]. Mononuclear Cu(II) complexes were prepared from three ONNO type Schiff base compounds in order to synthesize polynuclear Cu(II) complexes [18–20] (Fig. 1).
Mononuclear complexes tend to form dinuclear complexes with excess Cu(II) salts [21]. Thus excess Cu(II) and equivalent N3 − were added to the solution. A dark brown crystalline precipitate was obtained. The structure was analyzed using X-ray and characterized using IR, elemental analysis and thermogravimetry. Similar studies are reported in literature [22, 23]. Reports show linear polymeric structures to form with Hg as the central atom via end–end formation of the azide ions. In the case of Cu(II) end-on bonding occurs forming tetranuclear complexes [24]. The solvent molecules are reported to be coordinated to the terminal Cu(II) ions. Thus we have chosen THF, dioxane and DMF as the solvent media. The stochiometry of the complexes prepared obtained from the thermal and elemental analysis.
Molecular models were obtained using X-ray diffraction for [CuL·Cu(N3)2·DMF]2 (I) and [CuL·Cu(N3)2·dioxane]2 (III). As stated before TG is an efficient method in determination of complex stochiometry. Thus solvent molecules and stochiometry were derived using TG.
Experimental
Apparatus
IR spectrums were obtained with a Mattson FTIR 1000 with KBr disks. Cu analysis was performed by dissolving a few milligram of the samples in H2O2:HNO3 mixture, followed by analysis using a GBC Avanta PM model AAS. As azide ions tend to explode elemental analysis was not very efficient and thermogravimetry was used instead. TG studies were performed on a Shimadzu DTG-60H instrument within a temperature range of 25–850 °C in N2 atmosphere with a heating rate of 10 °C min−1. The exothermic heat evolved during decomposition of azide ions was measured on a DSC-60 instrument.
X-ray crystal structure analysis
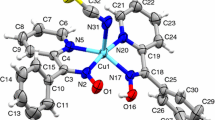
For the crystal structure determination, the single-crystal of the compounds I and II were used for data collection on a Enraf-Nonius CAD4 diffractometer [25] with MoKα (λ = 0.71073 Å) radiation using the ω/2θ scan mode. The cell parameters were determined from least-squares analysis using 25 centered reflections in the range 3.36° ≤ θ ≤ 25.21° for compound I and 2.51° ≤ θ ≤ 26.30° for compound II. Three standard reflections were periodically measured (every 120 min) during data collection and showed no significant intensity variations. The structures were solved by direct methods (SHELXS-97) [26] and non-H atoms were refined by full-matrix least-squares method with anisotropic temperature factors (SHELXL-97) [26] in the WinGX package [27]. A PLATON drawing [28] of the compounds I and II with 50% probability displacement thermal ellipsoids and atomic numbering scheme are shown in Fig. 2a, b. Crystal and experimental data, selected bond lengths and angles of the trimeric complexes I [CuL·Cu(N3)2·DMF]2 and II [CuL·Cu(N3)2·dioxane]2 are given in Tables 1, 2, 3, respectively.
Preparation of complexes
They were prepared in two steps. In the first step mononuclear complexes were prepared followed by the second step where the polynuclear complexes were synthesized.
First step: 0.005 moles of the Schiff base (1.41 g LH2, 1.515 g LDMH2 or 1.48 g LMH2) was dissolved in EtOH. One milliliter of Et3N was added to this solution followed by the addition of the solution of 0.005 mole CuCl2·2H2O in 50 mL of water. The mixture was left to stand for 2 days. The mononuclear complex crystals were filtered and dried in open air.
Second step: 0.001 mole of CuL, CuLDM or CuLM were dissolved in 30 mL DMF or 40 mL dioxane or 50 mL THF. To this was added to solutions of 0.001 mole CuCl2·2H2O in 20 mL hot MeOH and 0.002 mole (0.130 g) NaN3 in 5 mL hot water. The mixture was left to stand for 2–3 day and the precipitate filtered.
