Abstract
The interaction of ginkgolic acid (15:1, GA) with human serum albumin (HSA) was investigated by FT–IR, CD and fluorescence spectroscopic methods as well as molecular modeling. FT–IR and CD spectroscopic showed that complexation with the drug alters the protein’s conformation by a major reduction of α-helix from 54 % (free HSA) to 46–31 % (drug–complex), inducing a partial protein destabilization. Fluorescence emission spectra demonstrated that the fluorescence quenching of HSA by GA was by a static quenching process with binding constants on the order of 105 L·mol−1. The thermodynamic parameters (ΔH = −28.26 kJ·mol−1, ΔS = 11.55 J·mol−1·K−1) indicate that hydrophobic forces play a leading role in the formation of the GA–HSA complex. The ratio of GA and HSA in the complex is 1:1 and the binding distance between them was calculated as 2.2 nm based on the Förster theory, which indicates that the energy transfer from the tryptophan residue in HSA to GA occurs with high probability. On the other hand, molecular docking studies reveal that GA binds to Site II of HSA (sub-domain IIIA), and it also shows that several amino acids participate in drug–protein complexation, which is stabilized by H-bonding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Ginkgolic acids (GA) are a series of structurally related nalkyl phenolic acid compounds that exist extensively in the leaves, nuts and external seed coat of Ginkgo biloba L. [1]. It was reported that GAs and their related alkylphenols (such as cardanols and cardols) from G. biloba L. are hazardous compounds with suspected cytotoxic, allergenic, mutagenic and carcinogenic properties. However, besides their toxicities, people generally pay more attention to their physiological and pharmacological activities, including antitumor and antidepressant activities, etc. [2–4]. For example, it was revealed that GAs can significantly inhibit the growth of many tumorigenic cell lines, such as HeLa, S180, U937 and so on [2]. These results make it possible to treat several kinds of diseases using GAs. However, although a large number of studies have demonstrated that GAs have various effects on tissue and cell functions, the mechanisms of action of GAs still remain largely unknown, with a deeper understanding of the way they are absorbed, metabolized, transported and delivered to specific tissues being required.
Study of the interaction between drugs and plasma proteins has been an interesting area of research in chemical biology and pharmacology [5]. Serum albumins are the most abundant proteins in the circulatory system of a wide variety of organisms and have many physiological functions [6]. They play an important role in the transport and deposition of endogenous and exogenous ligands present in the blood. Studies on the binding of drugs to albumins elucidate the properties of the drug–protein complex, as they provide useful information of the structural features that determine the therapeutic effectiveness of drugs [7]. Therefore, it has become an important research field in the life sciences, chemistry, and clinical medicine.
Human serum albumin (HSA) has been one of the most extensively studied of this group of proteins. In the body, drug distribution is mainly controlled by HSA, because most drugs circulate in plasma and reach the target tissues by binding to HSA [8]. One of the most important factors affecting the distribution and the free, active concentration of many drugs is their binding affinity for HSA. Therefore, drug binding to proteins such as HSA has becomes an important determinant of pharmacokinetics, e.g. prolonging their in vivo half-life, restricting the unbound concentration and affecting distribution and elimination of the drug [9]. In this regard, HSA is the most extensively studied protein due to its lack of toxicity and immunogenicity that make it an ideal candidate for drug delivery [10]. Additionally, HSA is known to accumulate in tumors, being taken up by tumor cells at increased levels compared to normal cells, and serves as conjugate carrier of various anticancer drugs, viz, chlorambucil, paclitaxel and doxorubicin [11]. Obviously, a study on the binding of GAs to HSA well be helpful for the application of the GAs in cancer therapy.
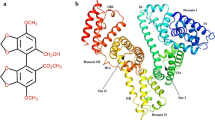
Crystal structure analysis revealed that HSA (Fig. 1b) has a 585 amino acid residue monomer and contains three structurally similar domains (I, II, and III), each containing two subdomains (A and B), is stabilized by 17 disulfide bridges, and has 67 % of α-helix. Based on crystallographic studies it is known that HSA contains two principal drug-binding sites for aromatic and heterocyclic ligands. They are located in subdomains IIA and IIIA and are named according to the pioneering work of Sudlow et al. as sites I and II [12, 13]. Seven binding sites for fatty acids are localized in subdomains IB, IIIA, IIIB and on the subdomain interfaces [12]. Hydrophobic, bulky and heterocyclic molecules with negative charge in the middle of the molecule bind to site I, while aromatic carboxylic acids with negative charge at one end of the molecule bind to site II [13]. HSA has only one tryptophan residue Trp-214 (Fig. 1b), which can be used as a fluorophore. Fluorescence quenching is considered a useful method for measuring binding affinities. Fluorescence quenching is the decrease of the quantum yield of fluorescence from a fluorophore induced by a variety of molecular interactions with a quencher molecule [14, 15]. Therefore, using the quenching of the intrinsic tryptophan fluorescence of HSA (Trp-214) as a probing tool allows us to study the interactions of doxorubicin and its derivative with serum proteins in an attempt to characterize the nature of drug–protein complexation.
