Abstract
Topotecan is a chemotherapeutic agent of choice for the second-line treatment of recurrent ovarian cancer. In this article, we have developed a physiologically based pharmacokinetic model to characterize and predict topotecan concentrations in mouse plasma and tissues. Single intravenous (IV) doses (5, 10 and 30 mg/kg) of topotecan were administered to male Swiss Webster mice, with plasma and tissue samples collected over 24 h, and with sample analysis by high performance liquid chromatography. Topotecan disposition in the lungs, heart, muscle, skin, spleen, gut, liver, brain and adipose was described by perfusion rate-limited compartments, whereas the testes and intraperitoneal (IP) fluid were described with permeability rate-limited compartments. The kidneys were modeled as a permeability rate-limited compartment with nonlinear efflux. The model included enterohepatic recycling of topotecan, with re-absorption of drug secreted in the bile and nonlinear bioavailability. Topotecan demonstrated dose-dependent, nonlinear pharmacokinetics and its elimination was described by nonlinear clearance from the liver and a parallel nonlinear and linear clearance from the kidneys. Mean tissue-to-plasma partition coefficients ranged from 0.123 (brain) to 55.3 (kidney). The model adequately characterized topotecan pharmacokinetics in plasma and tissue for all three doses. Additionally, the model provided good prediction of topotecan pharmacokinetics from several external data sets, including prediction of topotecan tissue pharmacokinetics following administration of 1 or 20 mg/kg IV, and prediction of plasma pharmacokinetics following doses of 1, 1.25, 15, 20 and 80 mg/kg IV and 20 mg/kg IP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Topotecan is a chemotherapeutic agent of choice for the second-line treatment of recurrent metastatic ovarian cancer [1, 2]. Although topotecan has been in clinical use for more than 10 years, there are several uncertainties and complexities associated with the disposition of the drug. Several preclinical and clinical investigations of topotecan plasma pharmacokinetics have been published, with some studies reporting linear kinetics [3–8] and with some studies showing dose dependent pharmacokinetics with saturable topotecan elimination [9, 10]. The major routes of elimination for topotecan are renal elimination and biliary secretion, with 26–80% of the dose recovered in urine and 18% of the dose eliminated in the bile [3, 11]. Although three topotecan metabolites have been identified (N-desmethyl topotecan, TPT-O-glucuronide and N-desmethyl TPT-O-glucuronide), [12, 13] only low concentrations of the metabolites have been found in blood, and the clinical relevance of topotecan metabolism appears to be limited [13]. Topotecan is a strong substrate for Breast Cancer Resistant Protein (BCRP) and a weak substrate for P-glycoprotein (P-gp) and Multidrug Resistance Protein 4 (MRP4) [14–17]. These drug efflux transporters mediate the biliary and renal secretion of topotecan. Given the capacity-limited nature of active secretion, it is possible that the reports of dose-dependent topotecan elimination relate to saturable biliary secretion and/or saturable renal secretion.
There have been few attempts to characterize the plasma pharmacokinetics of topotecan. Prior work has largely employed two-compartment open models of disposition, with linear or nonlinear elimination, to characterize data obtained from clinical and preclinical studies [7, 10, 18]. To the best of our knowledge, there have been no prior attempts to develop a physiologically-based pharmacokinetic (PBPK) model of the tissue disposition of topotecan. The purpose of this study was to establish a PBPK model to characterize the time-course of plasma and tissue topotecan exposure, following administration of different dosing levels in mice. The model was evaluated by comparison of model-predictions to external plasma and tissue topotecan concentration data, following administration of topotecan at doses of 1–80 mg/kg.
Materials and methods
Chemicals and animals
Topotecan, as the hydrochloride salt (SK&F S-104864-A), was obtained from Beta Pharma Inc. (New Haven, CT). Triethylamine, acridine, potassium dihydrogenphosphate, phosphoric acid and methanol were purchased from Sigma (St. Louis, MO, USA). The water used for HPLC was double distilled water. All other reagents were of analytical grade. Swiss Webster male mice (20–30 g, 4–5 weeks old, Harlan Laboratories, Inc., Indianapolis, IN) were used for PBPK studies. Protocols for animal use were approved by the Institutional Animal Care and Use Committee of the University at Buffalo. Mice were housed in a clean room on a standard light–dark cycle, with continuous access to food and water.
