Abstract
The purpose of this research is to study the thermal unfolding of high concentration bovine Immunoglobulin G (IgG) under 26 different experimental conditions by Fourier Transform Infrared spectroscopy with improved purge conditions and software calculations. When bovine IgG (25–200 mg/mL) was thermally denatured between pH 4.0 and 8.0, it was observed that at 25 mg/mL concentration, the protein exhibited maximum thermal stability at pH 6.0 and 7.0 as evident from the apparent Tm values. Increasing the concentration from 25 to 100 mg/mL at those pH values increased the thermal resistance of the protein by 2–3 °C. But, at 200 mg/mL, IgG showed a small decrease in its transition temperature. Presence of 100 mM Trehalose enhanced the Tm values at all conditions and possibly prevented the complete loss of IgG as insoluble aggregates at higher temperatures. Second derivative plots were constructed to explain the conformational changes of IgG during thermal unfolding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
More than 20 therapeutic monoclonal antibodies have been approved by Food and Drug Administration (FDA) and hundreds of others are currently in various stages of development [23]. All of the approved therapeutic monoclonal antibodies belong to Immunoglobulin G (IgG) class which is the major immunoglobulin type in human sera. Stabilization of IgG especially at higher concentrations poses a major formulation challenge for commercial usage [26]. Because of their ease of administration and the costs, liquid antibody dosage forms are usually preferred over lyophilized forms. Among all the commercial antibody products, above half are stable enough to be formulated in liquid form [33].
The major challenge in formulating liquid antibodies is to control the rate of antibody degradation, thereby increasing their efficacy and potency. When a high concentration liquid dosage is developed numerous factors such as pH, concentration, temperature, and excipients are the independent factors that can be balanced to achieve the required physical and chemical stability of the antibodies. Formulation scientists focus on maintaining the protein’s native conformation for maximum activity and safety.
Biophysical methods, including Fourier transform infrared (FTIR), Circular Dichroism (CD), Fluorescence, UV–Visible, Nuclear Magnetic Resonance (NMR) and Differential Scanning Calorimetry (DSC) have been used to characterize protein denaturation and thereby provide important information on the conformational and secondary structure changes and their causes. FTIR spectroscopy is one of the oldest and well established techniques and is now widely used in pharmaceutical industries [34]. The FTIR technique requires small amount of the samples, offers a rapid and easy use, and is applicable to turbid liquids and to different physical states of the samples [11]. An inherent problem of other traditional methods, avoided by FTIR, is the inability to analyze samples at high concentrations. As well, FTIR can often be used to monitor aggregation in proteins, which is necessary for the formulation development of high concentration protein samples.
Bovine serum and lacteal secretions contain three major classes of immunoglobulins: IgG, IgM, and IgA where serum and colostrum have higher IgG concentration than milk [7]. Traditional milk processing involves thermal treatments that include sterilization, pasteurization and concentration. High temperature processing (heating above 70 °C) and alterations in the pH values cause bovine IgG to loose its antigen binding activity [3, 4]. CD thermal melting at low concentration and neutral pH showed that heat treatment above 72 °C resulted in the loss of immuno activity of bovine IgG [17]. Activity of a protein is dependent on its conformational changes. Understanding why bovine IgG losses its immuno activity when subjected to thermal stress calls for a thorough investigation of its conformational changes under different experimental conditions. Here we attempted to study the effect of varying the pH and protein concentration during thermal treatment of bovine IgG with and without an excipient using FTIR spectroscopy. The lab protocol and data analysis for thermal melting with off-the-shelf equipment poses some challenges—water subtraction, evaporation of sample, purge times, biocell cleaning, and statistical accuracy. As Sect. 2.2 below describes, we developed some custom hardware and software to augment the commercially available equipment at our disposal.
2 Materials and Methods
2.1 Materials
Bovine IgG (≥95% pure) was purchased from Sigma Aldrich. For pH screening studies, 25 mg/mL of the protein was dialyzed against a buffer cocktail of 25 mM borate, citrate and phosphate (pH 4.0–8.0) with 100 mM NaCl as ionic strength. For excipient studies, the buffer also contained 100 mM Trehalose. After dialysis, all protein samples and buffers were filtered using 0.2 μm syringe filters (Millipore). The pH of the samples and buffers was confirmed by pH meter. The protein samples were then concentrated to the desired concentrations using microcon centrifuge tubes (MWCO 10,000 Da, Millipore) and the concentration was measured by UV-spectroscopy at the wavelength of 280 nm. The error in the concentration measurement was <2%. Freshly prepared samples were used for thermal unfolding studies.
