Abstract
Innate immune system is a primary line of defense in fish that protects it from the invading pathogens. Antimicrobial peptides (AMPs) are widely distributed in nature and are essential components of innate immunity. These molecules enable the host’s innate immune system to fight against a variety of infectious agents. One such AMP, hepcidin, is a cysteine rich amphipathic peptide. We have amplified, cloned and characterized hepcidin like AMP from Schizothorax richardsonii that inhabits one of the most difficult aquatic ecosystems in the Indian Himalayas. The cDNA encoding hepcidin like peptide was amplified as a 371 bp fragment with an open reading frame (ORF) of 279 nucleotides flanked by 5′ and 3′ UTRs of 70 and 22 bases respectively. This ORF encodes a peptide of 93 amino acids with a signal peptide of 24 amino acids and a mature peptide of 25 amino acids. The mature hepcidin like peptide of S. richardsonii has eight cystine residues that participate in the formation of four disulfide bonds, a unique feature of hepcidin like AMPs. A 3D model of hepcidin like mature peptide was generated using Modeller 9.10 which was validated using PROCHECK and ERRAT. Phylogenetic analysis of hepcidin like AMP from S. richardsonii revealed that it was closely related to hepcidin from olive barb (Puntius sarana).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Co-evolution of host and pathogens has developed a variety of versatile defense mechanisms that can be either acquired or germline-encoded [1]. Innate immune system, a key defense mechanism, provides first line of defense against pathogens [2]. Fish being poikilothermic, possess a limited antibody repertoire, affinity maturation and memory besides weak lymphocyte proliferation. This accounts for a poor acquired immune response compared with relatively temperature independent innate immune response [3]. Antimicrobial peptides (AMPs) one of the important mediators of innate immunity, are evolutionarily conserved elements and widely distributed in nature from invertebrates to mammals [4]. These molecules synthesized by host are involved in defense against a variety of infections [1]. A number of AMPs have been identified in fish for example hepcidin, defensins, misgurin, piscidin, parasin, moronecidin and daxin [1, 6, 7]. AMPs possess unique broad spectrum anti-microbial activity against a number of viruses, bacteria, fungi and protozoa. Besides being able to kill 99.9 % pathogens within 20 min [8] it has also been reported that AMPs can possess anticancer properties [9]. Synthetic tilapia hepcidin 2–3 [10] and TH-1-5 [11] have been demonstrated to possess antitumor activity in human cancer cells that may have an application in cancer therapy. Moreover AMPs are also effective against antibiotic resistant pathogens therefore, free from side effects associated with conventional antibiotics [12]. All these characteristics make AMPs attractive potential therapeutic agents that may find an application in both human and veterinary medicine [13].
Hepcidin is a 25 amino acid cysteine rich cationic peptide with hairpin structure in which two β-sheets are linked by four disulfide bonds. Initially this AMP was termed as liver expressed antimicrobial peptide (LEAP) as it was primarily detected in liver [14]. Two types of LEAPs, LEAP1 and LEAP2 are widespread in fish species and humans. There are eight and four cysteine residues in LEAP1 and LEAP2 resulting in the formation of four and two disulfide bonds [15]. In humans, three isoforms of hepcidin are known to exhibit antibacterial activities that contain 20, 22 and 25 amino acids [16]. The amino acid sequences are highly conserved among species and share a characteristic feature of six to eight cysteine residues conserved at positions that signify the importance of disulfide bonds in antibacterial activity [17, 18]. Hepcidin also acts as an important component of host innate immune system upon pathogenic stimuli. It has been reported in various studies that natural and synthetic hepcidin possess antibacterial activity against Gram-positive and Gram-negative bacteria besides having strong activity against other fish pathogens [19]. Antimicrobial activity of purified hepcidin-P1 from large yellow croker [20], synthetic hepcidin peptide from Japanese flounder [21], tilapia hepcidin TH-1-5 and TH-2-3 [22], gilthead seabream hepcidin [4], large yellow croaker PC-hep [19], orange-spotted grouper EC-hepcidin 1 [23], black porgy AS-hepc2 and AS-hepc6 [24] and medaka Om-hep1 [25], recombinant Japanese flounder hepcidin fusion peptide JFL4 [26] and recombinant medaka hepcidin Pro-Omhep1[25] have been well documented. Besides