Abstract
In Oryza sativa (rice) there are seven members in the NRAMP (natural resistance- associated macrophage protein) family of transporter proteins. They have been identified as OsNRAMP1, OsNRAMP2, OsNRAMP3, OsNRAMP4, OsNRAMP5, OsNRAMP6 and OsNRAMP7. Several metal ions like Zn2+, Mn2+, Fe2+, Cd2+ etc. have been studied to be transported via NRAMP transporter proteins in rice plant. In spite of this, very little information is available regarding these transporters. Hence it is important to computationally predict and characterize the OsNRAMP family of transporters for studying and understanding their molecular insights in future studies. For this purpose, various in silico methods and tools were used for the characterization of OsNRAMP family of transporter proteins. Physico-chemical properties of the protein sequences were calculated, putative transmembrane domains (TMDs) and conserved motif signatures were determined and their interaction partners were predicted. 3D models of all the members of OsNRAMP transporters were generated using online structure prediction tool followed by their analysis. In silico microarray analysis was done to understand the expression pattern of these transporters in rice plant. Currently, only limited knowledge is available about the structural and functional aspects of these transporters, hence this study would provide more theoretical information about them.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Transporters play a very important role in metal uptake and sequestration in plants. A lot of study has been done on plant transporters using heterologous mode of expression. Transporter proteins on the cell membranes work as carriers and channels for the transport of metals and other ions. One such family of transporters is the NRAMP (natural resistance-associated macrophage protein) family of transporters. NRAMP protein family functions as a divalent metal transporter in a wide range of living beings i.e. from bacteria to humans [1]. They share a remarkable protein sequence identity between yeast, plant, fly and mammals [2]. In plants, NRAMPs also transport a range of divalent metal cations including iron, manganese, cadmium, and zinc [3].
Rice is the majorly consumed food crop in many South East Asian countries. Unfortunately, rice is poor in many essential micronutrients and vitamins, and deficiencies in these micronutrients are common in developing countries [4]. In Oryza sativa (rice) 7 such NRAMP transporters have been identified viz., OsNRAMP1, OsNRAMP2, OsNRAMP3, OsNRAMP4, OsNRAMP5, OsNRAMP6 and OsNRAMP7.
Of the seven NRAMPs in rice, only five have been functionally characterized. The exact role of OsNRAMP1 is unknown but it is suggested to be involved in Cd uptake. OsNRAMP4 also known as Nrat1 (Nramp aluminum transporter1) is the first transporter in this family to be identified as the trivalent Al ion transporter [5]. In contrast to other rice NRAMP members, OsNRAMP4 does not show transport activity for divalent metal ions, like Zn, Mn, and Fe. It shares relatively low similarity with the other OsNRAMP members [5, 6]. In response to environmental changes, OsNRAMP3 differentially transports Mn [7]. It is reported that OsNRAMP5 plays a role in the uptake of Mn, Fe and also Cd from the soil in rice [8]. Recently OsNRAMP6 has been identified to be involved in uptake of Fe and Mn [9].
It has been previously reported that NRAMPs are highly hydrophobic membrane proteins with 10–12 putative TMDs with cytosolic N and C terminals. The consensus of transport residues lies on the intracellular loop between TMD-8 and 9 [2, 10]. In spite of its important role in rice plant, very little information is available about them and there is no available crystal structure for these transporters. Therefore, this study is focused on the study of characteristics of OsNRAMP family of transporters, prediction of their 3D structure and study of their expression pattern using bioinformatics tools.
2 Materials and Methods
2.1 Retrieval of NRAMP Transporter Protein Sequences
The protein sequences of OsNRAMP family transporters were obtained from Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/) [11, 12]. The protein sequence IDs are OsNRAMP1 (Os07g0258400), OsNRAMP2 (Os03g0208500), OsNRAMP3 (Os06g0676000), OsNRAMP4 (Os02g0131800), OsNRAMP5 (Os07g0257200), OsNRAMP6 (Os01g0503400), OsNRAMP7 (Os12g0581600).