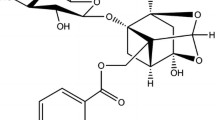
In this report, to determine the interaction between GAs and HSA, we investigated the cytotoxicity of one model compound, ginkgolic acid (15:1, GA) (Fig. 1a), using in vitro bioassay systems. We present spectroscopic analysis and docking studies of the interaction of GA with HSA in aqueous solution at physiological conditions, using constant protein concentration and various drug contents. The fluorescence quenching mechanism of HSA by GA was explored at different temperatures (298, 303, 308 and 313 K) and the binding constants (K A), binding site number (n), and corresponding thermodynamic parameters (ΔG, ΔH and ΔS) were calculated. The energy transfer and distance between the acceptor HSA and the donor GA were calculated by Förster’s theory. Moreover, conformational changes of HSA during the course of binding were ascertained by CD and FTIR measurements. These results can elucidate the properties of the GA–HSA complex, which will be helpful for understanding pharmacokinetics and the design of dosage forms of GA.
2 Materials and Methods
2.1 Materials
HAS fraction V powder (96–99 %, A1653) was purchased from Sigma Chemical Co. (St. Louise, MO). GA (15:1, 99.0 %) was a product of Shanghai Tauto Biotech Co., Ltd. Na2HPO4, NaH2PO4, NaCl and other reagents were of analytical grade. Solutions were prepared with Milli-Q water. The HSA solution (30 µmol·L−1) was prepared in PBS (50 mmol·L−1, pH = 7.4) and was kept at 4 °C until needed. The assays in the presented work were carried out at pH = 7.4.
2.2 Apparatus
Infrared spectra were recorded on a FT–IR spectrometer (Nicolet 6700), equipped with deuterated triglycine sulfate (DTGS) detector and KBr beam splitter, using AgBr windows. CD spectra of HSA and its GA complexes were recorded with an AVIV Model 400 circular dichroism spectrophotometer. For measurements in the far-UV region (190–260 nm), a quartz cell with a path length of 0.2 cm was used under a nitrogen atmosphere. All steady-state fluorescence spectra were recorded on a Hitachi F-4500 fluorescence spectrophotometer (Hitachi, Kyoto, Japan) equipped with a xenon lamp source and a water bath. The excitation and emission slits were 5 and 5 nm, respectively. The synchronous fluorescence spectra were obtained by setting the excitation and emission wavelength interval (Δλ) at 15 and 60 nm, respectively. The absorption spectra were obtained from a UV-2550 UV/vis spectrophotometer (Shimadzu, Japan). The pH measurement was made with a PHS-4C+ pH meter (SiChuan, China) with a combined glass–calomel electrode.
2.3 Procedure
For infrared spectra measurements, a solution of GA was added dropwise to the HSA solution with constant stirring to ensure the formation of homogeneous solution and to have GA concentrations of (15, 45 and 75) μmol·L−1 with a final protein concentration of 15 μmol·L−1 (1 mg·mL−1 in 50 mmol·L−1 PBS solution). Spectra were collected after 2 h incubation of the HSA with GA solution at room temperature, using hydrated films. Interferograms were accumulated over the spectral range 4,000–600 cm−1 with a nominal resolution of 4 cm−1 and 32 scans. The difference spectra [(protein solution + GA solution) − (protein solution)] were generated using the water combination mode around 2,300 cm−1 as standard [16]. When producing difference spectra this band was adjusted to the baseline level, in order to normalize the difference spectra. Fourier self-deconvolution and secondary derivatives were applied over the range of 1,600–1,700 cm−1 (amide I of HSA) to estimate the number, position and the width of component bands. Based on these parameters, a curve–fitting process was carried out using Origin 8.0 software of the OriginLab Corporation to get the optimal Gaussian-shaped curves to fit the original protein spectra [17]. After identification of the individual bands, the content of each representative structure of HSA was calculated using the area of their respective component bands.
The CD measurements of HSA in the absence and presence of GA (1:0, 1:1, 1:3, 1:5) were recorded in the wavelength range of 260–190 nm. Quartz cells having path lengths of 0.2 cm were used at a scanning speed of 50 nm·min−1. Sample temperature was maintained at 298 K and the solvent spectrum was subtracted. The contents of the secondary structures were estimated by CDNN software [18].
A quantitative analysis of the potential interaction between GA and HSA was performed by fluoremetric titration. A 300 μL solution containing 6.0 µmol·L−1 HSA was titrated by successive additions of GA solution. Titrations were done manually by using a microinjector. The fluorescence emission spectra were recorded at different temperatures (298, 303, 308 and 313 K) in the wavelength range of 300–500 nm with the excitation wavelength at 278 nm. The range of synchronous scanning was λ ex = 250, λ em(a) = 265, and λ em(b) = 310 nm, where the differences in the wavelengths (Δλ) are 15 and 60 nm. The fluorescence intensities were corrected to decrease the inner filter effect according to the following relationship [19]:
where F cor and F obs are corrected and observed fluorescence intensities, respectively, and A ex and A em are the absorption of the GA at the excitation and the emission wavelengths of HSA, respectively. The fluorescence intensities used in the next parts of this article are the thus corrected values.
The structure of free HSA (PDB id: 1AO6, chain A) obtained by X-ray crystallography was used as a template [20]. The 3D structure of GA was generated by Materials Studio 4.4. With the aid of AutoDock, the ligand root of GA was detected and rotatable bonds were defined. Essential hydrogen atoms and Kollman united atom type charges were added into the HSA protein model. AutoDock used the local search to search for the optimum binding site of small molecules to the protein. To recognize the binding sites in HSA, blind docking was carried out with the grid size set to 126, 126 and 126 Å along the X-, Y- and Z-axes with 0.375 Å grid spacing, respectively. The center of the grid was set to 13.9585, −1.2192 and −0.2388 Å. The conformation with the lowest binding free energy was used for further analysis using Accelrys-Discovery Studio.
