Abstract
MicroRNAs (miRNAs) are a class of naturally occurring and small non-coding RNA molecules of about 21–25 nucleotides in length. Their main function is to downregulate gene expression in different manners like translational repression, mRNA cleavage and epigenetic modification. To predict new miRNAs in plants different computational approaches have been developed. In the present study, an EST based approach has been used to identify novel miRNAs in horsegram. Identification of miRNAs was initiated by mining the EST database available at NCBI. Total of 989 ESTs were obtained for the identification of miRNAs. These ESTs were subjected to CAP3 assembly to remove the redundancy. This resulted in an output of 72 contigs and 606 singletons as non redundant datasets. The miRNAs were then predicted by using miRNA-finder. A total of eight potential miRNAs were predicted and named as hor-miR1 to hor-miR8. None of identified miRNAs showed significant homology with the previously reported in plants and therefore should be considered novel. These miRNAs were inputted to miRU2 program to predict their targets. The target mRNAs for these miRNAs mainly belong to zinc finger, chromosome condensation, protein kinase, abscisic acid-responsive, calcineurin-like phosphoesterase, disease resistance and transcriptional factor family proteins. These targets appeared to be involved in plant growth and development and environmental stress responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For years, RNA molecules have been thought to bear just two major functions in cells. The coding RNAs (messenger RNAs) are essential intermediaries in gene expression and non-coding RNAs (ribosomal and transfer RNAs) have structural, catalytic and information decoding roles in protein synthesis. The path breaking discovery of silencing of genes by non-coding RNAs known as RNA interference (RNAi) has changed the insight of people in this field [1]. Non-coding RNAs are abundant in eukaryotic cells. These small RNAs play central roles in important regulatory mechanisms mediating many biological processes in plants and animals.
The microRNAs (miRNAs) and small interfering RNAs (siRNAs) represent two major classes of small RNAs that regulate gene expression at the post-transcriptional level in plants [2, 3]. siRNAs are processed from long, double-stranded RNA precursors and direct gene silencing through both mRNA degradation and chromatin modification [4]. Though miRNAs are chemically and functionally similar to siRNAs they are derived from local stem-loop structures in the genome. The miRNAs should have following characteristic features: (a) miRNA should consist of 20–24 nt [2, 5], (b) all miRNA precursors should have a well predicted stem-loop hairpin structure with low free energy [6, 7], (c) usually mature miRNAs for specific functions are conserved in plants [2].
The miRNAs are classified into families. The miRNA family classification is based on the Rfam database. The basic idea behind family classification is that each family represents sequences that have evolved from a common ancestor. The biogenesis and the function of many miRNAs in various systems including plants have been worked out [8–11]. In plants, miRNAs originate mostly from independent transcriptional units and are transcribed by RNA polymerase II into long primary transcripts (pri-miRNAs). Subsequently, the pri-miRNA is cut into miRNA precursors (pre-miRNAs) with stem-loop (hairpin) structures. The loop region of the hairpin is removed by ribonuclease III-like enzyme Dicer (DCL1) and the remainder (miRNA-miRNA duplex) is exported to the cytoplasm by Hasty (plant ortholog of exportin). Further plant miRNA is methylated at 3′ end by HEN1 factor. One strand of the duplex becomes mature miRNA and gets incorporated into the RNA-induced silencing complex (RISC) and guides RISC to complementary mRNA targets. Eventually, the RISC inhibits translation elongation or triggers the degradation of target mRNA [12].
miRNAs are implicated in diverse aspects of plant growth and development, including leaf morphology and polarity, lateral root formation, hormone signaling, transition from juvenile to adult vegetative phase and vegetative to flowering phase, flowering time, floral organ identity and reproduction [13, 14]. Several miRNAs are regulated in response to diverse stress conditions, suggesting important role in plants to cope with the stresses. Identification of miRNAs in large number of diverse plant species is important to understand the evolution of miRNAs and miRNA-targeted gene regulations. The low abundance of some miRNAs and their time- and tissue-specific expression patterns make experimental miRNA identification difficult. Now-a-days, publicly available databases play central role in the in silico biology [13, 15–21].
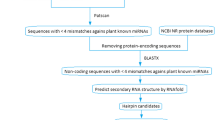
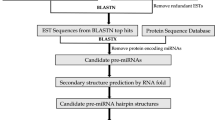
Horsegram is an important legume crop and source of proteins in vegetarian diet of many developing countries. It is known to be drought tolerant and possesses many neutraceutical properties [22]. The grain is used as human food and also as a concentrated feed for cattle. The US National Academy of Sciences has identified this legume as a potential food source for the future [23]. Till date no miRNA from this pulse crop has been reported. In this study, in silico approach has been used to identify potential miRNAs from the ESTs of horsegram. For this, we searched the EST databases to find ESTs matched with the previously known Arabidopsis miRNAs. Then we predicted the secondary structures of the identified ESTs in the first step using RNA MFOLD software. Finally, we identified new miRNAs. Further, the newly identified miRNAs have been used to find out targets that improve our understanding towards their possible regulatory roles in horsegram.