2.2 Analysis of NRAMP Family of Transporter Proteins
Physico-chemical features of OsNRAMP family transporter sequences were analysed by Protparam tool (http://web.expasy.org/protparam/) [13]. Protein domain families were searched in Pfam database (http://pfam.xfam.org/) [14]. Transmembrane domains (TMDs) were predicted by TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) [15]. Sub-cellular localizations were predicted by CELLO server (http://cello.life.nctu.edu.tw/) [16]. Motif analysis was performed using MEME tool (http://meme-suite.org/tools/meme) [17] with the following parameters; maximum number of motifs to find, 5; minimum width of motif, 6 and maximum width of motif, 50. Each motif was scanned with the protein sequence database (Swissprot) using FIMO (Find Individual Motif Occurrences) (http://meme-suite.org/tools/fimo) [18]. Other physicochemical features like net charge of the protein at pH7 was predicted using Innovagen’s Peptide Calculator (http://pepcalc.com/), its estimated solubility and hydropathy plot was predicted using Expasy ProtScale (https://web.expasy.org/protscale/) [13] with a window size of 19 amino acids. Using Clustal Omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/) [19] percent identity matrix was created. To infer the evolutionary relationship between the OsNRAMP transporters in rice, a phylogenetic tree was constructed using Clustal Omega Phylogenetic tree using default parameters. Also the alignment of rice NRAMP transporter protein sequences was carried out using Clustal omega with default parameters. Interactome network was generated using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (http://string-db.org) [20, 21]. It is a pre-computed database for the exploration and analysis of protein–protein associations.
2.3 3D Structure Prediction and Validation
The 3D structure of OsNRAMP transporters is unavailable in the RCSB protein data bank. Hence the models for all the 7 members of OsNRAMP transporter were predicted via I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) server [22] and Robetta (http://robetta.bakerlab.org/) [23]. I-TASSER (Iterative Threading ASSEmbly Refinement) is a hierarchical protein structure modelling method [24, 25]. The program retrieves template proteins of similar folds from the PDB library. If no similar structures are detected, I-TASSER models the whole structure ab initio. I-TASSER does not consider disulfide bond during simulation. In spite of that the predicted models are considered correct as it is judged by global topology, not local bonding. The structure is not only held together by disulfide bond. It also has hydrophobic interaction, salt bridge, van der Waals, and hydrogen bond. Force pairing can be done by giving additional restraints to I-TASSER by specifying contacts. Through the webserver, one can unfold “Option I: Assign additional restraints & templates to guide I-TASSER modelling.” and choose “Assign contact/distance restraints”.
For 3D structure generation, protein sequences of OsNRAMP transporters were used as input. Starting from an amino acid sequence, three-dimensional (3D) atomic models were generated from multiple threading alignments and iterative structural assembly simulations using I-TASSER. The models are ranked based on their confidence score (c-score), TM score, root mean square deviation (RMSD) and standard deviation. The accuracy of the predicted models by I-TASSER was provided based on the confidence score (C-score) of the modelling. The predicted structures were visualized using PyMOL [26].
The stereo-chemical quality of the final models was evaluated using Ramachandran plot obtained from Rampage (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) [27]. These are used to determine the statistical significance of a protein 3D model considering the spatial position of amino-acids and overall stability of the structure. The amino acid environment was assessed by using Errat [28] from the UCLA-DOE LAB- SAVES server (https://services.mbi.ucla.edu/SAVES/). Also ProQ-Protein Quality Predictor (http://www.sbc.su.se/~bjornw/ProQ/ProQ.cgi) [29] was used to determine the statistical significance of the 3D models considering the spatial position of amino-acids and overall stability of the structure. Structural comparison of the predicted models with the available structures in Protein Data Bank was done using Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server/start) [30]. I-TASSER models were compared to Robetta models using Dali pairwise comparison (http://ekhidna.biocenter.helsinki.fi/dali_lite/start) [31]. The Dali server reports significance of each match with an estimated Z-score which is the raw comparison score, normalized by the combined length of the proteins.
2.4 Gene Expression Data Analysis
In silico expression profile of OsNRAMP genes was analysed in tissue and organs in the entire developmental cycle specific libraries using RiceXPro (http://ricexpro.dna.affrc.go.jp/) [32, 33] by retrieving the expression values from the array database. To gain insight into expression profiles of OsNRAMP members in rice, transcript expressions were searched against RiceXpro databases using the Rice Annotation Project Database ID’s. Later, a heat map was generated with the software CIMMiner (http://discover.nci.nih.gov/cimminer) [34, 35].