The UV absorption spectra of GA, HSA and the GA–HSA system with the molar ratio of 1:1 were recorded in the range 200–700 nm at 298 K.
3 Results and Discussion
3.1 FT-IR Spectra of Drug Complex with HSA
The drug interactions with HSA were characterized by infrared spectroscopy and its derivative methods. The spectral shifting and intensity variations of protein amide I band at 1,652–1,650 cm−1 (mainly C=O stretch) and amide II band at 1,537–1,535 cm−1 (C–N stretching coupled with N–H bending modes) [21–23] were monitored upon interaction with the drug. The difference spectra [(protein solution + drug solution) − (protein solution)] were obtained, in order to monitor the intensity variations of these vibrations and the results are shown in Fig. 2. Similarly, the infrared spectra, self-deconvolution with second derivative resolution enhancement and curve-fitting procedures [24], were used to determine the protein’s secondary structures in the presence of the drug (Fig. 3; Table 1).
At low drug concentration (15 μmol·L−1), a decrease of intensity was observed for the protein amide I at 1,654–1,652 and amide II at 1,557–1,555 cm−1, in the difference spectra of the drug–HSA complexes (Fig. 2, diff. HSA–GA 1:1). The negative features located in the difference spectra for amide I and II bands at 1,653 and 1,556 cm−1 (GA–HSA) are due to the loss of intensity of amide I and amide II bands upon interaction with the drug. This reduction of the intensity for the amide I and amide II bands is due to drug binding to the protein C=O, C–N and N–H groups (hydrophilic contacts). Additional evidence to support the drug interactions with C–N and N–H groups comes from the shifting of the protein amide A band at 3,295 cm−1 (N–H stretching) in the free HSA to higher frequency at 3,315–3,320 cm−1 upon drug interaction (spectra not shown). As the drug concentration is increased to 75 μmol·L−1, strong negative features are observed for amide I band at 1,651 and 1,540 cm−1 (HSA–GA) upon drug complexation (Fig. 2, diff., HSA–GA 1:5). In addition, spectral shifting was observed for the amide I band at 1,655–1,652 and amide II at 1,541–1,539 cm−1 upon drug–protein complexation (Fig. 2, diff, HSA–GA 1:5). The observed spectral shifting amide I and amide II bands are due to drug binding to protein C–O and C–N groups, while the decrease in the intensity of the amide I band in the spectra of the drug–protein complexes suggests a major reduction of the protein α–helical structure at high drug concentrations [25].
A quantitative analysis of the protein’s secondary structure for the free HSA, HSA and their drug complexes in hydrated films has been carried out and the results are shown in Fig. 3 and Table 1. The free HSA contains α-helix 55 % (1,670–1,650 cm−1), β-sheet 19 % (1,633–1,613 cm−1), turn 8 % (1686 cm−1), β-antiparallel 1 % (1,696 cm−1) and random coil 17 % (1,641 and 1,606 cm−1) (Fig. 3a; Table 1), consistent with the spectroscopic studies of HSA [26, 27]. Upon drug interaction, a major decrease of α-helix from 55 % (free HSA) to 45–35 % (drug–complex) was observed along with an increase in the β–sheet structure (from 19 to 37 %) (Fig. 3b–d; Table 1).
3.2 CD Spectra
Far-UV CD spectroscopy is a very sensitive technique to investigate the secondary structural changes of HSA induced by the binding of small molecules [28–30]. Our previous research showed that the binding of GA can induce an increase of α-helix in BSA, and the decrease in α–helix is mainly balanced by an enhancement of the β-sheet [31]. Consequently, we carried out Far-UV CD measurements to explore whether GA can also induce changes in the secondary structure of HSA, and the results are shown in Fig. 4. The CD spectra (Fig. 4) display the characteristic peaks (intensive positive peak at around 192 nm and two negative double-humped peaks at 208 and 222 nm) of the high α-helical content in native HSA [28, 30]. The binding of GA to HSA induced decreases in the intensities of the CD signals at 192, 208 and 222 nm, especially at 192 nm, mainly indicating a decrease of the α-helical content in HSA [30]. Here, we used the CDNN software to estimate the secondary structures of HSA [18], and the results are shown in Table 2. The α-helical contents of free HSA calculated by CDNN is 55 %, which is approximately that found in previous research (54 %) [27]. Upon complexation with the drug, a major reduction of α-helix was observed from 55 % in free HSA to 46–32 % in the drug–HSA complex (Table 2). The major reduction of the α-helix with a major increase in the β-sheet structure (from 13 to 26 %) is consistent with the FT–IR results, indicating a partial protein destabilization (Tables 1, 2). These results indicate that similar structural changes are induced by the binding of GA to HSA as compared with the binding of GA to BSA [32]. Furthermore, the structural changes are greater for HSA–GA complexes in comparison to BSA–GA complexes.