2.2 Method
FTIR spectra were recorded using a Nicolet 550 Magna spectrometer. Samples were placed between CaF2 windows of 5 μm pathlength (Biocell, Biotools, Inc.). For each scan, a 16,384 point double sided interferogram was collected with 4 cm−1 resolution. The Nicolet was purged with dry nitrogen for 1 h prior to data collection. The biocell was then heated from 25 to 95 °C uniformly over 70 min. Such FTIR work faces three challenges: (1) good dry nitrogen purge, (2) subtraction of the strongly absorbing water-based buffer, and (3) repeating observations for assignment of confidence intervals (error bars). We solved the problems as described below.
2.2.1 Dry Nitrogen Purge
Low signal to noise in the second derivative of the protein spectra is dependent on a thorough dry nitrogen purge. The different purge conditions in scans of the buffer from the purge for scans of the sample lead to further undesirable spikes in the protein absorbance caused by the subtraction of the buffer and sample absorbancies. Venyaminov [30] found results improve with purges up to 24 h. However, such extended purge times would have greatly increased the time and cost of our study. We therefore followed Venyaminov’s [30] method and built a shuttle system consisting of a stepper motor to move the biocell into and out of the path of the light so the background and sample observations could be taken in the same purged chamber.
The following protocol was developed for sample measurements in the biocell:
-
1.
Purge for 60 min, and then begin continuous uniform heating for 70 min from 25 to 95 °C.
-
2.
Take 10 scans of the background and average the intensities. Call this Iback i .
-
3.
The stepper motor moves the biocell (without breaking the purge) into the light beam and then take 20 scans. Average the intensities, Isample i . Take the average temperature T i of the biocell for these scans. Compute the absorbance of the sample at temperature T i as log10 ((Iback i + Iback i+1 /2)/Isample i ). Note that we use the background scans twice so that as the water vapor spectrum changes with temperature, we reduce this effect by bracketing it around the sample scans.
-
4.
The stepper motor moves the biocell out of the light beam in preparation for the scans of the background. Go to step (2) until the temperature reaches 95 °C at 70 min, which results in about 1,500 interferogram scans for each thermal melting curve.
-
5.
Cool the biocell slowly over 45 min to avoid cracking of the CaF2 window.
We found we had to rigorously follow the biocell filling procedure of Venyaminov et al. [30] to get a seal to prevent evaporation of buffer during the thermal heating. Reference spectra were recorded under identical conditions with only the reference buffer (without sample) in the cell. Figure 1 demonstrates the power of this technique. Protein sample measured by different purge methods are plotted in solid and dash dotted traces in the amide I region (1,800–1,500 cm−1). The solid curve is the resulting unsmoothed protein spectrum from 20 sample scans and 20 background scans at 28 °C for each of a sample and its buffer, but all in separate purges for a heat ramp starting after 1 h of purge. The water vapor is not matched and this results in the many uneven segments of the protein absorbance curve. The dash dotted curve is the same protein spectrum from the same number of scans for each of the sample and buffer, but for each the shuttle system moves the biocell aside so the same purge is used for the background. The water vapor interference is almost non-existent in this curve because the absorbance effect for the sample and background is much better matched and are divided out in the sample absorbance calculation in step (3) above.