these fish species, hepcidin has been reported in other fish species including striped bass, Morone saxatilis × Morone chrysops [27], winter flounder Pseudopleuronectes americanus [28], Atlantic salmon Salmo salar [28], rainbow trout Oncorhynchus mykiss [28], medaka Oryzias latipes [28], zebrafish Danio rerio [29], olive flounder Paralichthys olivaceus [21, 30], red sea bream Chrysophrys major [31], channel catfish Ictalurus punctatus [32], black porgy Acanthopagrus schlegelii [33], turbot Scophthalmus maximus [34], Atlantic cod Gadus morhua [35] and sea bass Dicentrarchus labrax [36]. In humans [14, 16] and mice [37] expression of hepcidin mRNA has primarily been reported in liver [22, 23, 32] while in fish, expression of this AMP has been demonstrated from liver, spleen, gill, intestine, brain, stomach and skin. The mRNA transcript of hepcidin in early stage blastula is maternally derived, implying a major role of hepcidin in innate defence of embryos in turbot. Bacterial challenge increases hepcidin transcript expression in different fish tissues such as head kidney, heart, spleen, skin and gills [34]. In hybrid striped bass antimicrobial activity and the induction of hepcidin has been observed upon infection [27]. It has been observed that expression of mRNA hepcidin transcript increases by 4,500 fold upon bacterial infection [38] advocating its role in innate immunity against bacterial pathogens [39]. Apart from antimicrobial activity, hepcidin has important role in iron regulation in fish [5, 36] for example, ferroportin, an iron transporter present on cells of the intestinal duodenum, macrophages, and cells of the placenta bind to ferroportin and induces its internalization as well as degradation [40]. Besides bactericidal and antifungal activity, AMPs have immune-modulatory functions and play a key role in iron metabolism, inflammation, hypoxia and anemia [41–46].
Emergence of antibiotic resistant microbes, a matter of grave concern, has resulted due to injudicious use of antibiotics. Therefore, a quest for development of new broad spectrum antimicrobial agents remains a challenge. A solution could be AMPs, which are active against broad spectrum of infectious agents that are resistant to conventional drugs [47, 48]. Looking into the tremendous applications that AMPs may find in agriculture, pharmaceutical and food industry [49], we attempted to unravel an AMP from Indian snow trout, Schizothorax richardsonii, a fish which inhabits a unique ecological niche in the foot hills of the Himalayas. In this report identification, cloning, structural and phylogenetic analysis of an AMP resembling hepcidin like AMP from S. richardsonii is discussed.
2 Materials and Methods
2.1 Fish and Tissue Collection
Eleven snow trout (Schizothorax richardsonii) weighing about 10–15 g were collected from Kalsa stream, Chafi 29.37°N, 79.57°E located at an altitude of 1,238 m above sea level. The fish were euthanized by immersion in 100 ppm of clove oil [50] before being dissected. Tissues including liver, spleen, kidney, brain, heart, gill, intestine, skin and muscle were collected in Ribozol (Ameresco) and homogenized using a dounce homogenizer.
2.2 Amplification of Hepcidin Gene
Total RNA was isolated from the tissues using Ribozol (Ameresco) following manufacturer’s recommendations. The concentration of the purified RNA was estimated using Eon multimode spectrometer (BioTek) and integrity tested by agarose gel electrophoresis. Total RNA (2 μg) was used for the synthesis of cDNA using RevertAid Reverse transcriptase (Fermentas) and Oligo dT18 (Fermentas) primer. Clustal alignment of the mRNA encoding hepcidin from grass carp, common carp and zebra fish were carried out to design specific primers for the amplification of full length hepcidin cDNA. The cDNA was used as template to amplify hepcidin gene using gene specific primer sequence Hep1 for 5′ATCAGAGCCGAGCAGAAGA 3′ and Hep1 Rev 5′CAGCCTGCATTTATACCCG3′ corresponding to the conserved region. Amplification of hepcidin cDNA was carried out by initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation, annealing and extension at 94 °C for 30 s, 51.6 °C for 30 s and 72 °C for 30 s, with final extension at 72 °C for 10 min. PCR products were resolved by agarose gel electrophoresis on 1.5 % agarose gel and visualized under UV using GelDoc XR (BioRad). The amplified products were gel purified using sure trap gel extraction kit (Nucleopore). The purified products were ligated into the InsTA cloning vector (Thermo-scientific) and transfected into Escherichia coli DH-5α cells. The nucleotide sequences of the inserts were obtained by outsourcing (Eurofin India).
