Abstract
Tumor differentiation factor (TDF) is a protein produced by the pituitary and secreted into the blood stream. The mechanism of its action has still not been elucidated, although the associated protein receptor was identified. Furthermore, the TDF protein does not have any homology with other known proteins, and the crystal structure of TDF also is not available at this time. To gain some insight into the structure of this rather underexplored protein, we have performed a molecular dynamics simulation of a model TDF structure. The structural stability of this protein is evaluated as a function of time. The time dependent structural changes of four cysteine residues present in this structure also are explored.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Molecular dynamics (MD) simulations permit us to predict and analyze the static and dynamic properties of proteins. They also help in the determination of the structural stability and conformational changes of a protein and give insight into a protein’s molecular structure [24]. These simulations are evolving in their ability to convey molecular information and have become highly valuable for supplementing experimental methods. They are able to convey molecular events and transitions that are important for understanding the function and physiology of biological systems [24]. MD simulations can uncover three-dimensional structures and help elucidate the protein folding process [19]. In addition to the value of MD simulations for basic research, they are further valuable for application to rational drug design and hold great promise for the future of this field [20].
Tumor differentiation factor (TDF) is a protein that has not been extensively researched and is not well-understood [23, 27, 32]. The application of MD simulations therefore could help to expand knowledge of TDF. TDF was originally identified as a pituitary-derived factor that causes aggregation and differentiation of rat mammary tumor cells [17]. TDF protein has 108 amino acids and was isolated from a human cDNA library through expression cloning [18]. The molecular weight of TDF protein is 17 kDa. TDF is believed to be produced by the pituitary and secreted into the blood, with an identified putative receptor and unknown mechanism of action [28, 29].
Cellular localization of TDF to pituitary cells has recently been confirmed. Such cells are likely anterior pituitary cells, not posterior pituitary cells. TDF was also identified in select brain neurons, not astrocytes, predominantly but not exclusively in cells producing gamma aminobutyric acid (GABA), an inhibitory neurotransmitter [32]. The putative receptor has been identified in both breast cancer and prostate cells, including steroid-resistant and steroid-responsive forms. Therefore, TDF may be a non-steroidal novel hormone with the ability to differentiate both breast and prostate cells. TDF does not have any homologous structure in the protein database.
Our group is working to determine the 3D structure of TDF protein however the crystal or NMR structure of TDF protein are not available at this time. In prior work, we overexpressed and characterized recombinant recombinant TDF (rTDF) and investigated native, secreted TDF. We also evaluated possible disulfide connectivities via molecular modeling. We found that rTDF is mostly expressed as insoluble, monomeric, and dimeric forms [23]. Mass spectrometry analysis of the overexpressed rTDF identified a peptide that is a part of TDF protein. Mass spectrometry is a particularly useful method not only for identification of particular peptides that are part of a specific protein (i.e. TDF), but also for additional studies such as post-translational modification, protein–protein interactions, as well as structural studies [2, 3, 14]. TDF contains the following four Cys residues: Cys17, Cys70, Cys97, and Cys98. Investigation of TDF via molecular modeling indicated that Cys residues may form disulfide bridges between Cys17–Cys98 and Cys70–Cys97 [23].
To further corroborate the above findings, we have performed MD simulation of the model TDF structure. Using MD simulation we have projected the stability of this model protein and some time-dependent aspects of its properties. Since this protein does not yet have a known crystal structure, it is expected that the present study can help to gain some insight into its structure. The structural changes of the four Cys residues present in this model protein are also evaluated in this work.
2 Methods
For a MD simulation model, the TDF structure was used. This model TDF protein structure was predicted using Iterative Threading ASSEmbly Refinement server [22, 33]. Although experimental validation is required there are examples in which model structures are used and have been evaluated in MD simulation when crystal structures are unavailable [1, 4, 8, 11, 15].
Initially, the model TDF protein was solvated with water using TIP3P model in a cubic box. We have used Chimera software to solvate the model protein [16]. An explicit solvation method was used. The numbers of solvent molecules were 4,199. No counter ions were added to the system as the TDF-protein has a neutral pH. After solvation the energy was minimized using 5000 iteration. Then MD simulation was performed using Accelrys Discover’s MD module. MD simulation of the system was performed using thermodynamic canonical ensemble NVT, where the number of molecules (N), volume (V) and temperature (T) were fixed. The temperature was set to 298 K. The total number of dynamic steps was set to 145,000, therefore, the total dynamics time of MD simulation was 145 ps. The time-step was set as 1.0 femtosecond (1 femtosecond = 10−15 s). Frame output was set in every 100 steps. This generated 1,450 trajectories or conformation frames. These MD trajectories essentially represent the molecular movements (dynamics) as functions of time. Using these trajectories we determined the time dependent conformational changes of the TDF-Protein structure. The total CPU time for this 145 ps MD simulation was approximately 12 h. As part of this simulation, we also checked the role of protein-solvation in the overall calculated dynamics, and to do this, we performed another set of MD simulations without solvating the TDF protein. The CPU time used for the unsolvated TDF protein simulation was 67 minutes. Energy minimization and MD simulations were performed using Accelrys Materials Studio/Discover Studio module. Trajectory analyses were performed using Visual Molecular Dynamics [7] and figures were generated by Accelrys Materials studio and Discovery Studio Visualizer [26].