Unsmoothed absorbance spectra of the protein sample (50 mg/mL IgG, pH 4) with buffer subtracted, measured by two different purge methods: sample measured under separate purge conditions (solid trace) and using shuttle system (dash dotted trace) as explained in the Sect. 2.2 . The spectrum in solid trace is translated up in the Y-direction for this graph. The signal-to-noise ratio for the protein absorbance is greater than 9,000 to 1. This gives error bars (±2 standard deviations) that would not be visible on this chart. They are approximately 1/2,000 the magnitude of the maximum absorbance of the protein as shown
2.2.2 Water Subtraction Problem
The selection of a scaling factor to subtract the buffer solution from the sample solution to get the spectrum of the protein poses a challenge in FTIR work. A scaling factor is needed because the path lengths between sample and buffer will differ and because the protein displaces buffer. Our Nicolet 550 Magna spectrometer had insufficient signal-to-noise to use the technique of relative absorbencies in the band at 2,125 wave numbers where protein absorbance is negligible, see Rahmelow et al. [22]. We attempted the “zero slope” approach first espoused by Dong et al. [5] but could not make sense of the data because the result was often a negative absorbance. We wanted a method that could be automatically applied to the proteins at all concentrations for all experiments without user intervention. We settled on a technique of making the slope of the protein curve equal to −0.00007 between 1,750 and 1,800 wave numbers throughout all the experiments at each concentration and each temperature. Although this value was derived from user judgment to give a subtracted spectrum that matched that of a lyophilized sample, this single user-input value was used for the entire data set. Thus, we did not have the added subjectivity of manual adjustments for each melting curve that are often referred to as “baseline adjustments”.
2.2.3 Data Processing and Error Bars
We took the raw interferograms directly from the hardware (about 1,500 for the 70 min in the Spectrometer) and applied a Blackman window (alpha 0.16) and Fast Fourier Transform using the popular MATLAB software package (www.mathworks.com). The resultant protein spectra were smoothed using a nine point Savitsky–Golay function (four each side) and second derivative spectrum was then calculated. The thermal transition curves were plotted in Microcal Origin Software (version 7.0, Microcal Software Inc., Northampton, MA, USA) by using the absorbance values at 1,636 cm−1 (intramolecular β sheet of IgG) of the second derivative plots at each temperature.
We wanted to put error bars on the experimentation without repeating the time-consuming experimentation multiple times. These FTIR experiments with repeated scans are ideally suited to the re-sampling technique known as bootstrapping [6]. In this case, we took the individual spectra from each sample and by sampling the 10 background and 20 sample/buffer with replacement at each temperature we simulated the running of our experiments 100 times. We used the results from these 100 experiments to develop confidence intervals on the data which were ±2 standard deviations of the 100 simulated experiments. Each simulated experiment took 15 s to compute the results on a MacBook Pro with our custom MABLAB code.
3 Results and Discussion
3.1 pH Dependent Thermal Unfolding of IgG at 25 mg/mL Concentration
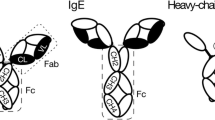
Bovine IgG at 25 mg/mL was prepared at pH 4.0–8.0 in the presence and absence of 100 mM Trehalose. The thermal unfolding of all samples between 25 and 95 °C was monitored by FTIR in the amide I region. Buffer subtraction and second derivative analysis were performed as described in Sect. 2.2. Assignment of Amide I protein peaks of bovine IgG according to the literature [8, 18, 21] is given in Table 1. The absorbance value at 1,636 cm−1 that corresponds to native β sheets of IgG was plotted against the temperature. The data points were fitted with sigmoid function according to Boltzmann equation using Origin software to calculate the apparent Tm values. A representative plot was shown for pH 7.0 (Fig. 2a) in the absence of Trehalose between 50 and 90 °C as the protein is thermally stable with subtle change in the secondary structure below 50 °C. All thermal unfolding curves were irreversible probably due to non two-state process of IgG. The calculated apparent Tm values are given in Table 2. A graphical representation of the comparison of the tabulated Tm values is given in Fig. 2b. Bovine IgG is relatively stable between pH 5.0 and 8.0 as evident from the transition temperatures. In the absence of Trehalose, the protein exhibited maximum thermal stability at pH 7.0 and minimum at pH 4.0. The decrease in the thermal stability at acidic pH could be attributed to the increased charge repulsions between the charged groups of IgG. The FTIR thermogram of bovine IgG displayed single main transition at all experimental conditions, the observation being consistent with similar study on another IgG [18]. Thermal denaturation of three humanized IgG1 antibodies and a monoclonal mouse anti-human antibody isotype 2 were conducted by DSC and CD which showed two main transitions between 60 and 80 °C that corresponded to the unfolding of Fab and Fc domains of IgG [12, 32].