2.3 Sequence Analysis of Hepcidin Like Peptide
Nucleotide sequence of hepcidin was analyzed for similarity with other known sequences using BLASTP [51] while the protein translation tool expert protein analysis system was used to predict amino acid sequence [52]. To predict the physiochemical properties of hepcidin such as molecular weight, theoretical pI, and grand average of hydropathy (Gravy) ProtParam tool [53] was used. The amino acid sequence of the predicted hepcidin like peptide of S. richardsonii was aligned to compare with hepcidin sequences of different species of fish and mammals (Table 1) using ClustalW [54].
2.4 Molecular Modelling of Hepcidin Mature Peptide
The 3D structural model of the hepcidin like mature peptide of S. richardsonii was constructed with the Modeller 9.10 [55]. The protein data bank (PDB) was searched for suitable template for active peptide using position specific iterated-basic local alignment search tool (PSI-BLAST). The alignment of the query- template was carried out using ClustalW program [56].
2.5 Model Optimization, Quality Assessment and Visualization
The model was viewed in YASARA view [56] and Swiss-Pdb Viewer 4.0.4 [57]. Hydrogen addition was done in Swiss-Pdb Viewer 4.0.4. Energy minimization was done in vacuo with GROMOS 96 implementation of Swiss-Pdb Viewer 4.0.4. The quality of the model was predicted by ERRAT [58]. PDBsum [59] was used for prediction of secondary structure. The stereo-chemical quality of the protein was analysed using PROCHECK. Superposition, alignment and RMSD of template and query were determined by YASARA View [56]. Structural alignment was done by MUSTANG implementation [60] of YASARA view.
2.6 Phylogenetic Analysis
MEGA 5 [61] was used to carry out phylogenetic and molecular evolutionary analyses [62]. Full length amino acid sequences of hepcidin from different species of fish and other mammals were retrieved from GenBank (Table 1) to construct phylogenetic tree by Neighbour-joining method.
3 Results
3.1 Characterization of S. richardsonii Hepcidin Like Antimicrobial Peptide
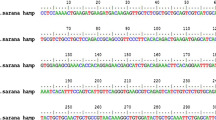
cDNA corresponding to hepcidin like AMP was amplified from the liver of S. richardsonii using the primers described earlier and a PCR product of 371 bp was obtained in all fish livers. Five PCR products were randomly sequenced and the deduced nucleotide sequence was submitted to Gen-Bank (Accession number KC894741). Amplified cDNA possesses an open reading frame (ORF) of 279 bp that encodes a predicted pre-prohepcidin of 93 amino acids. The cDNA of hepcidin like AMP from S. richardsonii has a 5′ UTR of 70 bp, an ORF followed by 3′ UTR of 19 bp. Using Signal P [63], a hydrophobic region rich in Val and Ala residues was determined in the signal peptide from S. richardsonii having a cleavage site between Ala 24 and Val 25. In S. richardsonii the mature peptide was predicted to possess 25 amino acid residues with a pro-domain of 44 amino acids. The molecular weight of hepcidin like mature peptide from S. richardsonii was predicted to be 2893D with theoretical pI 8.74. The mature hepcidin like AMP of S. richardsonii had a Gravy (grand average of hydropathicity) index of −0.204, exhibiting its hydrophilic nature. Intriguingly, we observed a substitution Gln30 in pro-domain, an unusual feature to be observed in cyprinids (Fig. 1).
3.2 Sequence Analysis of Hepcidin Like Peptide of Schizothorax richardsonii
BLASTP analysis revealed that the deduced amino acid sequence of S. richardsonii has 92, 87, 78, and 66 %, identity with Cyprinus carpio, Systomus sarana, Megalobrama amblycephala and D. rerio respectively. We observed eight cysteine residues in the mature peptide, motif [RX(K/R)R] for propeptide cleavage and (-CCR/KF) sequence at the C-termini of hepcidin like AMP of S. richardsonii (Fig. 1).
3.3 Molecular Modelling of Hepcidin Mature Peptide
3D model of the active peptide of hepcidin like AMP of S. richardsonii was generated by homology modelling that provides important information about its function. Upon a PSI-BLAST search against PDB 1S6 W from hybrid white striped bass hepcidin, best template with 70 % identity and 76 % query coverage was selected. The query and template were given as an input in Modeller 9.10 and the best model was selected based on the lowest discrete optimized protein energy. Upon predicting the 3D structure of the mature peptide using Modeller 9.10, it was revealed that hepcidin like mature peptide of S. richardsonii was a β-sheet structure with eight cysteine residues. All the eight cysteine residues were connected as Cys7–Cys23, Cys10–Cys22, Cys11–Cys19, Cys13–Cys14 to form four disulfide bonds (Fig. 2a) and stabilized hairpin shaped molecular structure.