Materials and methods
EST database mining and processing
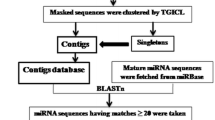
The ESTs of horsegram were retrieved from dbEST available at http://www.ncbi.nih.gov/dbEST/site. The redundancy of EST sequences was removed using the sequence assembly program CAP3 (http://pbil.univlyon1.fr/cap3.php). The overlapping sequences were clustered by CAP3 program as contigs and non-overlapping sequences as singleton.
Prediction of potential miRNA
The processed ESTs were used for the prediction of potential miRNAs with miRNAFinder (http://bioinfo3.noble.org/mirna/). miRNAFinder can accept three kinds of input sequences such as EST/cDNA, genomic sequence and small RNA. Therefore, miRNAFinder predicted potential intronic miRNA in intron regions of expressed genes (ESTs/cDNAs), find possible miRNA in genomic sequence or predict if the input small RNA is mature miRNA. The sequences are needed to be submitted in FASTA format. miRNAFinder execute back-end prediction pipeline and output a list of putative pri-miRNAs, their position information, and potential target genes. In this study, only ESTs data of horsegram was used. The processed ESTs of horsegram were submitted to miRNAFinder to produce output after comparative analysis with target ESTs library of Arabidopsis thaliana. False positive prediction of miRNAs was also removed using Oryza sativa ESTs library as a reference.
Prediction of targets for identified miRNAs
It has been documented that most of the known plant miRNAs bind to the protein coding region of mRNA targets with perfect or nearly perfect sequence complementarities [24, 25]. The targets were predicted with a plant miRNA potential target finder miRU2 available at http://bioinfo3.noble.org/miRNA/miRU.htm [26]. The Arabidopsis thaliana genome sequences were used as a base to predict the targets. Targets were predicted with potential complementarities in sequences against the submitted miRNAs with no gaps and <4 mismatches.
Prediction of secondary structures of miRNA precursor sequences
The secondary structures of miRNA precursor sequences were predicted with MFOLD software [6]. The parameters selected for predicting the secondary structures were as a fixed folding temperature of 37°C, 1 M NaCl ionic conditions with no divalent ions and rest of the parameters kept as default. For selecting the potential miRNAs or pre-miRNAs, various criteria have been considered as used in the previous studies [27–30]. Predicted mature miRNAs were allowed to have only 0–3 nucleotide mismatches in sequence with all previously known plant mature miRNAs. The pre-miRNAs sequence should be folded into an appropriate hairpin secondary structure. No loop or break in miRNA sequences was allowed. The MFEI was calculated using the following equation:
where MFE denotes the negative folding free energies (ΔG Kcal/mol).
Results and discussion
Identification of potential miRNAs from horsegram
The computational approaches based on the software which are used in this study have already been used for such miRNA analysis in various plant and animal systemts [13, 15–21]. In this study, a computational approach was used for searching the miRNAs from horsegram ESTs database following strict filtering criteria. From the available 989 ESTs, 72 contigs and 606 singletons were achieved as non redundant data using CAP3 program. In the first phase of CAP3 program, 5′ and 3′ poor regions of each read were identified and removed. Overlaps between reads were computed. False overlaps were identified and removed. In the second phase of CAP3 program, reads were joined to form contigs in decreasing order of overlap scores. Then, forward–reverse constraints were used to make corrections to contigs. In the third phase, a multiple sequence alignment of reads was constructed. During multiple sequence alignments a consensus sequence along with a quality value for each base was computed for each contig. A total of eight potential miRNAs were predicted from the processed data using miRNA-finder program and named as hor-miR1 to hor-miR8 (Table 1). The predicted miRNAs were either 20 or 21 nt in size. Majority of known miRNAs in other plants are of same size [2, 5, 27, 31]. The A + U content of predicted miRNAs ranged from 45 to 53%. The predicted miRNAs show higher negative minimum fold energies (MFEs). The MFEI is another useful criterion for distinguishing miRNAs from other types of coding and non-coding RNAs. The miRNA precursors with secondary structures had minimal free energy index (MFEIs) than other different types of RNAs. The newly identified miRNAs show MFEI in range of 0.45–0.75. The length of horsegram pre-miRNA varies from 97 to 110. These parameters are in agreement with the previously reported results for in silico predicted miRNAs [7, 28, 32].