3.2 Effect of 100 mM Trehalose on Thermal Unfolding of IgG at 25 mg/mL Concentration
Usually a therapeutic protein is formulated in various excipients that include amino acids, sugars, sugar alcohols, surfactants etc. to avoid major problems like aggregation and degradation that otherwise might lead to protein’s instability. Sugars are the most commonly used excipients to enhance the stability of drug candidates [2]. Out of various sugars, Trehalose is known to be an exceptional stabilizer of protein both in solution and in solid state [13]. The mechanism of protein structure stabilization by Trehalose can be explained by preferential exclusion phenomenon [16]. Protein is stabilized in the presence of Trehalose due to ordering of water molecules around it and Trehalose is preferentially excluded from protein surface’s vicinity. We used 100 mM Trehalose as an excipient in all protein samples and ran the thermal unfolding experiments. As can be evident from Fig. 1b, presence of Trehalose enhanced the thermal stability of IgG at all pHs. From Table 2, calculated ΔTm values for pHs 5.0–8.0 with and without trehalose were 1–2 °C while for pH 4.0 it was ~5 °C. Similar dominant effect of Trehalose at acidic pH was also observed in another study [14]. The reason for such observation could be explained as below. In general, a decrease in the pH is known to denature the protein and increase the hydrophobicity of the same. In the presence of an excipient (Trehalose, in our case) as protein’s hydrophobicity increases, more excipient molecules would be excluded from the macromolecule’s surface. This increased degree of exclusion would result in enhanced protein stability. Thus a relatively larger stability effect is seen at pH 4.0 for bovine IgG than at other pHs.
From pH screening, we observed that bovine IgG shows higher thermal stability at pH 7.0 (absence of Trehalose) and pH 6.0 (presence of Trehalose). Extreme acidic or basic solutions catalyze several degradation reactions like hydrolysis, aggregation, deamidation, racemization and β elimination of proteins. Hence slightly acidic pH of 6.0 is considered appropriate for IgG formulations to avoid such chemical and physical degradation [19]. We chose both pH 6.0 and 7.0 to further understand the effect of concentration on the thermal unfolding of Bovine IgG.
3.3 Concentration Dependent Thermal Unfolding of IgG at pH 6.0 and pH 7.0
The effect of increasing the protein concentration from 25 to 200 mg/mL on the thermal stability of IgG was studied at pH 6.0 and pH 7.0. The bar diagrams in Fig. 3 shows the effect of varying concentrations on the thermal stability of IgG. As the protein’s concentration was increased from 25 to 100 mg/mL, the Tm increased suggesting that higher concentrations have a positive impact on the thermal stability. This observation is consistent with the excluded volume theory which states that the unfolding of proteins at high concentrations should be reduced due to protein self association thereby increasing their conformational stability [20]. Such an increase in the transition temperature values also suggest that unfolding of secondary structure could dominate the thermal denaturation process. On the other hand, a small decrease in the Tm value at 200 mg/mL concentration required further analysis. Hence second derivative plots for the thermal unfolding curves of IgG were constructed. One such 3-D surface plot is shown in Fig. 4 for IgG at pH 7.0 and 100 mg/mL concentration. The plot was created using surface function (www.mathworks.com) in MATLAB. The clear shift in the wavenumber which in turn corresponds to the secondary structure is indicated by an arrow at higher temperatures. Similar plots at 100 and 200 mg/mL IgG concentration at pH 6.0 (Fig. 5a, b) were constructed for two temperatures 72 °C (solid trace) and 74 °C (dotted trace) around the transition zone. At both temperatures and concentrations, IgG existed as a mixture of aggregated β strands (1,625 cm−1), intramolecular β sheet (1,636 and 1,685 cm−1) and β turns (1,670 cm−1). At 74 °C, formation of intermolecular β sheet aggregates at 1,625 cm−1 is higher at 200 mg/mL that at 100 mg/mL IgG concentration. Enhancement in the appearance of aggregates at 200 mg/mL concentration may be used as an indication for the decrease in the thermal stability of the protein. Similar effect was seen at pH 7.0 (data not shown). Protein’s conformational and colloidal stability is a delicate balance between several factors such as concentration, pH, temperature, protein dynamics etc. [9, 10]. While a variety of biophysical techniques like spectroscopy, scattering and calorimetry have to be used to clearly understand the effect of concentrations on protein’s stability, in this particular study we have restricted to FTIR spectroscopy.