3 Results
3.1 Analysis of NRAMP Family of Transporter Proteins
For the molecular insight of OsNRAMP transporter proteins, their physicochemical characteristics were studied. All identified OsNRAMP’s contained the NRAMP (PF01566) family domain. They encode a protein of 518–550 amino acid residues long with their molecular weight ranging from 55.814 to 59.708 kDa and mainly demonstrated basic characteristics with 5.19–8.48 pI value. These NRAMP transporter proteins were predicted to be localised in the plasma membrane (Table 1). They contained 10–12 putative TMDs with cytoplasmic N- and C-terminal regions.
Hydropathy for all the seven protein sequences of OsNRAMP transporter was predicted using ExPASy ProtScale. It plots the hydropathy of each amino acid in the protein sequence as a graph using the Kyte–Doolittle algorithm, with a window size of 19 amino acids (Online Resource Fig. 1). All the protein sequences were predicted to have poor water solubility. For all the protein sequences their net charge at pH 7 was predicted using Innovagen’s Peptide Calculator to be as follows, OsNRAMP1 (1.3); OsNRAMP2 (-2); OsNRAMP3 (3.3); OsNRAMP4 (0.1); OsNRAMP5 (-0.4); OsNRAMP6 (5.5) and OsNRAMP7 (-9.7).
Multiple sequence alignment was done for all the 7 OsNRAMP protein sequences using Clustal Omega which uses seeded guide trees and HMM-profile techniques to generate alignment between multiple sequences as shown in Fig. 1. It is used to measure the similarity between sequences, examine patterns of conservation and variability and derive evolutionary relationship. Percent identity matrix of the OsNRAMP protein sequences (Table 2) indicate that OsNRAMP1 sequence is more identical to OsNRAMP5 sequence with 73.84% identity, OsNRAMP2 sequence is more identical to OsNRAMP7 sequence with 68.28% identity, OsNRAMP3 sequence is more identical to OsNRAMP5 sequence with 58.24% identity, OsNRAMP4 sequence is also more identical to OsNRAMP5 sequence with 59.13% identity and OsNRAMP6 sequence is more identical to OsNRAMP5 sequence with 53.85% identity.
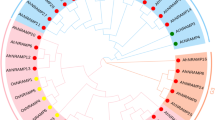
The phylogenetic tree shows the evolutionary relationship of OsNRAMP transporters using Neighbour-joining tree without distance corrections. In Fig. 2, we observe that we have three sister groups, one group being OsNRAMP1 and 5, another is OsNRAMP2 and 7 each having a common ancestor and the third one is a lone taxon OsNRAMP6. OsNRAMP3 shares a common ancestry with OsNRAMP2 and 7, and similarly OsNRAMP4 shares its ancestry with OsNRAMP3, 2 and 7.
3.2 Conserved Motif Analysis
The five most conserved motifs in identified NRAMP transporters were searched using MEME tool (Table 3). Motifs 1, 2 & 3 are 50 amino acid residues long, while motif 4 is 29 and motif 5 is 41 amino acids long. All the motifs are present in all the members of NRAMP transporter except motif 3 which is absent or is least conserved in OsNRAMP6 protein sequence. All motifs are related with NRAMP (PF01566) protein family. The presence of these long conserved residues in all sequences indicates the highly conserved structure among the rice NRAMP family of transporter proteins. The predicted motifs when scanned in the Swissprot (protein sequence database) using FIMO most of the top ten hits were NRAMP transporters in rice and Arabidopsis. The list of top ten hits for each motif sorted by increasing p-value is given in online resource Table 1 along with their q-value. The p-value of a motif occurrence is the probability of a random sequence of the same length as the motif matching that position of the sequence. The q-value of a motif occurrence is the rate of false discovery if the occurrence is accepted as significant. The top 10 hits for motif 2 and 3 also included NRAMP like transporters smf-2, smf-1and smf-3 from Caenorhabditis elegans. Motif 4 included divalent metal cation transporter from Pseudomonas aeruginosa, Xanthomonas axonopodis pv. Citri and Clostridium acetobutylicum while motif five included Rat NRAMP apart from NRAMP transporters from rice and Arabidopsis.