Results and discussion
IR data and Cu analysis results are given in Table 4. The νC=O vibration of the DMF molecules are seen between 1,654 and 1,649 cm−1, whereas the νC=N stretching of the ligands is seen between 1,620 and 1,627 cm−1. νC=N band seen at 1,640 cm−1 for the ligand shifts by 15 cm−1 to lower energy level [29–31]. N3 − ions are observed clearly at 2,200 cm−1. Azide ions form two types of μ-bridges 1,1 (end–on) and 1,3 (end–end). The double bond structure should be the case in both μ-bridge structures (Fig. 3).
The triple bond structure is observed at 2,200 cm−1, but the double bond structure shifts to lower energy values [9, 32]. These can be observed in the complexes. The \( \nu_{{{\text{N}}_{3} }} \) bonds show at least the presence of one μ-bridge as two vibration at 2,034 and 2,071 cm−1 are observed. But IR does not show the structure of the bridge. Literature states this bridge to be of 1.1 structure [32–34]. The Platon drawing of [CuLDM·Cu(N3)2·DMF]2 and [CuLDM·Cu(N3)2·dioxane]2 are given in Fig. 2a and b, respectively.
Figure 2 shows a tetranuclear structure with two CuLDM·Cu(N3)2 units bonded by a 1,1 μ-bridge and two free azide ions. A μ-bridge between CuLDM and Cu(N3)2 is found via the phenolic oxygens. In the terminal CuLDM unit the Cu(II) ion situated between the ON1N2O2 donor atom is coordinated by a THF molecule. The coordination sphere of the four Cu(II) ions is a square pyramid (Fig. 4).
The ideal structure of penta coordinated structures is measured by a τ value [30].
α and β are the largest angles in the vicinity of the central ions. If τ = 0 the coordination is an ideal square pyramid, if τ = 1 it is an ideal trigonal bipyramid. Crystal data of complex I and complex III are given in Table 1 whereas important coordinative bond lengths and bond angels are given in Tables 2 and 3. The α and β values of Cu1 are 164.0(2)° and 148.2(4)° and 169.9(2)° and 164.1(3)°, respectively for complex I. In this case τ value for Cu1 is 0.263 and 0.093 for Cu2. These values can calculate for complex III as 0.163 and 0.130, respectively and these values indicate the coordination sphere to be a distorted square pyramid. The basal plane of the Cu2 ion coordination sphere are the O1N1N2O2 atoms, because Cu2 is 0.1326 Å in distance from this plane for the complex I and 0.1104 Å for complex III. The Cu2–O3 distance is 2.362(5) Å and is clearly further than the Cu2–N1, Cu2–O2 and Cu2–N2 distances fort he complex I. This means the Cu2 coordination sphere has elongated towards the THF oxygen.
Thermoanalytical data of the complexes have been given in Table 5. TG and DTA curves are shown in Fig. 5a–e, respectively. The figures reveal three thermal reactions; mass loss between 120 and 170 °C, exothermic mass loss between 200 and 210 °C and degradation around 260 °C. The first reaction corresponds to separation of coordinated solvent molecules from the structure. The mass loss coincides with the mass loss of these molecules. THF leaves the structure at 120 °C, dioxane between 135 and 140 °C and DMF between 160 and 170 °C. The second thermal reaction is due to the explosive reaction of the azide ions according to the our recently work [35]. After this reaction the TG curves resembles that of CuL. Above 260 °C degradation of the compound continues. In all complexes the TG curves resemble that of CuL above 210 °C [36–38]. The explosive reaction does not destroy the whole molecule. The remaining CuL decomposes with increasing temperature. The mass loss during the explosive reaction is higher than expected and is probably due to scattering of the molecules. Also above 750 °C oxygen was purged into the system and residue converted to CuO, but the remaining mass loss of CuO does not correspond to the expected amount. The question “Does the tetranuclear complex decompose after separation of the solvent molecules?” crises. The terminal Schiff base-Cu(II) complex bonds to Cu(N3)2 via the phenolic oxygens, resulting in a decrease of electron density given to Cu(II) ions from the phenolic oxygens. The terminal Cu(II) ion completes the electron deficiency from solvent molecules by coordinating them. If this coordination is damaged then the μ-bridges can break.