Second derivative surface plot of IgG at pH 7.0 (100 mg/mL concentration) constructed using surface function in MATLAB. Totally 35 absorbance curves are shown as the temperature is increased from 25 to 95 °C. The arrow indicates the wavenumber at 1,625 cm−1 (intermolecular β) that shifted from 1,636 cm−1 (intramolecular β) at higher temperatures due to aggregation of IgG. There are much smaller shifts that can be seen at other wavenumbers as explained in Fig. 6
Second derivative plots of IgG at 100 mg/mL a and 200 mg/mL b at pH 6.0 in the absence of Trehalose. In both plots, the curves correspond to 72 °C (solid) and 74 °C (dotted). The major peaks are intramolecular β sheet (1,636 and 1,685 cm−1), aggregated β strands (1,625 cm−1) and β turns (1,670 cm−1)
3.4 Conformational Changes of IgG at Higher Temperatures
The second derivative plots of 200 mg/mL of IgG at pH 6.0 and 7.0 in the absence of Trehalose (Fig. 6a, b) explain the structural changes of the protein samples during thermal unfolding. At both pHs, the protein exhibited major peaks at 1,636 and 1,685 cm−1 (intramolecular β sheet), 1,670 cm−1 (β turns), and 1,611 cm−1 (side chain vibrations or β sheets) at 60 and 70 °C. But when they were heated beyond 70 °C new peaks at 1,625 and 1,692 cm−1 appeared that correspond to β strand aggregates. At 80 °C, absence of the peak at 1,636 cm−1 indicates that all of the intramolecular β sheets have aggregated. Protein aggregation is the major event of physical instability. Completely folded or unfolded proteins do not aggregate easily as the hydrophobic side chains are either mostly buried or randomly scattered. Aggregation is initiated by partially unfolded protein chains. This is evident from the second derivative plots of IgG (Fig. 6). The partial unfolding of IgG close to the Tm values was immediately followed by an aggregation step and all the IgG molecules seem to have aggregated even before reaching 80 °C.
Second derivative plots of 200 mg/mL of bovine IgG at pH 6.0 a and pH 7.0 b in the absence of Trehalose. The curves correspond to 60 °C (solid), 70 °C (dash), 80 °C (dot) and 90 °C (dash dot). The major peaks are intramolecular β sheet (1,636 and 1,685 cm−1), aggregated β strands (1,625 and 1,692 cm−1), β turns (1,670 cm−1) and side chain vibrations (1,611 cm−1)
3.5 Minor structural Change in IgG Induced by Trehalose at Higher Temperatures
Concentration dependent thermal unfolding of IgG was also monitored in the presence of Trehalose (100 mM). Table 3 gives the Tm values calculated for all concentrations at pH 6.0 and 7.0 with and without the excipient. In Trehalose, IgG showed increased thermal resistance at all concentrations by 1–2 °C when compared to samples in the absence of the same. Again, decrease in the Tm values from 100 to 200 mg/mL protein concentration is observed even in the presence of excipient. When we analyzed the second derivative plots of IgG in Trehalose (Fig. 7a, b), we found a minor structural change induced by the excipient at higher temperatures. Even after crossing the transition temperature, unlike as seen in other unfolding curves in the absence of excipient, not all of the IgG molecules have seem to be aggregated. Instead, partial structure that remains as intramolecular β sheets (1,641 cm−1) until 80 °C gets converted to random coil (1,651 cm−1) at 90 °C. Errors calculated from the statistical bootstrapping method for the thermal curves (Fig. 7a, b) show that these calculated errors are significantly smaller than the intensity of the peaks at 1,641 and 1,651 cm−1. One interesting observation was that after heat treatment at all pHs without the excipient, IgG precipitated out of solution. But in the presence of Trehalose, the protein solution (partly) was still clear implying that if aggregation had occurred, it is invisible to eye. This observation strongly correlates to the formation of random coil at 1,651 cm−1 in the second derivative plot. In general, the IR amide I peak at 1,651 cm−1 can be assigned to either α-helix or random coil of a protein. Induction of α-helix at higher temperatures during thermal denaturation of IgG has been observed in other studies [31]. In our studies, we have assigned this peak to random coil due to the following reasons. A) This IgG peak was seen when heated only in the presence of Trehalose B) Similar study on bovine IgG has assigned the peak at 1,650 cm−1 to unordered structure [8].