Structural analysis of mature hepcidin like AMP from S. richardsonii.
a 3D structure of mature peptide of hepcidin like AMP of S. richardsonii. Green colour in the model indicates the disulfide bonds. b Predicted secondary structure of mature peptide of hepcidin like AMP from S. richardsonii.
A indicates β-sheet with two β-strands (one strand at Arg8–Cys10 and second strand from Gly20 to Cys22).  ,
,  , β, γ represent disulfide bonds, β- hairpin, beta turn and gamma turns respectively. c Ramachandran plot showing plot statistics of the mature peptide comprising 25 amino acid residues of hepcidin like AMP from S. richardsonii (Color figure online)
, β, γ represent disulfide bonds, β- hairpin, beta turn and gamma turns respectively. c Ramachandran plot showing plot statistics of the mature peptide comprising 25 amino acid residues of hepcidin like AMP from S. richardsonii (Color figure online)
3.4 Model Optimization, Quality Assessment and Visualization
The model generated from Modeller 9.10 was subject to energy minimization using Swiss-Pdb Viewer 4.0.4 to repair distorted geometries by moving atoms to release local constraints. The generated model showed a quality factor of 100 % in ERRAT. For evaluation of the said structure, 3D model was submitted to PDBsum and the position of secondary structure elements was generated. The model of the active peptide was found to contain one β-sheet with two anti-parallel β-strands, four disulfide bonds, a β-hairpin, a β-turn and a γ-turn (Fig. 2b). Ramachandran plot (Fig. 2c) and RMSD were used to validate the model. Ramachandran plot showed 85.7 % residues in most favored regions, 14.3 % residues in allowed regions and 0 % residues in disallowed region. Overall G factor, a measure of a protein’s stereo-chemical property, was calculated to be −0.38 which is greater than the recommended value (−0.50). To further validate the model, it was compared with the respective crystal structure of the template, 1S6 W. The generated model and the template were superimposed for calculation of RMSD. RMSD of mature hepcidin like AMP from S. richardsonii was calculated by superimposition over the said template from hybrid white striped bass which was calculated to be 0.935 Å for 18 aligned out of 25 residues.
3.5 Phylogenetic Analysis
To reveal the phylogenetic relationship between hepcidin of S. richardsonii and other vertebrates, a phylogenetic tree was constructed using Nebigour-joining method [61]. In phylogenetic analysis, fish hepcidins separate in a different clade from the mammals, inferring that fish and mammalian hepcidins diversified from the same ortholog (Fig. 3). The fish clade could be further separated into three distinct clades. The hepcidin-like AMP of S. richardsonii reported herein closely resembles cyprinid hepcidin with olive barb (Puntius sarana) as its closest relative.