3 Results
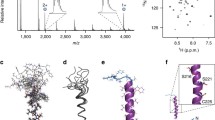
The hydrophobic surface of model TDF-protein is displayed in Fig. 1a and b. From Fig. 1b it is apparent that the hydrophobic residues are buried inside the TDF structure. The Connolly surface of the occupied volume is 13,595.39 Å3 and the surface area is 7,737.13 Å2. The Connolly surface of the TDF-structure is displayed in Fig. 1c. Figure 1d displays the secondary structure of the model TDF protein after solvation. TDF-protein conformations were analyzed for 145 ps dynamic simulation time. The structural and conformational stability of a protein can be measured by RMSD from its initial structure. The all atom RMSD of the simulated structure with time is shown in Fig. 2a. The protein backbone RMSD is displayed in Fig. 2b. RMSF is the standard deviation of atomic position from its initial structure. Figure 2c depicts the RMSF of the beginning and last trajectory structures. Figure 2d displays the regions of greater flexibility within the model TDF. SASA of initial and final model structures are plotted in Fig. 2e. The snapshots of superposed structures of TDF-protein at different simulation time with reference to its starting structure are portrayed in Fig. 3. Four Cys residues are present in the TDF structures. RMSD verses time for each of these Cys residues are plotted in Fig. 4. Figure 5a displays the comparative RMSF values of Cys residues from initial and last trajectory. Figure 5b shows the SASA values of four Cys residues from first and last snapshots. Superposed Cys configurations with time are displayed in Fig. 6. Figures 1, 2, 3, 4, 5, 6 are from the solvated TDF simulations. Figure 7 displays the all atom RMSD graph of unsolvated TDF simulation along with the backbone results, both as functions of time.
Comparative structural analysis of model TDF protein with time. a, b. The RMSD of TDF-protein as functions of time (with regards to the starting trajectory structure) is depicted here. a All atom RMSD (red). b Backbone RMSD (blue). The timescale in ps is plotted along X axis and RMSD values are plotted along Y axis. c Comparative RMSF in residues of initial and final TDF proteins structure. The residue numbers are plotted along X axis and the RMSF values are plotted along Y axis. d Regions of greater flexibility on TDF model protein. Residues between 23–31, 48–61 and 83–96 are displayed in green, brown and yellow respectively. e Relative solvent accessible surface area (SASA) versus residue number from the initial and last trajectory structures of MD simulation. The residue numbers are plotted along X axis and the SASA values are plotted along Y axis (Color figure online)
Superposition of TDF-protein structures with the initial structure during MD simulation. The first trajectory structure is colored in red. Super position of structures with initial structure at time a 25 ps (green), b 50 ps (yellow), c 75 ps (pink), d 100 ps (orange), e 125 ps (violet). f Superposition of initial structure (red) with the last trajectory structure (blue) is displayed (Color figure online)
Structural superposition of Cys residues at different times during MD simulation. Initial and final trajectory structures of Cys residues are colored in red and blue respectively. Structures of Cys at 25, 50, 75, 100, and 125 ps are colored in green, yellow, pink, orange, and violet respectively (Color figure online)
4 Discussion
Tumor differentiation factor protein is a novel protein and very little is known about its structure. In our previous experiments we over-expressed and characterized recombinant TDF and described the structure of the model TDF protein [23]. We classified four Cys residues, predicted their disulphide connectivities, and discussed the possible interactions between Cys residues and their neighboring amino acids [23]. Our present study focuses on the time-dependent structural stability of this model structure using MD simulation. The stability of the four Cys residues present in this protein is measured and compared using MD simulation. In addition, we have examined the role of protein solvation in the calculated dynamics by comparing the stabilities of solvated and unsolvated systems.