3.3 Fluorescence Spectra and Stability of Drug Complexes with HSA
Human serum albumin contains a single polypeptide of 585 amino acids with only one tryptophan (Trp-214) located in subdomain II A. The intrinsic fluorescence of HSA is very sensitive to its microenvironment. Actually, the intrinsic fluorescence of HSA is mostly contributed by the Trp residue alone. When small molecules interact with HAS, its intrinsic fluorescence will obviously be weaken [33]. Figure 5a shows the fluorescence emission spectra of HSA with various amounts of GA at 298 K. Obviously, HSA had a strong fluorescence emission band at 340 nm when excited at 278 nm, which is mainly due to the fluorescence emission of tryptophan residues. Also, the fluorescence intensity of HSA decreases regularly with increasing concentrations of GA. These results are very similar to the binding of GA to BSA [32], suggesting that there are strong interactions between GA and HSA, which are responsible for quenching the fluorescence of HSA. We also found that the fluorescence intensity of GA at 418 nm was significantly enhanced once it was added to the HSA solution, which also indicates the strong interactions between GA and HSA.
Fluorescence emission spectra of GA–HSA systems in 50 mmol·L−1 PBS buffer pH = 7.4 at 298 K. a Fluorescence emission spectra for HSA with GA, (1) free HSA (3 μmol·L−1), (2–7) with GA = (1.5, 3, 4.5, 6, 9, and 12) μmol·L−1, (8) GA only (12 μmol·L−1); b Stern–Volmer plots of fluorescence quenching constant (k q) for the GA–HSA complexes at different drug concentrations; c plot of log10 [(F 0 − F)/F] as a function of the log10 (GA concentration) and the number of bound drug molecules per protein (n); d van’t Hoff plot
The quenching process can be induced by a collision process or by formation of a complex between quencher and fluorophore, the former being referred to as a dynamic quenching mechanism and the latter as a static quenching mechanism. The two possible mechanisms can be distinguished from each other by differences in the temperature dependent behavior of the system. For dynamic quenching, a higher temperature leads to higher energy and larger amounts of collisional quenching, thus the bimolecular quenching is expected to become greater with increasing temperature [23, 34]. In contrast, increasing the temperature is likely to result in decreased stability of the complex, and thus lower the slope for the static quenching. Our previous research indicates that the main fluorescence quenching mechanism of BSA by GA is a static quenching process by formation of a BSA–GA complex [32]. In order to verify the presence of static or dynamic quenching in drug–protein complexes, the fluorescence quenching of HSA at different temperatures (298, 303, 308 and 313 K) were determined using the Stern–Volmer equation, and the results are show in Fig. 5b; furthermore, the corresponding quenching constant K sv and the correlation coefficient R are listed in Table 3. The Stern–Volmer equation was applied as [31, 35–37]:
where F 0 and F are the fluorescence intensities in the absence and presence of quencher, [Q] is the quencher concentration, and K sv is the Stern–Volmer quenching constant (k q) which can be written as K sv = k q τ 0 where k q is the bimolecular quenching rate constant and τ 0 is the lifetime of the fluorophore in the absence of quencher (5.6 ns for HSA [15]). Therefore, Eq. 2 was applied to determine K sv by linear regression of a plot of F 0/F versus [Q].
As shown in the Stern–Volmer plots in Fig. 5b, like the binding of GA to BSA, the probable quenching mechanism of fluorescence of HAS by GA is a static quenching procedure, because the quenching constant K sv decreases with increasing temperature. Moreover, the k q values for the quenching of HSA caused by GA are much greater than 2.0 × 1010 L·mol−1·s−1, the maximum scatter collision quenching constant of various kinds of quenchers with a biopolymer [15]. These results prove that the main fluorescence quenching mechanism of HSA by GA should also be a static quenching process by forming a HSA–GA complex, as with the quenching mechanism of BSA by GA [32, 38, 39].
3.4 Determinations of Binding Constants and Number of Binding Sites
For a static quenching process, the results from fluorescence measurements can be used to estimate the binding constant of the GA–protein complex according to a modified form of the Stern–Volmer equation [40]:
where K A is the apparent binding constant and n is the number of binding sites per HSA molecule. The corresponding linear regression equations and results at four different temperatures are shown in Fig. 5c and Table 3. The K A values given here are averages of three replicate runs for drug–protein systems, each run involving several different drug concentrations. The binding constants (K A) obtained is 3.57 × 105 mol·L−1 at the temperature 298 K. The association constants calculated for the drug complexes suggest strong affinity for drug–protein binding, compared to the other ligand–protein adducts [41, 42]. Besides, Table 3 also shows that the decreasing trend of K A with increasing temperature is in accordance with the K sv dependence on temperature, implying that the HSA–GA complex partly decomposes at higher temperatures [43, 44].
In addition, the n values from the slope of the straight line are 0.92, 0.91, 0.93 and 0.93 at 298, 303, 308 and 313 K, respectively, for the drug–protein complexes, suggesting that n ≈ 1. These results indicate that there is one independent class of binding sites on HSA for GA [45, 46], which is remarkably consistent with the binding of GA to BSA [32].