Dosing and sampling
Single IV bolus doses of topotecan (1, 5, 10, and 30 mg/kg) were given to mice via penile vein injection. For all four groups, three mice were killed and samples were collected at 10, 30 min, 1, 2, 4, 6, 8, 12 and 20 h. For the 10 and 30 mg/kg groups, samples were also collected at 24 h. At each time point, blood, lungs, heart, muscle, skin, spleen, gut, liver, kidney, brain, adipose, testes and IP fluid were collected. Plasma was obtained from blood by centrifugation at 10,000×g for 2 min. Where feasible, entire organs were collected and analyzed. Both lungs, testes, and kidneys were utilized for drug quantification. While collecting the liver, the gall bladder was carefully dissected and removed to avoid contamination liver samples with bile. The whole heart, spleen, liver and brain were used for drug quantification. Representative samples for the skin, skeletal muscle, and adipose tissues were collected. Tissue samples from the large and small intestines were obtained and, after careful removal of gut contents, combined for analysis. Samples of peritoneal fluid were collected with pre-weighed filter paper, and the volume was determined by weight, assuming a density of 1 g/ml. Plasma and all tissues were collected, weighed and stored at −20°C until further analysis.
Sample analysis by HPLC
Topotecan concentrations in mouse plasma, peritoneal fluid, and tissues were determined using a validated reversed-phase high-performance liquid chromatographic assay [19]. This assay utilizes an Agilent 1200 Series LC System, and measures the total topotecan concentration as the sum of the lactone and carboxylate forms of the drug. Separation was performed with a Nova-Pak C18 (3.9 × 150 mm, 5 μm) column heated to 35°C (Waters Corporation). Fluorescence detection was carried out at an excitation wavelength of 361 nm and an emission wavelength of 527 nm. The isocratic mobile phase consisted of a mixture of 10 mM KH2PO4 water–methanol–triethylamine (72:26:2, v/v/v, pH is adjusted to 3.5 by phosphoric acid). The flow rate was 1.0 ml/min and the injection volume was 10 μl. For preparing the calibration curve, 20 μl mouse blank plasma samples were spiked with 2 μl of the topotecan working solution and 2 μl of the internal standard (acridine) stock solution. The samples were then mixed with 56 μl of ice-cold methanol to precipitate protein, and acidified with 20 μl of 100 mM H3PO4 to convert topotecan carboxylate to topotecan lactone. All components of the mixture were vortexed for approximately 10 s and centrifuged at 10,000×g for 10 min at 4°C. The calibration curve was prepared using standards of 1, 5, 10, 50, 100, 500, 1000 and 2000 ng/ml. For sample analysis, 20 μl sample were mixed with 2 μl of the internal standard, 56 μl ice-cold methanol and 20 μl 100 mM H3PO4, and processed as described above. Tissue samples were prepared by homogenization into a known volume of saline using a Tissue Tearor® (Biospec Products, Bartlesville, OK; Model#398, 985370-395). IP fluid samples were prepared by homogenizing filter paper, which had been used to absorb IP fluid, into a known volume of 100 mM phosphoric acid. Fresh standard curves were prepared for each analytical run. The lower limit of quantitation was 1 ng/ml and recovery from tissue samples was more than 98%. Within- and between-day CV% were less than 13%, and the assay was linear over a concentration range of 1–2000 ng/ml.
Noncompartmental data analysis
All samples collected for development of the PBPK model were obtained via terminal sampling. Noncompartmental analysis of data, which included concentration measurements from three doses (5, 10 and 30 mg/kg), was carried out using the sparse sampling computation option available in WinNonlin (version 5.0, Pharsight Corporation, Palo Alto, CA). This approach calculates areas under the drug concentration versus time curve (AUC), and its standard error, via the Bailer-Satterthwaite method, which was developed for calculation of confidence interval for AUC calculated from destructive sampling [20, 21]. The tissue-to-plasma partition coefficients (K p) for each tissue, for each dose level, were calculated using the ratio of AUC for the tissue to the AUC of plasma. The variance of AUC ratio was calculated using the method of propagation of error:
where SEMTissue and SEMPlasma represent the standard error of the tissue and plasma AUC values obtained from noncompartmental analysis of the data. The K p (calculated as tissue-to-plasma AUC ratios) between different dose levels were compared using Student’s t-test.