Second derivative plots of 200 mg/mL of bovine IgG at pH 6.0 a and pH 7.0 b in the presence of Trehalose. The curves correspond to 60 °C (solid), 70 °C (dash), 80 °C (dot) and 90 °C (dash dot). The peaks are intramolecular β sheet (1,636, 1,641 and 1,685 cm−1), aggregated β strands (1,625 and 1,692 cm−1), β turns (1,670 cm−1), side chain vibrations (1,611 cm−1) and random coil (1,651 cm−1). The arrows indicate the two new peaks at 1,641 and 1,651 cm−1. The error bars for all the curves are calculated from 100 bootstrap simulations (±2 standard deviations)
According to the preferential exclusion theory, cosolvents like Trehalose are excluded from protein’s vicinity which results in the increased hydration of IgG by solvent molecules. This hydration effect could be responsible for not aggregating all of IgG molecules. If this assumption is true, it’s tempting to speculate that either increasing the concentration of Trehalose or trying other excipients like Sucrose might help to prevent the complete aggregation of IgG depending on protein’s concentration. In one of the recent efforts, lyophilized human growth hormone demonstrated a decreased amount of insoluble aggregates in sucrose than in trehalose formulation [25]. Protein aggregates specifically insoluble ones have profound effect on immunogenicity [24] and pose high risks in inducing immune responses associated with adverse clinical effects to the therapeutic protein product. Hence, it is particularly important to screen for excipients that could inhibit the formation of insoluble aggregates. Also, the shift in the frequency (blue shift) from 1,636 to 1,641 cm−1 at higher temperatures of IgG indicates the change in the structure of native β sheets associated with weak hydrogen bonding of the C=O group. These β sheets at 1,641 cm−1 (higher frequency) are less solvent exposed and more stable than those at 1,636 cm−1 (lower frequency) and hence require higher temperatures to denature them. Second derivative plots at all other conditions were similar implying that the concentration and pH had little or no impact over the alteration of the secondary structures of the protein.
Bovine Immunoglobulins provide antimicrobial protection, confer a passive immunity and protect infants from intestinal infections [1, 15, 27]. It was already shown that traditional milk processing that involves thermal treatments makes bovine IgG to loose its stability and activity [17]. Conformational changes are responsible for a protein to alter its properties. To understand the effect of pH, concentration and temperature on the conformational changes of bovine IgG, we used FTIR spectroscopy by modifying the routinely used hardware and software as described in Sect. 2.2.
FTIR is a valuable tool to study the conformational integrity of proteins at higher concentrations. In the Table 4 and chart (Fig. 8), we have compared the FTIR method with the two most popular techniques for measuring thermal melting curves, DSC and CD spectroscopy. As the exhibits show, the amount of sample used and concentration requirements often dictate the most appropriate method to use. At concentrations above 20 mg/mL (~0.13 mM for IgG), FTIR is often the only method available to labs with off-the-shelf instrumentation. We found that with the modified equipment and custom MATLAB software, a technician could take 24 FTIR thermal melting curves including analysis in 10 working days. This included software to automatically generate the error bars through bootstrapping. Once the single slope constant for water subtraction was selected the protocol did not have manual (subjective) changes (often called “corrections”) to the equipment readings. Through the modified FTIR method, we were able to understand what happens to the secondary structure of Bovine IgG when it is subjected to thermal stress under 26 different experimental conditions. From pH screening studies, we observed that the protein showed greater stability between pH 6.0 and pH 7.0. This finding of ours correlates well with a recent study where the average pH value of various bovine milk samples is measured and found to be 6.63 [29]. Also, this pH range is convenient for formulation studies that are widely carried out in the pharmaceutical industries. Presence of 100 mM Trehalose in all the protein samples increased their thermal stability and prevented the complete loss of IgG as insoluble aggregates. Such kinds of excipients suggest the advantage of preserving IgG’s activity during thermal processing thereby enhancing its therapeutic potential. An important point to be noted is that the protein aggregated immediately after crossing the transition temperature irrespective of the pH and concentration. Partial unfolding along with aggregation of bovine IgG could be the reason for losing its activity when heated above 70 °C as seen in other studies.