Phylogenetic analysis of hepcidin like AMP from snow trout and other vertebrates. Phylogenetic tree was constructed using Mega 5. Neighbor-joining method was used to show the relationship between hepcidins of different fish families and other vertebrates. The GenBank accession numbers for these hepcidin sequences are shown in Table 1. The tree is divided into three clades A, B and C which represent hepcidin peptide from different fish families along with bird and mammals. The numbers at tree nodes indicate the percentage of 1,000 bootstrap samples. The scale bar refers to a phylogenetic distance of 0.1 amino acid substitutions per site
4 Discussion
AMPs are an important component of innate immune system in fish. In this study we amplified an ORF of 279 bp from S. richardsonii that encodes a protein of 93 amino acids and has a high similarity with fish and mammalian hepcidins. It is known that the number of amino acids residues in hepcidin ranges between 83–96 in fish and mammals. For example in gilthead seabream (Sparus aurata) an ORF of 255 bp has been reported that codes for pre-prohepcidin of 84 amino acid residues [4] while in blunt snout bream (M. amblycephala) an ORF of 285 bp codes for a polypeptide of 94 residues [15]. Mature hepcidin is known to possess 20, 22, 24, 25 and 26 residues in different fish species [4] while in humans three isoforms of 20, 22 and 25 amino acid residues have been reported [16]. It is known that eight cysteine residues constitute molecular signatures for most hepcidins [29] while some species have six. These eight cysteine residues are responsible for the formation of four disulfide bonds in mature peptide. The eight cysteine residues in the mature peptides are highly conserved among fish species including turbot, S. maximus [34], zebra fish, D. rerio [29], common carp, C. carpio [39], gilthead seabream, Sparus aurata [4], olive flounder, P. olivaceus [30], and humans [16] while in some fish species including Japanese flounder II and winter flounder II [30], six cysteine residues have been reported. The predicted mature peptide of hepcidin like AMP of S. richardsonii consists of 25 amino acid residues. We too have observed eight cysteine residues at conserved positions in hepcidin like AMP from S. richardsonii involved in formation of four disulfide bonds. The predicted molecular weight of S. richardsonii is comparable with blunt snout bream molecular weight 2,892.46 Da and pI 8.34. It has been well documented in several studies that positive charge is a prerequisite factor for interaction between the AMPs and bacterial lipid membrane that leads to pore formation. The hepcidin like mature peptide of S. richardsonii too is positively charged (pI 8.74) and may be involved in increasing the membrane permeability thus facilitating the destruction of microbes [64, 65]. We predicted Gravy (grand average of hydropathicity) index −0.204 value of mature hepcidin like peptide of S. richardsonii which revealed its hydrophobic nature. The hydrophilic amino acids participate in the formation of hydrogen bond with polar head groups in lipid bilayer and membrane proteins of bacterial cell membrane [66]. In silico analysis of the mature peptide suggests that the amino acid sequence of the predicted peptide is similar to most hepcidins and it may thus possess antimicrobial capability [4] something, that needs to be elucidated. We observed unique amino acid Gln30 in the prodomain of hepcidin like AMP of S. richardsonii which is absent in other cyprinids.
The amino acid sequences of hepcidin like AMP are highly conserved among fish species rather than between fish and other vertebrates [28]. Douglas et al. [28] reported that prepro-hepcidin like peptide of fish and mammals possess high similarity in the amino acids of their mature peptide while among fish, amino acids are highly conserved in signal peptide, pro-domain and mature peptide regions. Our findings are similar to the findings of Douglas et al. [28] as alignment of amino-acid sequence of S. richardsonii prepro-hepcidin like peptide were highly conserved in signal peptide, pro-peptide and mature peptide regions among the fish species (Fig. 1) while, the deduced amino acid sequence of the pre-propeptide was different from other mammals. Based on the amino acid sequence of human hepcidin [14, 16] and close proximity of [RX(K/R)R] motif, necessary for pro-peptide cleavage, the amino termini of hepcidin like peptides was allocated. C-termini (-CCR/KF) of mature peptide of prepro-hepcidin like peptide is conserved among fish species [5] and similar sequence at the C-terminus of S. richardsonii has been reported by us.
In 3D modeling, we predicted β-sheet structure of hepcidin like AMP of S. richardsonii in which eight cysteine residues are involved in formation of four disulfide bonds in a discrete pattern Cys7–Cys23, Cys10–Cys22, Cys11–Cys19, Cys13–Cys14. Same pattern of disulfide bonds have been reported in hepcidin from humans (25 or 20 amino acids) [17] and hybrid striped bass (M. chrysops × M. saxatilis) (21 amino acids) [67]. In the mature peptide of S. richardsonii the vicinal disulfide bonds between Cys13 and Cys14 located at the turn of the hairpin points may also be responsible for the activity of the molecule as reported earlier by Hunter et al. [17]. A quality factor of 100 % was observed for this model by ERRAT. Models having sequence similarity between template and query of more than 50 % are reliable, while similarity between 30 and 50 % and below 30 % results in errors such as mis-prediction of basic folds [68]. We observed 70 % sequence identity in the predicted 3D model of mature peptide of hepcidin like AMP of S. richardsonii therefore the predicted 3D model is reliable. To further evaluate the reliability of predicted 3D model, we used Ramachandran plot and RMSD structural assessment methods. In Ramachandran plot we observed 85.7 % residues in most favored region therefore the predicted 3D model of mature peptide of hepcidin like AMP of S. richardsonii is a good model. G factor provides a measure of unusualness in the stereo-chemical property of a molecule. The values below −0.5 shows the unusual property while the value below −1.0 high unusualness. The value of G factor for predicted model is −0.38. Similarity between two 3D models is indicated by RMSD value. Lower the value of RMSD, higher the similarity between the two 3D models. The calculated RMSD value for the mature peptide of hepcidin like AMP from S. richardsonii is 0.935 Å. This indicates that the 3D models of the mature peptides derived from 1S6 W (hybrid white striped bass) and hepcidin like AMP of S. richardsonii are quite similar. Therefore, all the structure assessment methods Ramachandran plot, G-factor, RMSD value and quality factor confirm the quality of the predicted model.