The RMSD values of the TDF protein were calculated and the all atom RMSD of the simulated solvated TDF protein is displayed in Fig. 2a. For the solvated TDF, the maximum, minimum and average values of all atom RMSD values are 0.89, 0.19 and 0.79 Å, respectively, with a standard deviation RMSD of 0.08 Å. The backbone RMSD of the simulated solvated TDF structure as a function of time is displayed in Fig. 2b. The maximum and minimum value of backbone RMSD are 0.32 and 0.10 Å and the average value of backbone RMSD is 0.28 Å and standard deviation RMSD is 0.02 Å. The changes in the RMSD values of the initial and final trajectory structures of solvated TDF model during the MD simulation are insignificant. RMSF values are used to describe the displacement of the final structure from the original one. Structural flexibility of the model structure can be assessed by analyzing RMSF. So the residues with a higher RMSF values usually seem to be more flexible [4]. RMSF of initial and final trajectory snapshots of the solvated TSD are displayed in Fig. 2c. Here the RMSF values are plotted as a function of a residue number. The final trajectory structure shows a larger flexibility compared to the initial structure. Three areas in the TDF protein show this increased flexibility. These are residues between 23–31, 48–61 and 83–96. These regions are colored differently in Fig. 2d. Figure 2e shows illustrative representation of SASA values of solvated TDF. From this SASA plot we can see that the last trajectory SASA values are relatively higher than the initial trajectory values but there are no extensive changes in the SASA values during the simulation timeframe. From the representative model structures of solvated TDF model protein during MD simulation (Fig. 3), it is possible to infer that, no major conformational changes between the protein structures are observed, and that the deviations are minimal between the initial and last trajectory structures. For Cys17 the maximum value of all atom RMSD is 1.27 Å and the lower RMSD value is 0.08 Å. Cys70 has an all atom RMSD maximum value 1.30 Å. The lower RMSD value is 0.34 Å. For Cys97 the maximum value of all atom RMSD is 0.40 Å. The lower RMSD value is 0.13 Å. For Cys98 the maximum value of all atom RMSD is 0.95 Å. The lower RMSD value is 0.16 Å. These values indicate that the Cys97 is the most stable Cys residue in the TDF model structure during the period of simulation; the order in which the stabilities decrease is as follows: Cys97 > Cys98 > Cys17 > Cys70. Figure 5a, b display the RMSF and SASA values for the Cys residues in solvated TDF structure. RMSF values of initial and final trajectories are almost same for Cys17 and Cys70. For Cys97 and Cys98 the RMSF values change marginally. Although there is a slight increase in the SASA values for the first three Cys residues, the SASA values for Cys98 are decreased between the first and the last trajectory structure. The superposed Cys residues during the time of MD simulation are displayed in Fig. 6. The stability of Cys97 is manifested in consistently overlying residues taken from different times. All these above analyses are based on a solvated TDF system. To further quantify the simulation results, we examined to what extent the protein’s solvation would contribute to these results. As indicated in the published literature, the standard approach for carrying out such a test is to execute a baseline MD simulation using an unsolvated protein system, and to compare the results with those for the solvated system [9, 12, 13, 21, 25, 30, 31]. In case of unsolvated TDF protein the maximum value of all atom RMSD is 1.148 Å. The lower RMSD value is 0.25 Å. The maximum and minimum values of unsolvated TDF backbone RMSD are 0.55 and 0.11 Å respectively. Figure 7 shows the all-atom and backbone RMSD of the unsolvated TDF structure as a function of time. The results are somewhat different from that of the solvated structure and suggest that, the solvated TDF protein is slightly more stable than the unsolvated one during the time period of MD simulation.
5 Conclusion
As the backbone RMSD values between the initial and final MD trajectory structures of the solvated protein do not show any significant variation we can state that the TDF protein structure does not change significantly with time. A comparison of the RMSD values obtained for solvated and unsolvated TDF proteins suggest only a moderately increased stability in the former case. In conclusion, this exploratory MD experiment was designed to examine the overall trend of the simulation involving such systems. We believe this work lays the groundwork for further future studies for a long time run of TDF structure in a nano-second timescale. Such longer runs of MD simulation using various diverse systems can give additional insight into the conformational changes (flexibility/folding-unfolding) of the protein. The solvent effects on proteins stability and dynamics can also be studied in more detail by extending the simulation period [5, 6]. Furthermore, upon coupling with docking, MD simulation can predict the ligand–receptor interaction at the interface, and this may provide additional useful information about ligand-induced conformational changes and structural reordering. Long time dynamic studies may also help to predict protein functions in a quantitative and systematic approach [10].