3.3 Interaction Partners
Interaction partners of all the 7 members of OsNRAMP were predicted using STRING. Potential interaction partners with high confidence score (≥ 0.700) for OsNRAMP1, OsNRAMP2, OsNRAMP3, OsNRAMP4, OsNRAMP5, OsNRAMP6 and OsNRAMP7 were predicted using STRING. OsNRAMP1 was predicted to have interaction with putatively expressed tubulin/FtsZ domain containing protein (Uniprot entry Q75GI3), expressed protein (Uniprot entry Q2QTV2) and Cadmium/Zinc transporting ATPase (Uniprot entry Q8H384) with confidence scores 0.713, 0.705 and 0.704 respectively. OsNRAMP2 was predicted to have interaction with putatively expressed Metal cation transporter (Zinc transporter) that may mediate zinc uptake from the rhizosphere and may be responsible for the translocation of zinc within the plant (Uniprot entry A3BI11); putatively expressed Pleiotropic drug resistance protein, it may be a general defense protein (Uniprot entry Q8GU92); putatively expressed Metal cation transporter (Zinc transporter) that may be involved in zinc uptake from the rhizosphere (Uniprot entry Q6L8F9); and putatively expressed Cyclin dependent kinase G-1 (Uniprot entry Q6K5F8) with confidence scores 0.786, 0.779, 0.737 and 0.716 respectively.
OsNRAMP3 was predicted to have only one interaction partner with a confidence score of 0.769, it is a putatively expressed unclassified protein which may be involved in the transport of nicotianamine chelated metals (Uniprot entry Q9FTU1). OsNRAMP4 was predicted to have interaction with expressed ZOS12-02-C2H2 Zinc finger protein which is a transcriptional activator that regulates the expression of genes involved in Al tolerance (Uniprot entry Q2QX40); putatively expressed ABC transporter, ATP binding protein which is required for detoxification of Al in roots and can specifically transport UDP- glucose (Uniprot entry QOD9V6); putatively expressed ABC transporter, membrane spanning/permease subunit (Uniprot entry Q5W7C1); and a hypothetical protein (Uniprot entry Q5VRD) with confidence scores 0.865, 0.864, 0.856 and 0.717 respectively. OsNRAMP5 was predicted to have interaction with putative Cd/Zn transporting ATPase (Uniprot entry Q8H384) and putatively expressed Cyclin dependent Kinase G1 (Uniprot entry Q6K5F8) with confidence scores 0.809 and 0.718 respectively. Interaction partners predicted for OsNRAMP6 and OsNRAMP7 had confidence scores < 0.700 hence they were not taken into consideration. Interactome analysis indicates that OsNramp transporters could be related to Cd/Zn transporters and also be involved in Al tolerance.
3.4 3D Structure Prediction and Validation of NRAMP Family of Transporters
There exists no clear knowledge about the exact structure of the OsNRAMP protein, as there is no crystal structure data in the protein database. Hence, the protein sequences of all the available rice NRAMP family of transporters were retrieved and subjected to protein–protein BLAST (blastp) to find suitable templates for building their 3D structure. All the hits obtained were of either predicted or hypothetical protein sequences, which cannot be used for homology modelling. And when blasted against PDB Protein database their sequence identity was < 40%. Due to lack of suitable templates, the sequences were modelled via I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) server and Robetta for comparison using Dali pairwise comparison. I-TASSER provides 5 different models for each of the protein sequences (OsNRAMP1, OsNRAMP2, OsNRAMP3, OsNRAMP4, OsNRAMP5, OsNRAMP6 and OsNRAMP7). Confidence score is used as a scale for estimating the accuracy of predicted models. It ranges from − 5 to 2, where a model with high c score signifies high confidence and vice versa. The models with the high confidence