In the remaining mixture Cu(N3)2 undergoes an explosive reaction and the remaining CuL decomposes between 260 and 300 °C. TG measurements do not explain this. With the explosion of Cu(N3)2 a portion of CuL disintegrates. The remaining mass after 300 °C is a random mass. We think it is comprised randomly of a mixture CuO and carbonized ports, because the mass loss in the vicinity of 300 °C is the larger than the CuO amount. DSC measurements show the explosion to spread throughout the molecule. For reference Cu(N3)2 was prepared and the heat values measured; 1.32 ± 0.04 kJ g−1 or 194.76 kJ mol−1. If after separation of the solvent molecules Cu(N3)2 is formed the value of heat obtained from DSC should equal the mass fraction of Cu(N3)2 in the complex and the explosion heat of this compound. But the values obtained are higher than expected. The mass ratio of Cu(N3)2 in [CuL·Cu(N3)2·dioxane]2 and [CuL·Cu(N3)2·DMF]2 are 0.254 and 0.261, respectively. The expected heat value for [CuL·Cu(N3)2·dioxane]2 is 335.25 and 349.52 J g−1 for [CuL·Cu(N3)2·DMF]2. but DSC data were 607.21 and 558.04 J g−1, respectively. This shows the complex does not decompose after separation of the solvent molecules. Cu(N3)2 has spread its explosive characteristic throughout the molecule. This can be seen from the DTA peak. The peak maximum of Cu(N3)2 is between 199 and 201 °C, but within the complex this value shifts to 210–222 °C. This shows that when μ-bridges are formed with azide ions the explosive character does not decrease within the complex structure but, all of the complex structure tends to become an explosive material alternative.
References
Charlot MF, Kahn O, Chaillet M, Larrieu C. Interaction between copper(II) ions through the azido bridge: concept of spin polarization and ab initio calculations on model systems. J Am Chem Soc. 1986;108:2574–81.
Cortes R, de Larramendi JIR, Lezama L, Rojo T, Urtiaga K, Arriortua MI. Synthesis, structural, spectroscopic and magnetic studies of two azido and thiocyanato nickel(II) dinuclear complexes with ferromagnetic interactions. JCS Dalton Trans. 1992; 2723–8.
Julve M, Verdaguer M, de Munno D, Real JA, Bruno G. Synthesis, crystal structure, and magnetic properties of (μ-bipyrimidine)(cyanato)copper(II) and -(thiocyanato)copper(II) complexes. Inorg Chem. 1993;32:795–802.
Ribas J, Escuer A, Monfort M, Vicente R, Cortes R, Lezama L, et al. Polynuclear NiII and MnII azido bridging complexes. Structural trends and magnetic behavior. Coord Chem Rev. 1999;193–195:1027–68.
Goher MAS, Escuer A, Mautner FA, Al-Salem NA. Synthesis, spectral, magnetic and crystal structural characterization of two new copper(II) azido complexes: catena-[μ(N3)Cu(pyridine)3] n (PF6) n and dimeric [Cu(4-ethylpyridine)(N3)2]2. Polyhedron. 2001;20:2971–7.
Gao EQ, Bai SQ, Wang CF, Yue YF, Yan CH. Structural and magnetic properties of three one-dimensional azido-bridged copper(II) and manganese(II) coordination polymers. Inorg Chem. 2003;42:8456–64.
Cabort A, Therrien B, Bernauer K, Süss-Fink G. Copper(II) azido complexes containing trinitrogen ligands: [Cu(η3-L)(N3)]2[Cu2Cl2(N3)4] [L=2,6-bis(3,4-dihydro-2H-pyrrol-5-yl)pyridine], a tridimensional network of cationic and anionic copper complexes. Inorg Chim Acta. 2003;349:78–84.