3.5 Thermodynamic Analysis and Nature of the Binding Force
Essentially, there are four main types of non-covalent interactions existing in the binding of a ligand to proteins, hydrogen bonding, van der Waals forces and hydrophobic and electrostatic interactions [39]. The thermodynamic parameters enthalpy change (ΔH), entropy change (ΔS) and Gibbs energy change (ΔG) are the main evidence for confirming the binding modes. The previous study showed that the hydrophobic force plays a major role in the binding of GA to BSA [32]. In order to elucidate the binding forces between GA and HSA, the thermodynamic parameters of the system were calculated from the van’t Hoff equation [47]:
where R is the gas constant, T is the temperatures, and K A is the binding constants at the corresponding temperature. The values of ∆H, ΔS and ΔG were obtained from the linear relationship between ln K A (Fig. 5d) and the reciprocal absolute temperature, and the results are summarized in Table 4. The negative value of ΔG reveals that the interaction process is spontaneous, and the positive ΔS (11.55 J·mol−1·K−1) value is frequently treated as a typical proof for hydrophobic interaction. The negative enthalpy (ΔH = −28.26 kJ·mol−1) reveals the existence of hydrogen bonds in the association reaction and the formation of the GA–HSA complex is exothermic, which means that higher temperatures are disadvantageous for binding. As it is unreasonable to account for the thermodynamic parameters of the GA–HSA complex totally based on a single intermolecular force model, we think that, like GA binding to BSA [32], the hydrophobic force plays a leading role in the binding of GA to HSA but hydrogen bonding cannot be ruled out.
3.6 Energy Transfer from HSA to GA
The Förster resonance energy transfer in biochemistry is important because it can evaluate the distance between drugs and tryptophan residues responsible for the natural intrinsic fluorescence of a protein. The binding distance (r) between a protein residue (donor) and a bound drug molecule (acceptor) can be calculated by Förster’s non-radiative energy transfer theory [48]. According to Förster, the resonance energy transfer (FRET), the efficiency of energy transfer (E), is given by:
where E is the efficiency of energy transfer, F and F 0 are the fluorescence intensities of HSA in the presence and absence of quencher, r is the distance between acceptor and donor and R 0 is the critical distance when the transfer efficiency is 50 %. The value of R 0 was calculated using the following equation:
where k 2 is the spatial orientation factor between the emission dipole of the donor and the absorption dipole of the acceptor, N is the refractive index of the medium, Φ is the fluorescence quantum yield of the donor and J is the overlap integral of the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor, which was calculated by the following equation:
where F(λ) is the corrected fluorescence intensity of the donor at wavelength λ and Δ(λ) is the molar absorption coefficient of the acceptor at wavelength λ. Under the experimental condition, the values are K 2 = 2/3, N = 1.336, and Φ = 0.188 [49].
Fluorescence energy transfer occurs via overlapping of emission spectrum of a fluorophore (donor) with the absorption spectrum of another molecule (acceptor). The overlap of the absorption spectrum of GA with the fluorescence emission spectra of free HSA is shown in Fig. 6. The rate of energy transfer depends on: (i) the extent of overlapping between the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor, (ii) the relative orientation of the donor and acceptor dipoles, and (iii) the distance between the donor and the acceptor. In the GA–BSA complex, the distance between the donor (BSA) and the acceptor (GA) is 1.63 nm, indicating that the fluorescence quenching of BSA by GA is a non-radiation transfer process and the energy transfer from BSA to GA occurs with high probability [32]. Here the donor and acceptor are HSA and the GA, respectively. From Eqs. 6–8, the values of E, R 0, r and J were calculated and found to be 0.493, 2.4 nm, 2.2 nm and 5.7 × 10−15 mol−1·cm3, respectively. The values of r (2.2 nm) indicated that the donor and acceptor are close to each other and hence have strong binding interactions between them [50]. Further, the donor-to-acceptor distance r < 7 nm indicates that the non-radiative energy transfer from the tryptophan residue in HSA to GA occurs with high probability [47, 51, 52]. This is also in accordance with the conditions of FRET, indicating a static quenching interaction between GA and HSA.
3.7 The Results of Synchronous Fluorescence Spectra
It was ascertained that binding of GA to HSA causes the fluorescence quenching of HSA, but it is still a puzzle if this binding affects the conformation and/or the molecular environment in the vicinity of HSA. The synchronous fluorescence spectra present information about the molecular microenvironment in the vicinity of the tyrosine residues and the tryptophan residues in HSA when the Δ value between excitation and emission wavelength is stabilized at 15 or 60 nm, respectively [51]. The synchronous fluorescence spectra for HSA in the absence and presence of GA are shown in Fig. 7. With increasing concentration of GA, the fluorescence intensities decreased regularly when ∆λ = 15 nm and ∆λ = 60 nm, which further demonstrates the occurrence of fluorescence quenching during the binding process. A high concentration of drug makes the protein molecules extend, thus reducing the energy transfer between amino acids thereby reducing the fluorescence intensity. It is more important that the shift in the maximum emission wavelength (λ max) of the synchronous fluorescence spectra is related to the changes in the polarity around tryptophan residues [30]. As shown in Fig. 7, no significant shift in the maximum emission wavelength (λ max) occurs at the investigated concentrations range when ∆λ = 60 nm (Fig. 7a) or with ∆λ = 15 nm (Fig. 7b), indicating that the interaction of GA with HSA does not affect the conformation of the region around the Tyr or Trp residues of HSA. In other words, GA is relatively far from the Tyr or Trp residue of HSA. Since HSA has only one tryptophan residue (Trp-214) in subdomain IIA [53], this probably indicates that the GA molecule is not located within the hydrophobic cavity of active site I in subdomain IIA of the HSA.