score and other parameters discussed below were selected for the OsNRAMP transporters 3D structures. They are OsNRAMP1 (model 3C-score = − 1.5), OsNRAMP2 (model 1C-score = 0.34), OsNRAMP3 (model 2C-score = − 1.17), OsNRAMP4 (model 5C-score = − 2.31), OsNRAMP5 (model 4C-score = − 2.50), OsNRAMP6 (model 2C-score = − 1.98) and OsNRAMP7 (model 5C-score = − 2.48) (Fig. 3). The stereo chemical property of the modelled NRAMP family of transporters was evaluated by Ramachandran plot (RAMPAGE) and it shows the percentage of amino acids in the favoured region, in the allowed region and in the outlier region. These values for the respective selected transporters are as follows OsNRAMP1 (85.1% in favoured region, 11% in allowed region, 3.9% in outlier region), OsNRAMP2 (84.5% in favoured region, 10.7% in allowed region, 4.8% in outlier region), OsNRAMP3 (81.2% in favoured region, 12.8% in allowed region, 6% in outlier region), OsNRAMP4 (80.7% in favoured region, 11.6% in allowed region, 7.7% in outlier region), OsNRAMP5 (83.6% in favoured region, 12.9% in allowed region, 3.5% in outlier region), OsNRAMP6 (80.3% in favoured region, 13.3% in allowed region, 6.4% in outlier region), OsNRAMP7 (82.9% in favoured region, 10.9% in allowed region, 6.1% in outlier region) (Online Resource Fig. 2). Similarly Robetta provides 5 models for each OsNRAMP protein sequences; one among them was selected after evaluating the percentage of amino acids in the favoured region, in the allowed region and in the outlier region using RAMPAGE. They are OsNRAMP1 (model 3), OsNRAMP2 (model 4), OsNRAMP3 (model 5), OsNRAMP4 (model 4), OsNRAMP5 (model 2), OsNRAMP6 (model 5) and OsNRAMP7 (model 4).
3D structure for OsNRAMP transporters a OsNRAMP1 (Os07g0258400), b OsNRAMP2 (Os03g0208500), c OsNRAMP3 (Os06g0676000), d OsNRAMP4 (Os02g0131800), e OsNRAMP5 (Os07g0257200), f OsNRAMP6 (Os01g0503400), g OsNRAMP7 (Os12g0581600) with their motifs highlighted in red (motif 1), cyan (motif 2), green (motif 3), blue (motif 4), orange (motif 5) predicted using I-TASSER
Errat plot estimates the arrangement of different types of atoms with respect to each other in protein models (Online Resource Fig. 3). Errat is a sensitive technique, hence is good for identifying incorrectly-folded regions in preliminary protein models. ERRAT is ‘‘overall quality factor’’ for non-bonded atomic interactions, and higher scores mean higher quality. The normally accepted range is > 50 for a high quality model [27]. The ERRAT overall quality factor for the models predicted using I-TASSER are OsNRAMP1 (90.588), OsNRAMP2 (92.621), OsNRAMP3 (91.513), OsNRAMP4 (87.896), OsNRAMP5 (87.736), OsNRAMP6 (83.026), OsNRAMP7 (80.488).
ProQ is a neural network based tool that based on a number of structural features predicts the quality of a protein model. ProQ is optimized to find correct models in contrast to other methods which are optimized to find native structures. Different ranges of quality can be predicted based on ProQ LG score and MaxSub. LG score > 1.5 indicates fairly good quality of the model, LG score > 2.5 indicates very good model and LG score > 4 indicates extremely good model. Similarly ProQ MaxSub > 0.1 indicates fairly good model, MaxSub > 0.5 very good model and MaxSub > 0.8 extremely good model [36]. The results from the ProQ server demonstrates a ProQ LG score and a ProQ MaxSub score of the I-TASSER models as OsNRAMP1 (5.705; 0.128), OsNRAMP2 (5.214; 0.137), OsNRAMP3 (5.705; 0.147), OsNRAMP4 (5.462; 0.131), OsNRAMP5 (5.229; 0.121), OsNRAMP6, (6.147; 0.234) OsNRAMP7 (5.807; 0.145). The overall results obtained from the above analyses reveal that these plausible 3D models of the rice NRAMP family of transporters, as shown in Fig. 3 are robust and satisfactory for further analysis.