Generally, miRNAs are distinguished from other RNAs on the basis of their surrounding sequences ability to adopt the hair-pin structure [5]. Therefore, secondary structures of all the identified miRNAs were predicted (Fig. 1). The identified miRNAs are found to vary in their locations in precursor sequences. The hor-miR1, hor-miR3, hor-miR5 and hor-miR7 are located at the 5′ end of their precursor sequences, whereas hor-miR2, hor-miR4, hor-miR6 and hor-miR8 are located at the 3′ end of their precursor sequences.
Prediction of targets for newly identified miRNAs and their putative role
The functional importance of miRNAs can de understood or described well by gaining insight into the miRNA targets. The predicted targets for the identified miRNAs are shown in Table 2. Targets were predicted for miRNAs sequences by using miRU2 software. Most of the predicted targets are involved in the regulation of plant growth and development and are functionally crucial for the plant physiology. It has been observed that one miRNA can target more than one regulatory gene [7, 28, 33]. In this study, hor-miR5, hor-miR6 and hor-miR7 are found to target 13, 22 and 6 sequences, respectively.
Earlier studies have documented that most of the miRNAs largely target transcription factors, signal transduction factors and metabolic transporters [28–30, 32]. In complementation with earlier studies, hor-miR1 and hor-miR6 are found to target zinc finger family protein. Such proteins are involved in numerous cellular processes including transcription, signal transduction, and recombination [34]. Most zinc finger proteins are E3 ubiquitin ligases [35] that mediate the transfer of ubiquitin to target proteins and play important roles in diverse aspects of cellular regulations in plants [36].
In an attempt to delve into the functional importance of the newly identified miRNAs, their targets were studied extensively. The hor-miR2 is found to target chromosome condensation proteins which play an important role in transcriptional gene silencing during cell cycle [37]. Furthermore, the hor-miR5 targets RNA recognition motif which are apparently known to control the post transcriptional gene expression. These RNA binding proteins either directly bind or indirectly control the expression by modulating other regulatory factors. The post-transcriptional regulatory events are pretty crucial in plant development [38]. The transcription factor B3 family protein targeted by the hor-miR7, has been very well characterized and found to have significant functional and evolutionary roles in plant development [39]. The hor-miR3 targets coatomer protein complex which are involving in trafficking of secretory proteins between the endoplasmic reticulum (ER) and the Golgi apparatus [40]. The hor-miR3 also targets WD-domain containing proteins which are essentially involved in plant growth and development [41, 42].
The hor-miR5 targets abscisic acid-responsive (ABA) family protein which is another example of transcriptional control under abiotic stress conditions in plants. The MADS-box proteins targeted by hor-miR5 are found to be a diverse class of transcription factors in the seed plants, playing an important role in establishment of certain reproductive structures [43]. Hence, similar to earlier known miRNAs, the newly identified miRNAs in horsegram are mostly targeting transcriptional factors. The hor-miR5 is also found to target S-locus protein kinase. S-locus is responsible for evolutionary transition among flowering plants i.e. the switch from outbreeding to an inbreeding mode of mating [44]. The functional role of hor-miR5 may unwind the intricacies of the above evolutionary transition and could provide a new understanding in the plant growth and development.
In addition, miRNAs have been documented to regulate cell signaling. The hor-miR6 targets mRNA coding for SecY translocase protein. The latter is involved in the insertion of signal transducing and recognizing proteins in the inner cytoplasmic membrane [45]. Interestingly, the disease resistance proteins are also targeted by the newly identified miRNA such as hor-miR6. The leucine-rich-repeat (LRR) domain containing disease resistance proteins have been particularly spotted by the hor-miR4 and hor-miR6. The LRR domain provides the platform for the recognition of pathogen [46] and more so, they are important determinants of specificity [47]. The hor-miR6 also shows complementarities to the sequences encoding fasciclin-like arabinogalactan proteins (FLAs). FLAs are a subclass of arabinogalactan proteins (AGPs) that contain putative cell adhesion domains known as fasciclin domain. These domains are critical for the cell-to-cell interactions and communication as well as for providing key structural, positional, and environmental signals during plant development [48].
Syntaxins are also reported to be targeted by miRNAs. Syntaxins are usually contributing to the plant resistance against bacteria [49]. In this study, hor-miR6 is found to be targeting syntaxin SYP132 transcript. Therefore, hor-miR6 could be an important miRNA to understand the regulation of plant defense system. Similarly, hor-miR7 is targeting some lipases which are involved in the hydrolysis of phospholipids, particularly phospholipases playing an important role in the plant responses to biotic stress [50]. We did not find any targets for hor-miR8. This may be due to incomplete coverage of mRNA in the horsegram database. Possibly, number of targets for miRNAs could not be identified because of their poor expression and stability or because of temporal and location specific expression.