Hepcidins from different fish species, bird and mammals are divided into three clades as evident from the phylogenetic analysis. All the fish hepcidins are clustered into clade A, while bird and mammalian hepcidins are clustered into clade B and C respectively. Therefore, phylogenetic analysis revealed distant evolutionary relationship between fish and mammalian hepcidins.
5 Conclusion
AMPs are the key mediators of innate immune system which play a vital role in host defense against microbial invasion. Hepcidin is multifunctional cysteine rich small cationic peptide. In this study, we identified and cloned hepcidin like AMP from S. richardsonii and a 3D model was predicted besides conducting the phylogenetic analysis. Hepcidin like AMP from S. richardsonii is 93 residues long with a signal peptide of 24 amino acid residues having alanine and valine rich hydrophobic region. Mature peptide consists of 25 amino acids with a stretch of eight cysteine residues at conserved positions that form the signature of hepcidins. 3D modeling predicted that the hepcidin like AMP from S. richardsonii has β-sheet structure in which all eight cysteine residues are involved in formation of disulfide bonds in a discrete pattern: Cys7−Cys23, Cys10−Cys22, Cys11−Cys19, Cys13−Cys14. The mature peptide of S. richardsonii has a vicinal disulfide bond (Cys13–Cys14) at the turn of the hairpin points that might be crucial domain in the active molecule. A unique amino acid Gln30 in propeptide of S. richardsonii was observed which is absent in other cyprinids. This amino acid might have an effect on function of pro-domain. A 3D model for hepcidin like mature peptide of S. richardsonii was generated, that may be employed to predict the functional behavior of the protein.
References
Mookherjee N, Hancock REW (2007) Cationic host defence peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell Mol Life Sci Rev 64:922–933
Janeway CA, Medzhitov R (2002) Innate immune recognition. Ann Rev Immunol 20:197–216
Magnadottir B (2006) Innate immunity of fish (overview). Fish Shellfish Immunol 20:137–151
Cuesta A, Mesegure J, Esteban MA (2008) The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish gilthead seabream. Mol Immunol 45:2333–2342
Shi J, Camus AC (2006) Hepcidins in amphibians and fishes: antimicrobial peptides or iron-regulatory hormones? Dev Comp Immunol 30:746–755
Yang M, Wang KL, Chen JH, Qu HD, Li SJ (2007) Genomic organization and tissue expression analysis of hepcidin-like genes from black porgy (Acanthopagrus schlegelii B.). Fish Shellfish Immunol 23:1060–1071
Falco A, Chico V, Marroqui L, Perez L, Coll JM, Estepa A (2008) Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol Immunol 45:757–765
Piers KL, Brown MH, Hancock RE (1994) Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by C-terminal modification. Antimicrob Agents Chemother 38:2311–2316
Rinaldi AC (2002) Antimicrobial peptides from amphibian skin: an expanding scenario. Curr Opin Bio 6:82–88
Chen JY, Lin WJ, Lin TL (2009) A fish antimicrobial peptide, tilapia hepcidin TH2-3, shows potent antitumor activity against human fibrosarcoma cells. Peptides 30:1636–1642
Chang WT, Pan CY, Rajanbabu V, Cheng CW, Chen JY (2011) Tilipia (Oreochomis mossambicus) antimicrobial peptide, hepcidin 1–5, shows antitumor activity in cancer cells. Peptides 32:342–352
Patrzykat A, Douglas SE (2003) Gone gene fishing: how to catch noval marine antimicrobials. Trends Biotechnol 21:362–369
Bao B, Peatman E, Xu P, Li P, Zeng H, He C, Liu Z (2006) The catfish liver-expressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Mol Immonol 43:367–377
Krause A, Neitz S, Magert AS, Forssmann WG, Schulz-Knappe P, Adermann K (2000) LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 480:147–150
Liang T, Ji W, Zhang GR, Wei KJ, Feng K, Wang WM, Zou GW (2013) Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol 35:553–563
Park CH, Valore EV, Waring AJ, Ganz T (2001) Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276:7806–7810
Hunter HN, Fulton DB, Ganz T, Vogel HJ (2002) The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J Biol Chem 277:37597–37603
Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME (2005) Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem 280:9272–9282
Wang KJ, Cai JJ, Cai L, Qu HD, Yang M, Zhang M (2009) Cloning and expression of a hepcidin gene from a marine fish (Pseudosciaena crocea) and the antimicrobial activity of its synthetic peptide. Peptides 30:638–646
Zhang J, Yan Q, Ji R, Zou W, Guo G (2009) Isolation and characterization of a hepcidin peptide from the head kidney of large yellow croaker, Pseudosciaena crocea. Fish Shellfish Immunol 26:864–870
Hirono I, Hwang JY, Ono Y, Kurobe T, Ohira T, Nozaki R, Aoki T (2005) Two different types of hepcidins from the Japanese flounder Paralichthys olivaceus. FEBS J 2272:5257–5264
Huang PH, Chen JY, Kuo CM (2007) Three different hepcidins from tilapia, Oreochromis mossambicus: analysis of their expressions and biological functions. Mol Immunol 44:1922–1934
Zhou JG, Wei JG, Xu D, Cui HC, Yan Y, Ou-Yang ZL, Huang XH, Huang YH, Qin QW (2011) Molecular cloning and characterization of two novel hepcidins from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol 30:559–568
Yang M, Chen B, Cai JJ, Peng H, Ling C, Yuan JJ, Wang KJ (2011) Molecular characterization of hepcidin AS-hepc2 and AS-hepc6 in black porgy (Acanthopagrus schlegelii): expression pattern responded to bacterial challenge and in vitro antimicrobial activity. Comp Biochem Physiol B: Biochem Mol Biol 158:155–163
Cai L, Cai JJ, Liu HP, Fan DQ, Peng H, Wang KJ (2012) Recombinant medaka (Oryzias melastigmus) pro-hepcidin: multifunctional characterization. Comp Biochem Physiol B 161:140–147
Srinivasulu B, Syvitski R, Seo JK, Mattatall NR, Knickle LC, Douglas SE (2008) Expression, purification and structural characterization of recombinant hepcidin, an antimicrobial peptide identified in Japanese flounder, Paralichthys olivaceus. Protein Expr Purif 61:36–44
Shike H, Lauth X, Westerman ME, Ostlnd VE, Carlberg JM, Van Olst JC, Shimizu C, Bulet P, Burns JC (2002) Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur J Biochem 269:2232–2237
Douglas SE, Gallant JW, Liebscher RS, Dacanay A, Tosi SCM (2003) Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev Comp Immunol 27:598–601
Shike H, Shimizu C, Lauth X, Burns JC (2004) Organization and expression analysis of the zebrafish hepcidin gene, an antimicrobial peptide gene conserved among vertebrates. Dev Comp Immunol 28:747–754
Kim YO, Hong S, Nam BH, Lee JH, Kim KK, Lee SJ (2005) Molecular cloning and expression analysis of two hepcidin genes from olive flounder (Paralichthys olivaceus). Biosci Biotechnol Biochem 69(7):1411–1414
Chen SL, Xu MY, Ji XS, Yu GC, Liu Y (2005) Cloning, characterization, and expression analysis of hepcidin gene from red sea bream (Chrysophrys major). Antimicrob Agents Chemother 49:1608–1612
Bao B, Peatman E, Li P, He C, Liu Z (2005) Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specific upregulation after bacterial infection. Dev Comp Immunol 29:939–950
Wang Q, Wang Y, Xu P, Liu Z (2006) NK-lysin of channel catfish: gene triplication, sequence variation, and expression analysis. Mol Immunol 43:1676–1686
Chen SL, Li W, Meng L, Sha ZX, Wang ZJ, Ren GC (2007) Molecular cloning and expression analysis of a hepcidin antimicrobial peptide gene from turbot (Scophthalmus maximus). Fish Shellfish Immunol 22:172–181
Solstad T, Larsen AN, Seppola M, Jorgensen TO (2008) Identification, cloning and expression analysis of a hepcidin cDNA of the Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol 25:298–310
Rodrigues PN, Vazquez-Dorado S, Neves JV, Wilson JM (2006) Dual function of fish hepcidin: response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev Comp Immunol 30:1156–1167
Pigeon C, Ilyin B, Courselaud P, Leroyer P, Turlin B, Brissot P, Loreal O (2001) A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276:7811–7819
Ganz T (2002) The role of hepcidin in iron sequestration during infections and in the pathogenesis of anemia of chronic disease. IMAJ 4:1043–1045
Li H, Zhang F, Guo H, Zhu Y, Yuan J, Yang G, An L (2013) Molecular characterization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol 35:1030–1038
Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J (2004) Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306(5704):2090–2093