The models predicted using I-TASSER and Robetta were structurally compared with each other using Dali pairwise comparison and with Protein databank using Dali server. Structural conservation was computed within the selected I-Tasser and Robetta models of OsNRAMP transporters by superposing the models using Dali pairwise comparison. Upon Dali pairwise comparison it was found that Robetta and I-TASSER models for OsNRAMP showed the following Z score, rmsd and number of aligned position (lali); OsNRAMP1 (Z score = 48, rmsd = 1.5, lali = 459), OsNRAMP2 (Z score = 49.3, rmsd = 1.5, lali = 462), OsNRAMP3 (Z score = 42.8, rmsd = 2.5, lali = 479), OsNRAMP4 (Z score = 46.2, rmsd = 1.7, lali = 469), OsNRAMP5 (Z score = 53.5, rmsd = 1.5, lali = 466), OsNRAMP6 (Z score = 44.7, rmsd = 1.6, lali = 470), OsNRAMP7 (Z score = 45.2, rmsd = 2.0, lali = 437) (Online Resource Fig. 4). From Dali result it was identified that all the OsNRAMP transporters models predicted using I-TASSER share structural similarity with crystal structure of Eremococcus coleocola manganese transporter (EcoDMT) with PDB entry 5 m87 with the following rmsd values OsNRAMP 1 (0.9), OsNRAMP 2 (0.9), OsNRAMP 3 (1.0), OsNRAMP 4 (0.7), OsNRAMP 5 (0.5), OsNRAMP 6 (0.7) and OsNRAMP 7 (1.7). To show their structural similarity they were superposed as shown in Online Resource Fig. 5. List of first 10 proteins for each of the 7 OsNRAMP I-TASSER models with high structural similarity has been included in Online Resource Fig. 6 (with Z scores over 2). OsNRAMP models predicted using Robetta for OsNRAMP2, 4, 5 and 6 also showed top most similarity to EcoDMT with PDB entry 5M87 with rmsd values 1.3, 1.5, 1.1 and 1.3; except for models of OsNRAMP1, 3 and 7. Robetta models for OsNRAMP1and 3 showed top most similarity to crystal structure of E. coleocola manganese transporter mutant E129Q with PDB entry 5M8K with rmsd values 1.3 and 1.2. While OsNRAMP7 showed top most similarity to crystal structure of E. coleocola manganese transporter mutant H236A with PDB entry 5M8J with rmsd value 1.4. To show their structural similarity they were superposed as shown in Online Resource Fig. 7. List of first 10 proteins for each of the 7 OsNRAMP Robetta models with high structural similarity has been included in Online Resource Fig. 8.
3.5 Gene Expression Analysis
Tissue/Organ-specific expression levels of the 7 OsNRAMP transporters under normal growth conditions in the field were obtained from the RiceXpro database (http://ricexpro.dna.affrc.go.jp/). The data was obtained as OsNRAMP expression level in different tissues of rice plant which was retrieved as a heat map using CIMMiner (Fig. 4). In silico gene expression analysis revealed that all the OsNRAMP genes are expressed in various tissues/organs throughout the plant life. OsNRAMP3 has a uniform expression pattern in almost all the tissues and organs of rice. OsNRAMP6 showed no significant transcript expression in various tissues of the plant throughout its entire growth in the field. Expression of OsNRAMP2, 3 and 7 seems to be higher in comparison to the remaining OsNRAMP transporters. Expression studies in normal plant tissue have indicated that the expression of OsNRAMP1 in roots is more than in leaves and OsNRAMP2, 3 is more in leaves than roots [37], a similar observation has been made in this in silico gene expression analysis. Expression of OsNRAMP genes in the root vegetative state probably indicates their involvement in metal nutrient uptake from the soil. Further study will be required to identify the roles of all the members of OsNRAMP transporter during the vegetative state in the root.