3.8 Molecular Docking Study of the HSA–GA Interaction
In order to understand the efficacy of a drug as a therapeutic agent, it is necessary to explore the binding site of that drug in proteins. Descriptions of the 3D structure of crystalline albumin have revealed that that HSA contains three predominantly helical structural domains, each of which is made up of two subdomains having specific structural and functional characteristics. The principal region of drug binding sites of HSA are located in hydrophobic cavities in subdomains IIA and IIIA, which correspond to sites I and II, respectively, and tryptophan residue (Trp-214) of HSA in subdomain IIA [53]. Site I is known as the warfarin binding site, and prefers to bind large heterocyclic and negatively charged compounds, while site II is suitable for small aromatic carboxylic acids [12, 53].
The best energy ranked results of the binding mode between GA and HSA are shown in Fig. 8. It can be seen that the optimal bonding model is the insertion of one arm of GA into the hydrophobic cavity of active site II in subdomain IIIA of the protein HSA, which is in good accordance with the results obtained by the synchronous fluorescence spectra described above. The models show that GA is surrounded by Asp-108, Tyr-148, His-146, Ser-193, Lys-190, Lys-436, Lys-432, Tyr-452, Asn-429, Val-456, Glu-425, Gln-459, Leu-463 and Arg-197, suggesting the existence of hydrophobic interaction between them. Hence, this finding provides a good structural basis to explain the efficient fluorescence quenching of HSA emission in the presence of the GA. Furthermore, Fig. 8c shows that there are hydrogen interactions between the two oxygens of GA and the residues His-146 and Asp-108 of HSA, indicated that there are also a number of weak electrostatic interactions and hydrogen bond interactions between GA and HSA. These results suggest that the formation of hydrogen bonds decreases the hydrophilicity and increases the hydrophobicity to maintain stability in the GA–HSA system. The results obtained from molecular docking indicate that the interaction between GA and HSA is dominated by hydrophobic forces as well as hydrogen bonds, which is in good consistency with the binding mode study.
Best docked conformations of GA–HSA complexes: a HSA in cartoon ribbons with GA; b GA (stick representation) located within the hydrophobic pocket in binding site II of HSA; c the interaction mode between GA and HSA. The residues of HSA are represented by using lines and the ligand structure is represented by using the stick model
4 Conclusion
In the present work, the interaction between GA and HSA was investigated by employing different spectroscopic (FT–IR, CD, fluorescence, UV) and molecular docking techniques. The experimental results indicate that the binding of GA to HSA induces a conformational change of HSA. The experimental results also show that GA binds to HSA with moderate affinity and the intrinsic fluorescence of HSA is quenched through a static quenching mechanism. The binding parameters indicate that hydrophobic and hydrogen bonding interactions play a major role in the binding process by forming a GA–HSA complex. Molecular modeling demonstrated that GA binds to Site II of HSA within sub-domain IIIA. This work may provide valuable information to understand the interaction mechanism of the physiologically important protein HSA with GAs at a molecular level, which may be valuable in understanding the mechanistic pathway of drug delivery and pharmacological behavior of this drug.
References
Xia, H.J., Wang, X.D., Li, L., Wang, S.J., Guo, C.C., Liu, Y., Yu, L.S., Jiang, H.D., Zeng, S.: Development of high performance liquid chromatography/electrospray ionization mass spectrometry for assay of ginkgolic acid (15:1) in rat plasma and its application to pharmacokinetics study. J. Chromatogr. B 878, 2701–2706 (2010)
Zhou, C.C., Li, X.Y., Du, W., Feng, Y., Kong, X.L., Li, Y., Xiao, L.Y., Zhang, P.: Antitumor effects of ginkgolic acid in human cancer cell occur via cell cycle arrest and decrease the bcl-2/bax ratio to induce apoptosis. Chemotherapy 56, 393–402 (2010)
Smith, J.V., Luo, Y.: Studies on molecular mechanisms of Ginkgo biloba extract. Appl. Microbiol. Biotechnol. 64, 465–472 (2004)
Van Beek, T.A., Montoro, P.: Chemical analysis and quality control of Ginkgo biloba leaves, extracts, and phytopharmaceuticals. J. Chromatogr. A. 1216, 2002–2032 (2009)
Tian, M.Y., Zhang, X.F., Xie, L., Xiang, J.F., Tang, Y.L., Zhao, C.Q.: The effect of Cu2+ on the interaction between an antitumor drug-mitoxantrone and human serum albumin. J. Mol. Struct. 892, 20–24 (2008)