Deoghoria S, Sain S, Sola M, Wong WJ, Christov G, Bera SK, et al. Synthesis, crystal structure and magnetic properties of a new ferromagnetic nickel(II) dimer derived from a hexadentate Schiff base ligand. Polyhedron. 2003;22:257–62.
Jia HP, Li W, Ju ZF, Zhang J. Synthesis, structure, and magnetic properties of a novel mixed-bridged heterometal tetranuclear complex [Mn2Ni2(MeOSalen)2(μ1,1-N3)2(N3)2]. Inorg Chem Commun. 2007;10:397–400.
Liu CM, Zhang DQ, Zhu DB. A copper(II) coordination polymer with alternating double EO-azido bridges and mixed EO-azido/alkoxo double bridges. Inorg Chim Acta. 2009;362:1383–6.
Fukuhara C, Tsuneyoshi K, Matsumoto N, Kida S, Mikuriya M, Mori M. Synthesis and characterization of trinuclear Schiff-base complexes containing sulphur dioxide or hydrogen-sulphite ions as bridging groups. Crystal structure of [Zn{(μ-CH3CO2)(salpd-μ-O,O′)Cu}2][salpd = propane-1,3-diylbis(salicylideneiminate)]. JCS Dalton Trans 1990:3473–9.
Uhlenbrock S, Wegner R, Krebs B. Syntheses and characterization of novel tri- and hexa-nuclear zinc complexes with biomimetic chelate ligands. JCS Dalton Trans 1996: 3731–6.
Mikuriya M, Tsuru N, Ikemi S, Ikenoue S. High nuclearity in a zinc(II) complex with 1,3-bis(salicylamino)-2-propanol. Chem Lett 1998;9:879–80.
Mikuriya M, Ikenoue S, Nukada R, Lim JW. Synthesis and structural characterization of tetranuclear zinc(II) complexes with a linear array. Bull Chem Soc Jpn. 2001;74:101–2.
Atakol O, Durmuş S, Durmuş Z, Arıcı C, Çiçek B. Investigations, on some heterotrinuclear complexes of nickel(II) and copper(II). Synth React Inorg Met Org Chem. 2001;31:1689–704.
Reglinski J, Taylor MK, Kennedy AR. Hydrogenated Schiff base ligands: towards the controlled organisation of open metal frameworks. Inorg Chem Commun. 2006;9:736–9.
Shi DH, You ZL, Xu C, Zhang Q, Zhu HL. Synthesis, crystal structure and urease inhibitory activities of Schiff base metal complexes. Inorg Chem Commun. 2007;10:404–6.
Drew MGB, Prasad RN, Sharma RP. Structures of (N,N′-trimethylenedisalicylideneaminato) nickel(II) (1) and (N,N′-trimethylenedisalicylideneaminato) copper(II) (2). Acta Crystallogr C. 1985;C41:1755–8.
Arıcı C, Ercan F, Kurtaran R, Atakol O. [N,N′-bis(salicylidene)-2,2-dimethyl-1,3-propanediaminato] nickel(II) and [N,N′-bis(salicylidene)-2,2-dimethyl-1,3-propanediaminato] copper(II). Acta Crystallogr C. 2001;C57:812–4.
Yua HH, Lo JM, Chen BH, Lu TH. [N,N′-bis(salicylidene)-1,4-diiminobutane] copper(II). Acta Crystallogr C. 1997;C53:1012–3.
Butcher RJ, Sinn E. Relation between magnetic, spectroscopic, and structural properties of bis[chloro(N-isopropyl-2-hydroxybenzylidene)aminato-μ-O-copper(II)] and bis(N-isopropyl-2-hydroxybenzylideneaminato)copper(II). Inorg Chem. 1976;15:1604–8.
Kurtaran R, Emregül KC, Arıcı C, Ercan F, Catalano VC, Atakol O. Synthesis and crystal structure of linear chain homotetranuclear complexes with N3 −. Synth React Inorg Met Org Chem. 2003;33:281–96.