Conclusions
This work presents the prediction of miRNAs and their targets from the available 989 ESTs of horsegram (Macrotyloma uniflorum (Lam.) Verdc.). None of the predicted hor-miRNAs showed identity with the previously reported miRNAs in plants. Therefore, these can be considered as novel and grouped in a new family. It is observed that most of the pooled targets predicted are very essential for the plant growth and development. They have been identified to play important role in variety of biological processes including plant defense, transcriptional regulation, stress defense, metabolic processes and structural development of plants.
References
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Nature 391:806–811
Bartel DP (2004) Cell 116:281–297
He L, Hannon GJ (2004) Nat Rev Genet 5:522–531
Brodersen P, Voinnet O (2006) Trend Genet 22:268–280
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) Genes Dev 16:1616–1626
Zuker M (2003) Nucleic Acid Res 31:3406–3415
Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Bioinformatics 20:2911–2917
Chapman EJ, Carrington JC (2007) Nat Rev Genet 8:884–896
Sanan-Mishra N, Mukherjee SK (2007) Open Plant Sci J 1:1–9
Jin H (2008) FEBS Lett 582:2679–2684
Zhu JK (2008) Proc Natl Acad Sci USA 105:9851–9852
Yin Z, Li C, Han X, Shen F (2008) Gene 414:60–66
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Trends Plant Sci 12:301–309
Mallory AC, Vaucheret H (2006) Nat Genet 38:S31–S36
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) PLoS One 2:e219
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Plant Cell 18:412–421
Jones-Rhoades MW, Bartel DP, Bartel B (2006) Annu Rev Plant Biol 57:19–53
Sunkar R, Kapoor A, Zhu JK (2006) Plant Cell 18:2051–2065
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) Curr Biol 15:2038–2043
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang WL (2005) Plant Cell 17:2186–2203
Jones-Rhoades MW, Bartel DP (2004) Mol Cell 14:787–799
Jeswani LM, Baldev B (1990) Advances in pulse production technology publication and information division. Indian Council of Agricultural Research, New Delhi
Yadava ND, Vyas NL (1994) Arid legumes. Agro publishers, India
Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Gen Biol 5:R65
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Science 297:2053–2056
Zhang Y (2005) Nucleic Acid Res 33:W701–W704
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003) RNA 9:277–279
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Plant J 46:243–259
Zhang B, Pan X, Cobb GP, Anderson TA (2006) Dev Biol 289:3–16
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Biochem Biophys Res Commun 354:585–590
Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) Curr Biol 13:807–818
Zhang B, Pan X, Anderson TA (2006) FEBS Lett 580:3753–3762
Zheng Y, Hsu W, Lee M-Li, Wong L (2006) VDMB 4316:131–145
Shi Y, Berg JM (1996) Biochemistry 35:3845–3848
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Plant Physiol 137:13–30
Ciechanover A (1998) EMBO J 17:7151–7160
Francis NJ, Kingston RE, Woodcock CL (2004) Science 306:1574–1577
Lorkovic ZJ, Barta A (2002) Nucleic Acids Res 30:623–635
Romanel EA, Schrago CG, Counago RM, Russo CA, Alves-Ferreira M (2009) PLoS One 4:e5791
Stefano G, Renna L, Chatre L, Hanton SL, Moreau P, Hawes C, Brandizzi F (2006) Plant J 46:95–110
Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman J, Sieburth LE (2003) Development 130:6577–6588
Zhong R, Ye ZH (2004) Plant Cell Physiol 45:1720–1728
Theissen G, Kim JT, Saedler H (1996) J Mol Evol 43:484–516
Boggs NA, Nasrallah JB, Nasrallah ME (2009) PLoS Genet 5:e1000426
Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J (2000) EMBO J 19:542–549
Kobe B, Deisenhofer J (2002) Nature 374:183–1866
Ellis J, Lawrence G, Ayliffe M, Anderson P, Collins N, Finnegan J, Frost D, Luck J, Pryor T (1997) Annu Rev Phytopathol 35:271–291
Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) Plant Physiol 133:1911–1925
Kalde M, Nuhse TS, Findlay K, Peck SC (2007) Proc Natl Acad Sci USA 104:11850–11855
Shah J (2005) Annu Rev Phytopathol 43:229–260
Acknowledgments
Authors are thankful to Dr. P. S. Ahuja, Director, IHBT for his valuable suggestions and guidance to conduct this work. We would like to thank the financial support from Council of Scientific and Industrial Research (CSIR) and Department of Science and Technology (DST), Govt of India. HM is thankful to CSIR for providing research fellowship in the form of JRF.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jyoti Bhardwaj and Hasan Mohammad contributed equally to this manuscript.
Rights and permissions
About this article
Cite this article
Bhardwaj, J., Mohammad, H. & Yadav, S.K. Computational identification of microRNAs and their targets from the expressed sequence tags of horsegram (Macrotyloma uniflorum (Lam.) Verdc.). J Struct Funct Genomics 11, 233–240 (2010). https://doi.org/10.1007/s10969-010-9098-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10969-010-9098-3