Abbreviations
- FTIR spectroscopy:
-
Fourier transform infrared spectroscopy
- Immunoglobulin G:
-
IgG
- CD:
-
Circular dichroism
- DSC:
-
Differential scanning calorimetry
- NMR:
-
Nuclear magnetic resonance
References
Davidson GP, Whyte PB, Daniels E, Franklin K, Nunan H, McCloud PI, Moore AG, Moore DJ (1989) Lancet 2:709–712
Devi VS, Chidi OO, Coleman D (2009) Spectroscopy 23:265–270
Dominguez E, Perez MD, Calvo M (1997) J Dairy Sci 80:3182–3187
Dominguez E, Perez MD, Puyol P, Sanchez L, Calvo M (2001) J Dairy Res 68:511–518
Dong A, Huang P, Caughey WS (1990) Biochemistry 29:3303–3308
Efron B, Tibshirani RJ (1993) In: Hall C (ed) An introduction to the bootstrap. Chapman and Hall/CRC Press, Boca Raton, pp 1–456
Gapper LW, Copestake DE, Otter DE, Indyk HE (2007) Anal Bioanal Chem 389:93–109
Gorga JC, Dong A, Manning MC, Woody RW, Caughey WS, Strominger JL (1989) Proc Natl Acad Sci USA 86:2321–2325
Guo J, Harn N, Robbins A, Dougherty R, Middaugh CR (2006) Biochemistry 45:8686–8696
Harn N, Allan C, Oliver C, Middaugh CR (2007) J Pharm Sci 96:532–546
Herron JN, Jiskoot W, Crommelin DJA (eds) (1995) Physical methods to characterize pharmaceutical proteins. Plenum Press, New York, pp 1–380
Ionescu RM, Vlasak J, Price C, Kirchmeier M (2008) J Pharm Sci 97:1414–1426
Jain NK, Roy I (2009) Protein Sci 18:24–36
Kaushik JK, Bhat R (2003) J Biol Chem 278:26458–26465
Korhonen H, Marnila P, Gill HS (2000) Br J Nutr 84(Suppl 1):S135–S146
Lee JC, Timasheff SN (1981) J Biol Chem 256:7193–7201
Li SQ, Bomser JA, Zhang QH (2005) J Agric Food Chem 53:663–670
Matheus S, Friess W, Mahler HC (2006) Pharm Res 23:1350–1363
Matheus S, Mahler HC, Friess W (2006) Pharm Res 23:1617–1627
Minton AP (2005) J Pharm Sci 94:1668–1675
Pelton JT, McLean LR (2000) Anal Biochem 277:167–176
Rahmelow K, Hubner W (1997) Appl Spectrosc 51:160–170
Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC (2005) Nat Biotechnol 23:1073–1078
Rosenberg AS (2006) AAPS J 8:E501–E507
Salnikova MS, Middaugh CR, Rytting JH (2008) Int J Pharm 358:108–113
Shire SJ, Shahrokh Z, Liu J (2004) J Pharm Sci 93:1390–1402
Tacket CO, Losonsky G, Link H, Hoang Y, Guesry P, Hilpert H, Levine MM (1988) N Engl J Med 318:1240–1243
Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC (1997) Nature 388:539–547
Tsioulpas A, Lewis MJ, Grandison AS (2007) Int J Dairy Technol 60:96–97
Venyaminov S, Prendergast FG (1997) Anal Biochem 248:234–245
Vermeer AW, Norde W (2000) Biophys J 78:394–404
Vermeer AW, Norde W, van Amerongen A (2000) Biophys J 79:2150–2154
Wang W, Singh S, Zeng DL, King K, Nema S (2007) J Pharm Sci 96:1–26
Wartewig S, Neubert RH (2005) Adv Drug Deliv Rev 57:1144–1170
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sathya Devi, V., Coleman, D.R. & Truntzer, J. Thermal Unfolding Curves of High Concentration Bovine IgG Measured by FTIR Spectroscopy. Protein J 30, 395–403 (2011). https://doi.org/10.1007/s10930-011-9344-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-011-9344-y