Koner S, Saha S, Okamato KI, Tuchagues JP. A novel tetranuclear copper(II) complex with alternating μ1,1-azido and phenoxo bridges: synthesis, structure, and magnetic properties of [Cu4(μ-salen)2(μ1,1-N3)2(N3)2]. Inorg Chem. 2003;42:4668–72.
Reddy KR, Rajasekharan MV, Tuchagues JP. Synthesis, structure, and magnetic properties of Mn(salpn)N3, a helical polymer, and Fe(salpn)N3, a ferromagnetically coupled dimer (salpnH2 = N,N′-bis(salicylidene)-1,3-diaminopropane). Inorg Chem. 1998;37:5978–82.
Enraf Nonius:Cad-4 Express Version 1.1. Delft, The Netherlands; 1993.
Sheldrick GM. SHELXS97 and SHEXL97. Program for crystal structure solution and refinement. University of Gottingen, Germany; 1997.
Farrugia LJ. WinGX. Program for crystallography package. J Appl Crystallogr. 1999;32:837.
Spek AL. PLATON. Program for crystal molecular drawing. The Netherlands: University of Ultrech; 2000.
Banerjee S, Ray A, Sen S, Mitra S, Hughes DL, Butcher RJ, et al. Pseudohalide-induced structural variations in hydrazone-based metal complexes: syntheses, electrochemical studies and structural aspects. Inorg Chim Acta. 2008;361:2692–700.
Addison AW, Rao TN, Reedijk J, van Rijn J, Verschoor GC. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. JCS Dalton Trans 1984:1349–56.
Wang QL, Yang C, Qi L, Liao DZ, Yang GM, Ren HX. A trinuclear nickel(II) complex with dissimilar bridges: synthesis, crystal structure, spectroscopy and magnetism. J Mol Struct. 2008;892:88–92.
Khalaji AD, Amirnasr M, Triki S. New coordination polymer based on salpn Schiff base and azide bridging ligands: synthesis and structural characterization of {Na[CoIII(μ-salpn)(μ1,1-N3)2]} n (H2salpn = N,N′-bis(salicylidene)-1,3-diaminopropane). Inorg Chim Acta. 2009;362:587–90.
Liu Z, Zhang T, Zhang J, Wang S. Studies on three-dimensional coordination polymer [Cd2(N2H4)2(N3)4] n : crystal structure, thermal decomposition mechanism and explosive properties. J Hazard Mater. 2008;154:832–8.
Demeshko S, Leibeling G, Marringgele W, Meyer F, Mennerich C, Klauss HH, et al. Structural variety and magnetic properties of tetranuclear nickel(II) complexes with a central μ4-azide. Inorg Chem. 2005;44:519–28.
Dinçer Kaya FN, Svoboda I, Atakol O, Ergun Ü, Kenar A, Sarı M, et al. Nickel(II) complexes prepared from NNN type ligands and pseudohalogens—synthesis, structure and thermal decomposition. J Therm Anal Calorim. 2008;92:617–24.
Soliman AA, Linert W. Investigations on new transition metal chelates of the 3-methoxy-salicylidene-2-aminothiophenol Schiff base. Thermochim Acta. 1999;338:67–75.
El-Said AI. Studies on some nickel(II) and cobalt(II) mixed ligand complexes of arylsalicyl-aldimine and other ligands. J Therm Anal Calorim. 2002;68:917–29.
Aranha PE, Souza JM, Romero S, Ramos LA, dos Santos MP, Dockal ER, et al. Thermal behavior of vanadyl complexes with Schiff bases derived from trans-N,N′-bis(salicylidene)-1,2-cyclohexadiamine (t-Salcn). Thermochim Acta. 2007;453:9–13.
Acknowledgements
The authors wish to acknowledge the financial support of the Ankara University Research Fund (Project no. 07B4240001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Öz, S., Kunduracı, M., Kurtaran, R. et al. Thermal decomposition of linear tetranuclear copper(II) complexes including μ-azido bridges. J Therm Anal Calorim 101, 221–227 (2010). https://doi.org/10.1007/s10973-009-0394-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10973-009-0394-8