Heat map showing differential expression profile of rice OsNRAMP genes OsNRAMP1 (Os07g0258400), OsNRAMP2 (Os03g0208500), OsNRAMP3 (Os06g0676000), OsNRAMP4 (Os02g0131800), OsNRAMP5 (Os07g0257200), OsNRAMP6 (Os01g0503400), OsNRAMP7 (Os12g0581600) in various tissues. Red indicates higher levels and green indicates lower levels of transcript accumulation. Each column represents the average of three biological replicates. Root veg root vegetative, Root rep root reproductive, LS veg leaf sheet vegetative, LS rep leaf sheet reproductive, Stem rep stem reproductive, LB rep leaf blade reproductive, Stem ripe stem ripening, LB veg leaf blade vegetative, LB ripe leaf blade ripening
4 Discussion
We report bioinformatics analysis of all 7 OsNRAMP transporters identified in rice plant. This study has given us a deeper insight of their physiochemical and molecular characteristics. Their localisation on plasma membrane indicates that they might function in transmembrane transport of metal ions. Also their poor water solubility predicted from the hydropathy plots confirms that they are membrane localised proteins. GQSSTITGTYAGQY(/F)V(/I)MQGFLD(/E/N) is the consensus transport motif (CTM) commonly present among NRAMP proteins. It is highly conserved in the OsNRAMP1, 2, 3, 5 and 7 and is partially conserved in other members of OsNRAMP family of transporter proteins. It is least conserved in OsNRAMP6 protein sequence. DPGN is the signature sequence of NRAMP transporter family [2] and mutation in these residues causes impairment of the transporter function [38, 39]. In OsNRAMP protein sequences it is present in motif 2. In bacterial NRAMP transporters DPGN motif has been identified to act as a binding pocket in which these amino acid residues coordinate with divalent metal ions. Hence this motif in OsNRAMP transporters could also play a role in metal binding.
We also report plausible 3D structure models for all the seven OsNRAMP transporters retrieved using I-TASSER and Robetta. All the highlighted motifs on their I-TASSER predicted structures show similarity in their location indicating that the models are robust. After the structure assembly simulation, I-TASSER uses the TM-align structural alignment program to match I-TASSER model to all structures in the PDB library and reports the top ten proteins from the PDB that have the closest structural similarity, i.e. the highest TM-score, to the predicted I-TASSER model. Due to the structural similarity, these proteins often have similar function to the target. All the seven OsNRAMP transporter models predicted using both the tools have structural similarity with crystal structure of E. coleocola manganese transporter (EcoDMT) which has been recently determined [40]. It is 511 amino acids long protein with 12 transmembrane helixes and is involved in Mn2+ transport. Dali server results also indicate that the models are structurally similar to crystal structure of EcoDMT. It can be seen from the number of matched positions (lali) in Online Resource Figs. 6 and 8 that most matches are almost covering the query sequence. The Dali pairwise comparison of the models predicted using Robetta and I-TASSER have rmsd < 3 indicating that they are closely homologous protein structures.
OsNRAMP interactome analysis indicates that these transporters could be related to Cd/Zn transporters and also be involved in Al tolerance. In reality, a cell is a dynamic structure hence a single molecule can interact in a different manner with several other molecules in coordination with various internal and external stimuli. Validation of all the models by RAMPAGE, Errat and ProQ; study shows their possible reliability as well as acceptance as good quality predicted models.
Eventually, expression profile through microarray result in root shows the prospect of NRAMP family of transporters in metal uptake from soil and their corresponding expression patterns throughout the plant. The results presented here is the detailed study to understand the NRAMP family of transporters in rice using an in silico approach. NRAMP family of transporters are proton coupled metal ion transporters. Till now the exact mechanism of these proton coupled transporters are not thoroughly understood though they are abundantly present in both prokaryotes and eukaryotes [40]. Thus understanding their structure would help us in identifying the accurate region or residues involved in metal binding and transport. With further studies, like simulation and other biological experiments involving yeast mutants and gene knockout plants, it would be useful to understand the transporter functioning mechanism associated with metal ion transport in rice plant and also help predict the metal ions that are transported by the transporter whose substrate specificity has not yet been studied.