Abbreviations
- MD:
-
Molecular dynamics
- TDF:
-
Tumor differentiation factor
- rTDF:
-
Recombinant TDF
- Cys:
-
Cysteine
- Ps:
-
Picosecond
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- SASA:
-
Solvent accessible surface area
References
Blessia TF, Sharmila DJS, Samian MR, Arsad H, Jamil NF (2012) Int Res J Pharm I(3):251–256
Darie CC (2013) Aust J Chem 66:1–2
Darie CC, Litscher ES, Wassarman PM (2008) Structure, processing, and polymerization of rainbow trout egg vitelline envelope proteins. Springer-Verlag, Düsseldorf
De Rosa MC, Pirolli D, Bozzi M, Sciandra F, Giardina B, Brancaccio A (2011) J Mol Graph Model 29:1015–1024
Dodson G, Verma CS (2006) Cell Mol Life Sci 63:207–219
Hinsen K, Kneller GR (2008) Proteins 70:1235–1242
Humphrey W, Dalke A, Schulten K (1996) J Mol Graph 14:33–38
Hussain MR, Shaik NA, Al-Aama JY, Asfour HZ, Khan FS, Masoodi TA, Khan MA, Shaik NS (2012) Gene 508:188–196
Iavarone AT, Patriksson A, Van der Spoel D, Parks JH (2007) J Am Chem Soc 129:6726–6735
Karplus M, Kuriyan J (2005) Proc Natl Acad Sci USA 102:6679–6685
Kemege KE, Hickey JM, Lovell S, Battaile KP, Zhang Y, Hefty PS (2011) J Bacteriol 193:6517–6528
Khairudin NBA, Wahab HA (2012) Int J Mod Phys Conf Ser 9:193–198
Mao Y, Woenckhaus J, Kolafa J, Ratner MA, Jarrold MF (1999) J Am Chem Soc 121:2712–2721
Ngounou Wetie AG, Sokolowska I, Wormwood K, Michel TM, Thome J, Darie CC, Woods AG et al (2013) J Mol Psychiatry 1:8. doi:10.1186/2049-9256-1-8
Nordling E, Abraham-Nordling M (2012) Comput Biol Chem 39:29–34
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) J Comput Chem 25:1605–1612
Platica M, Chen HZ, Ciurea D, Gil J, Mandeli J, Hollander VP (1992) Endocrinology 131:2573–2580
Platica M, Ivan E, Holland JF, Ionescu A, Chen S, Mandeli J, Unger PD, Platica O (2004) Proc Natl Acad Sci USA 101:1560–1565
Rizzuti B, Daggett V (2013) Arch Biochem Biophys 531:128–135
Rosas-Trigueros JL, Ilizaliturri-Flores I, Benitez-Cardoza CG, Correa-Basurto J, Zamorano-Carrillo A (2012) Curr Med Chem 19:6081–6094
Rossky PJ, Karplus M (1979) J Am Chem Soc 101:1913–1937
Roy A, Kucukural A, Zhang Y (2010) Nat Protoc 5:725–738
Roy U, Sokolowska I, Woods AG, Darie CC (2012) Biotechnol Appl Biochem 59:445–450
Shaikh SA, Li J, Enkavi G, Wen PC, Huang Z, Tajkhorshid E (2013) Biochemistry 52:569–587
Shi W, Inamdar MV, Sastry AM, Lastoskie CM (2007) J Phys Chem C 111:15642–15652
Software: Accelrys Software Inc. (2012) Discovery studio modeling environment, Release 3.5. Accelrys Software Inc., San Diego
Sokolowska I, Woods AG, Gawinowicz MA, Roy U, Darie CC (2013) Cell Mol Life Sci 70(16):2835–2848. doi:10.1007/s00018-012-1185-0
Sokolowska I, Woods AG, Gawinowicz MA, Roy U, Darie CC (2012) FEBS J 279:2579–2594
Sokolowska I, Woods AG, Gawinowicz MA, Roy U, Darie CC (2012) J Biol Chem 287:1719–1733
Thirumuruganandham SP, Urbassek HM (2010) Bioche Res Int 2010:6
Van Gunsteren WF, Karplus M (1982) Biochemistry 21:2259–2274
Woods AG, Sokolowska I, Deinhardt K, Sandu C, Darie CC (2013) Brain Struct Funct. doi:10.1007/s00429-013-0571-1
Zhang Y (2008) BMC Bioinformatics 9:40
Acknowledgments
We are thankful to Professor John B. McLaughlin for allowing us access to the Materials Studio software. This work was supported in part by the Keep A Breast Foundation (KEABF-375-35054) and by support from the U.S. Army research office through the Defense University Research Instrumentation Program (DURIP Grant #W911NF-11-1-0304). This work was also supported in part by Mary Joyce, Kenneth Sandler, Robert Matloff, and by the SciFund challenge contributors. CCD was supported during the summer of 2013 by the David A. Walsh’67 fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roy, U., Woods, A.G., Sokolowska, I. et al. Structural Evaluation and Analyses of Tumor Differentiation Factor. Protein J 32, 512–518 (2013). https://doi.org/10.1007/s10930-013-9510-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-013-9510-5