Carter, D.C., Ho, J.X.: Structure of serum albumin. Adv. Protein Chem. 45, 153–203 (1994)
Faridbod, F., Ganjali, M.R., Larijani, B., Riahi, S., Saboury, A.A., Hosseini, M., Norouzi, P., Pillip, C.: Interaction study of pioglitazone with albumin by fluorescence spectroscopy and molecular docking. Spectrochim. Acta A 78, 96–101 (2011)
Shahabadi, N., Maghsudi, M.: Binding studies of a new copper(II) complex containing mixed aliphatic and aromatic dinitrogen ligands with bovine serum albumin using different instrumental methods. J. Mol. Struct. 929, 193–199 (2009)
Peters, T.: All About Albumin. Academic Press, San Diego, CA (1996)
Ang, W.H., Daldini, E., Juillerat-Jeanneret, L., Dyson, P.J.: Strategy to tether organometallic ruthenium–arene anticancer compounds to recombinant human serum albumin. Inorg. Chem. 46, 9048–9050 (2007)
Chuang, V.T.G., Kragh-Hansen, U., Otagiri, M.: Pharmaceutical strategies utilizing recombinant human serum albumin. Pharm. Res. 19, 569–577 (2002)
He, X.M., Carter, D.C.: Atomic structure and chemistry of human serum albumin. Nature 358, 209–215 (1992)
Peters Jr, T.: Serum albumin. Adv. Protein. Chem. 37, 161–245 (1985)
Tayeh, N., Rungassamy, T., Albani, J.R.: Fluorescence spectral resolution of tryptophan residues in bovine and human serum albumins. J. Pharm. Biomed. Anal. 50, 107–116 (2009)
Lakowicz, J.R.: Principles of Fluorescence Spectroscopy, 3rd edn. Springer, New York (2006)
Dousseau, F., Therrien, M., Pezolet, M.: On the spectral subtraction of water from the FT–IR spectra of aqueous solutions of proteins. Appl. Spectrosc. 43, 538–542 (1989)
Wang, J.Z., Lin, T., Teng, T., Xie, S.S., Zhu, G.F., Du, L.F.: Spectroscopic studies on the irreversible heat-induced structural transition of Pin1. Spectrochim. Acta A 78, 142–147 (2011)
Wang, J.Z., Lin, T., Zhu, G.F., Du, L.F.: Stability of Pin1 as revealed by thermal and spectroscopic studies. J. Mol. Struct. 975, 310–316 (2010)
Xu, Y., Wang, J.Z., Li, J.S., Huang, X.H., Xing, Z.H., Du, L.F.: Heat treatment-induced functional and structural aspects of Mus musculus TAp63γ. J. Mol. Struct. 996, 42–47 (2011)
Sugio, S., Kashima, A., Mochizuki, S., Noda, M., Kobayashi, K.: Crystal structure of human serum albumin at 2.5 Å resolution. Protein. Eng. 12, 439–446 (1999)
Beauchemin, R., N’soukpoe-Kossi, C.N., Thomas, T.J., Thomas, T., Carpentier, R., Tajmir-Riahi, H.A.: Polyamine analogues bind human serum albumin. Biomacromolecules 8, 3177–3183 (2007)
Tang, J.H., Liang, G.B., Zheng, C.Z., Lian, N.: Investigation on the binding behavior of ellagic acid to human serum albumin in aqueous solution. J. Solution Chem. 42, 226–238 (2013)
Krimm, S., Bandekar, J.: Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 38, 181–364 (1986)
Byler, D.M., Susi, H.: Examination of the secondary structure of protein by deconvoluted FTIR spectra. Biopolymers 25, 469–487 (1986)
Ahmed-Ouameur, A., Diamantoglou, S., Sedaghat-Herati, M.R., Nafisi, S.H., Carpentier, R., Tajmir-Riahi, H.A.: An overview of drug binding to human serum albumin: Protein folding and unfolding. Cell Biochem. Biophys. 45, 203–213 (2006)
Froehlich, E., Mandeville, J.S., Jennings, C.J., Sedaghat-Herati, R., Tajmir-Riahi, H.A.: Dendrimers bind human serum albumin. J. Phys. Chem. B 113, 6986–6993 (2009)
Agudelo, D., Bourassa, P., Bruneau, J., Bérubé, G., Asselin, E., Tajmir-Riahi, H.A.: Probing the binding sites of antibiotic drugs doxorubicin and n-(trifluoroacetyl) doxorubicin with human and bovine serum albumins. PLOS One 8, 43814 (2012)
Ju, P., Fan, H., Liu, T., Cui, L., Ai, S.: Probing the interaction of flower-like CdSe nanostructure particles targeted to bovine serum albumin using spectroscopic techniques. J. Lumin. 131, 1724–1730 (2011)
Tian, J., Xie, Y., Zhao, Y., Li, C., Zhao, S.: Spectroscopy characterization of the interaction between brevifolin carboxylic acid and bovine serum albumin. Luminescence 26, 296–304 (2011)
Shi, X., Li, X., Gui, M., Zhou, H., Yang, R., Zhang, H., Jin, Y.: Studies on interaction between flavonoids and bovine serum albumin by spectral methods. J. Lumin. 130, 637–644 (2010)
Zhang, J., Sun, H.H., Zhang, Y.Z., Yang, L.Y., Dai, J., Liu, Y.: Interaction of human serum albumin with indomethacin: spectroscopic and molecular modeling studies. J. Solution Chem. 41, 422–435 (2012)
Du, W., Teng, T., Zhou, C.C., Xi, L., Wang, J.Z.: Spectroscopic studies on the interaction of bovine serum albumin with ginkgolic acid: Binding characteristics and structural analysis. J. Lumin. 132, 1207–1214 (2012)