References
Nelson N (1999) Metal ion transporters and homeostasis. EMBO J 18(16):4361–4371
Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P (1995) Nramp defines a family of membrane proteins. Proc Natl Acad Sci USA 92(22):10089–10093
Migeon A, Blaudez D, Wilkins O, Montanini B, Campbell MM, Richaud P, Thomine S, Chalot M (2010) Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell Mol Life Sci 67(22):3763–3784
Narayanan NN, Vasconcellos MW, Grusak MA (2007) Expression profiling of Oryza sativa metal homeostasis genes in different rice cultivars using a cDNA macroarray. Plant Physiol Biochem 45:277–286
Xia J, Yamaji N, Kasai T, Ma JF (2010) Plasma membrane localized transporter for aluminum in rice. Proc Natl Acad Sci USA 107:18381–18385
Xia J, Yamaji N, Ma JF (2011) Further characterization of an aluminum influx transporter in rice. Plant Signal Behav 6:160–163
Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF (2013) Anode based switch for preferential distribution of manganese in rice. Nat Commun 4:2442
Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24:2155–2167
Peris-Peris C, Serra-Cardona A, Sánchez-Sanuy F, Campo S, Ariño J, San Segundo B (2016) Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol Plant Microbe Interact. https://doi.org/10.1094/MPMI-01-17-0005-R
Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97:4991–4996
Sakai H, Lee SS et al (2013) Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54(2):e6
Kawahara Y, Bastide M et al (2013) Improvement of the Oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice 6:4
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana, Louisville, pp 571–607
Sonnhammer EL, Eddy SR, Durbin R (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420
Krogh A, Larsson B, Heijne GV, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305:567–580
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Timothy L, Mikael B, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Grant CE, Timothy L, Bailey, Noble WS (2011) FIMO: Scanning for occurrences of a given motif, Bioinformatics 27(7):1017–1018
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P (2005) STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 33:D433-437
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (2009) STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738
David E, Kim D, Chivian, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32(Web Server issue): W526–W531. https://doi.org/10.1093/nar/gkh468
Wu S, Skolnick J, Zhang Y (2007) Ab initio modelling of small proteins by iterative TASSER simulations. BMC Biol 5:17. https://doi.org/10.1186/1741-7007-5-17
Zang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. https://doi.org/10.1186/1471-2105-9-40
DeLano WL (2002) The PyMOL molecular graphics system
Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM et al (2003) Structure validation by C-alpha geometry: phi, psi and beta deviation. Proteins 50:437–450
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of non bonded atomic interactions. Protein Sci 2:1511–1519
Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12:1073–1086
Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucl. Acids Res 38:W545-549
Hasegawa H, Holm L (2009) Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol 19:341–348
Sato Y, Antonio B, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y (2011) RiceXpro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res 39:D1141–D1148
Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio B, Nagamura Y (2013) RiceXPro Version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41:D1206–D1213
Weinstein JN et al (1994) Predictive statistics and artificial intelligence in the U.S. National cancer institute’s drug discovery program for cancer and AIDS. Stem Cells 12:13–22
Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, Buolamwini JK, van Osdol WW, Monks AP, Scudiero DA, Sausville EA, Zaharevitz DW, Bunow B, Viswanadhan VN, Johnson GS, Wittes RE, Paull KD (1997) An information intensive approach to the molecular pharmacology of cancer. Science 17:343–349
Cristobal S, Zemla A, Fischer D, Rychlewski L, Elofsson A (2001) A study of quality measures for protein threading models. BMC Bioinformatics 2(1):5
Belouchi A, Kwan T, Gros P (1997) Cloning and characterization of the OsNRAMP family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Mol Bio 33(6):1085–1092
Courville P et al (2008) Solute carrier 11 cation symport requires distinct residues in transmembrane helices 1 and 6. J Biol Chem 283:9651–9658
Haemig HA, Brooker RJ (2004) Importance of conserved acidic residues in mntH, the Nramp homolog of Escherichia coli. J Membr Biol 201:97–107
Ehrnstorfer IA, Manatschal C, Arnold FM, Laederach J, Dutzler R (2017) Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat Commun 8:14033. https://doi.org/10.1038/ncomms14033
Acknowledgements
AM would like to thank T. Manonanthini, Bioinformatics lab, AU-KBC Research Centre for her help. AM would like to thank Anna University for providing Anna Centenary Research Fellowship during the research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Both the authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mani, A., Sankaranarayanan, K. In Silico Analysis of Natural Resistance-Associated Macrophage Protein (NRAMP) Family of Transporters in Rice. Protein J 37, 237–247 (2018). https://doi.org/10.1007/s10930-018-9773-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-018-9773-y