Ashrafian H (2003) Hepcidin: the missing link between hemochromatosis and infections. Infect Immun 71:6693–6700
Rossi E (2005) Hepcidin- the iron regulatory hormone. Clin Biochem Rev 26:47–49
Kemna E, Pickkers P, Nemeth E, van der Hoeven H, Swinkels D (2005) Time course analysis of hepcidin, serum iron and plasma cytokine levels in human injected with LPS. Blood 106:1864–1866
Hugman A (2006) Hepcidin: an important new regulator of iron homeostasis. Clin Lab Haematol 28:75–83
De Domenico I, Zhang TY, Koening C, Branch RW, London N, Lo E, Daynes AR, Kushner JP, Li D, Ward DM, Kaplan J (2010) Hepcidin mediates transcriptional changes that modulate acute cytokine-induced inflammatory responses in mice. J Clin Invest 120:2395–2405
Rajanbabu V, Pan CY, Lee SC, Lin CC, Li CL (2010) Tilapia hepcidin 2-3 peptide modulates lipopolysaccharide-induced cytokines and inhibits tumor necrosis factor-alpha through cyclooxygenase-2 and phosphodiesterase 4D. J Biol Chem 285:30577–30586
Mangoni ML (2011) Host-defense peptides: from biology to therapeutic strategies. Cell Mol Life Sci 68:2157–2159
Yeung ATY, Gellatly SL, Hancock REW (2011) Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci 68:2161–2176
Keymanesh K, Soltani S, Soroush S (2009) Application of antimicrobial peptides in agriculture and food industry. World J Microbiol Biotechnol 25:933–944
Veliek J, Svobodova Z, Piakova V (2005) Effects of clove oil anaesthesia on rainbow trout (Oncorhynchus mykiss). Acta Vet Brno 74:139–146
Altschul SF, Gish W, Miller W, Myers E, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASY: the proteomics server for in depth protein knowledge and analysis. Nucleic Acid Res 31:3748–3788
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy Server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, NJ, pp 571–607. http://springerlink.bibliotecabuap.elogim.com/protocol/10.1385%2F1-59259-890-0%3A571
Thompson JD, Higgins DJ, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Sali A (2011) MODELLER: a program for protein structure modeling release 9.10, r8346. Modeller. http://salilab.org/modeller/
Krieger E, Koralmann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA—a self parameterizing force field. Proteins 47:393–402
Guex N, Peltsch MC (1997) SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative modelling. Electrophoresis 18:2714–2723
Colovos C, Yeates TO (1993) Verification protein structures: patterns of non bonded atomic interactions. Protein Sci 2:1511–1519
Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM (1997) PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci 22:488–490
Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM (2006) MUSTANG: a multiple structural alignment algorithm. Proteins 64:559–574
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of procariotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Sitaram N, Nagaraj R (1999) Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochim Biophys Acta 1462:29–54
Dathe M, Schumann M, Wieprecht T, Winkler A, Beyermann M, Krause E, Matsuzaki K, Murase O, Bienert M (1996) Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 35:12612–12622
Hancock REW, Rozek A (2002) Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett 206:143–149
Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME (2004) Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism and in vivo hepatic response to bacterial infection. J Biol Chem 280:9272–9282
Baker D, Sali A (2001) Protein structure prediction and structural genomics. Science 294(5540):93–96
Acknowledgments
The authors are grateful to the Director, Directorate of Coldwater Fisheries Research for providing the necessary facilities and Department of Biotechnology, Government of India for the Grant BT/PR12760/AAQ/03/477/2009.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chaturvedi, P., Dhanik, M. & Pande, A. Characterization and Structural Analysis of Hepcidin Like Antimicrobial Peptide From Schizothorax richardsonii (Gray). Protein J 33, 1–10 (2014). https://doi.org/10.1007/s10930-013-9530-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-013-9530-1