Zhao, X., Sheng, F., Zheng, J., Liu, R.: Composition and stability of anthocyanins from purple Solanum tuberosum and their protective influence on Cr(VI) targeted to bovine serum albumin. J. Agric. Food Chem. 59, 7902–7909 (2011)
Katrahalli, U., Kalalbandi, V.K.A., Jaldappagari, S.: The effect of anti-tubercular drug, ethionamide on the secondary structure of serum albumins: a biophysical study. J. Pharm. Biomed. Anal. 59, 102–108 (2012)
Cheng, Z.-J.: Comparative studies on the interactions of honokiol and magnolol with human serum albumin. J. Pharm. Biomed. Anal. 66, 240–251 (2012)
Sudha, N., Enoch, I.M.V.: Interaction of curculigosides and their β-cyclodextrin complexes with bovine serum albumin: a fluorescence spectroscopic study. J. Solution Chem. 40, 1755–1768 (2011)
Dubeau, S., Bourassa, P., Thomas, T.J., Tajmir-Riahi, H.A.: Biogenic and synthetic polyamines bind bovine serum albumin. Biomacromolecules 11, 1507–1515 (2010)
Yang, Y., Yu, X.Y., Tong, W.H., Lu, S.Y., Liu, H.T., Yao, Q., Zhou, H.: Investigation of the interaction between novel spiro thiazolo[3,2-a] [1, 3, 5] triazines and bovine serum albumin by spectroscopic methods. J. Solution Chem. 42, 666–675 (2013)
Zhou, N., Liang, Y.Z., Wang, P.: Characterization of the interaction between furosemide and bovine serum albumin. J. Mol. Struct. 872, 190–196 (2008)
Kumar, R.S., van den Bergh, H., Wagnières, G.: Probing the interaction between a surfactant–cobalt(III) complex and bovine serum albumin. J. Solution Chem. 41, 294–306 (2012)
Yue, Y.Y., Liu, J.M., Fan, J., Yao, X.J.: Binding studies of phloridzin with human serum albumin and its effect on the conformation of protein. J. Pharm. Biomed. Anal. 56, 336–342 (2011)
Kratochwil, N.A., Huber, W., Muller, F., Kansy, M., Gerber, P.R.: Predicting plasma protein binding of drugs: a new approach. Biochem. Pharmacol. 64, 1355–1374 (2002)
Sheng, L.J., Ding, H., Wang, Z., Song, G.W.: Binding of amphiphilic p(DMAEMA79-b-AZOM5) diblock copolymer with bovine serum albumin—a spectroscopic investigation with warfarin and ibuprofen as site markers. J. Mol. Struct. 979, 152–159 (2010)
Li, S., Yao, D., Bian, H.D., Chen, Z.F., Yu, J., Yu, Q., Liang, H.: Interaction between plumbagin and human serum albumin by fluorescence spectroscopy. J. Solution Chem. 40, 709–718 (2011)
Yu, X.Y., Yao, Q., Li, W., Liao, X.C., Yang, Y., Liu, H.T., Li, X.F., Yi, P.G.: Investigation of the interaction between n-benzyl piperidones and bovine serum albumin by spectroscopic approaches. J. Solution Chem. 41, 1747–1758 (2012)
Zhang, W.J., Xiong, X.J., Wang, F., Ge, Y.S., Liu, Y.: Studies of the interaction between ronidazole and human serum albumin by spectroscopic and molecular docking methods. J. Solution Chem. 42, 1194–1206 (2013)
Bi, S., Sun, Y., Qiao, C., Zhang, H., Liu, C.: Binding of several anti-tumor drugs to bovine serum albumin: fluorescence study. J. Lumin. 129, 541–547 (2009)
Förster, T., Sinanoglu, O. (eds.): Modern Quantum Chemistry, vol. 3, pp. 93–138. Academic Press, New York (1996)
Cyril, L., Earl, J.K., Sperry, W.M.: Biochemists’ Handbook, pp. 84–88. E & FN Epon Press Ltd., London (1961)
Jiang, C.Q., Gao, M.X., Meng, X.Z.: Study of the interaction between daunorubicin and human serum albumin, and the determination of daunorubicin in blood serum samples. Spectrochim. Acta A 59, 1605–1610 (2003)
Wang, J., Guo, Y.W., Liu, B., Cheng, C.P., Wang, Z.Q., Han, G.X., Gao, J.Q., Zhang, X.D.: Spectroscopic analyses on interaction of bovine serum albumin (BSA) with toluidine blue (TB) and its sonodynamic damage under ultrasonic irradiation. J. Lumin. 131, 231–237 (2011)
Carter, D.C., He, X.M., Munson, S.H., Twigg, P.D., Gernert, K.M., Broom, M.B., Miller, T.Y.: Three-dimensional structure of human serum albumin Science 244, 1195–1198 (1989)
Dockal, M., Carter, D.C., Ruker, F.: Conformational transitions of the three recombinant domains of human serum albumin depending on pH. J. Biol. Chem. 275, 3042–3050 (2000)
Acknowledgments
The authors thank Zhi-Hua Xing from the Institute for Nanobiomedical Technology and Membrane Biology (Sichuan University, Chengdu, P.R. China) for her kind help with the CD measurements. This work was financially supported by the National Key Technology R&D program of China (no. 2006BAF07B01) and 985 program from Sichuan University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, Gf., Wang, Y., Liu, J. et al. Interaction Between Ginkgolic Acid and Human Serum Albumin by Spectroscopy and Molecular Modeling Methods. J Solution Chem 43, 1232–1249 (2014). https://doi.org/10.1007/s10953-014-0200-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10953-014-0200-5