Theoretical
Model development
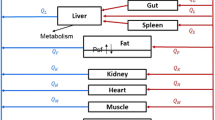
The PBPK model consists of 13 anatomical compartments (blood, lungs, heart, muscle, skin, spleen, gut, liver, kidney, brain, adipose, testes and IP fluid) connected to each other in an anatomical fashion via blood flow. The model structure is illustrated in Fig. 1 and all the physiological parameters are listed in Table 1. Attempts were made to develop a minimalistic model; the final model structure was based on visual inspection of the fit to the data, the value of Negative Log Likelihood, and the Akaike Information Criterion.
The whole body PBPK model for topotecan in mice. All organs are represented by a rectangular compartment and connected in an anatomical manner with blood flow represented by solid lines. The arrows represent the direction of the flow and the dashed line in the tissue compartment represents separation between vascular and extra vascular sub-compartments
It was assumed that topotecan distributes quickly within the blood, and plasma concentration was assumed to be equal to the blood concentration (i.e., no red blood cell compartment was added). Plasma protein binding of topotecan and the transition between the lactone and carboxylate forms of topotecan were not considered in the modeling effort. Topotecan was assumed to distribute rapidly to several tissues, including the lungs, heart, muscle, skin, spleen, gut, liver, brain and adipose. For these tissues, topotecan distribution kinetics were described by simple perfusion rate limited distribution models. Topotecan concentrations in IP fluid compartment were related to blood concentrations via a linear distributional clearance (CLDIP), which was estimated via model fitting. The fractional bioavailability for absorption of topotecan from the peritoneal compartment (BAIP) was fixed at 62%, based on our previous experimental results [19]. Topotecan concentration in the testes did not show a parallel profile with plasma concentrations, suggesting that topotecan uptake by testes is restricted by a blood-testis diffusion barrier. To account for the apparent distribution delay, a linear membrane permeability-limited sub-model was used to characterize topotecan concentrations in testes. The testes compartment was divided into vascular and extravascular sub-compartments, and theses two compartments were connected via a permeability-surface area product (PSTe), which was estimated via model fitting. Similarly, the kidney was also divided into vascular and extravascular sub-compartments. In order to complete the circulation circuit, venous returns from all tissues (except spleen and gut) were entered into the blood compartment followed by the lung compartment, which served as the source for arterial blood for all other tissue compartments.
Topotecan elimination was modeled from the kidney and liver via linear and non-linear processes. Linear elimination from the vascular compartment of the kidney was fixed to the value of the glomerular filtration rate (GFR) for mice, 0.015 l/h. Non-linear kidney elimination was described with a Michaelis-Menten function, and the associated parameters (\( V_{{{\text{m}}_{\text{Ki}} }} \) and \( K_{{{\text{m}}_{\text{Ki}} }} \)) were estimated via model fitting. Topotecan in the vascular compartment of the kidney was allowed to diffuse to the extravascular compartment, as described by the parameter CLDKi, which was estimated by model fitting. Topotecan in the extravascular kidney compartment was returned to vascular compartment via a nonlinear pathway, with parameters (\( V_{{{\text{m2}}_{\text{Ki}} }} \) and \( K_{{{\text{m2}}_{\text{Ki}} }} \)) estimated via model fitting.
Drug distribution to the liver was modeled with a perfusion rate-limited process, and venous returns from the gut and spleen were incorporated within the input function for the liver. The tissue-to-plasma partition coefficient for liver was described as a nonlinear function, with parameters (\( K_{{ 1_{\text{LiKp}} }} \), \( K_{{ 2_{\text{LiKp}} }} \)) estimated by model fitting. Topotecan elimination from the liver was assumed to proceed via Michaelis-Menten elimination kinetics, as described with the parameters \( V_{{{\text{m}}_{\text{Li}} }} \) and \( K_{{{\text{m}}_{\text{Li}} }} \). Topotecan eliminated from the liver was considered to be available for enterohepatic cycling. Briefly, drug eliminated from the liver enters a bile compartment, which empties into the gut lumen compartment. The bile and gut lumen were described as a series of transduction compartments, and the transit time (Tau) was estimated via model fitting. The partial absorption of topotecan was described by a nonlinear bioavailability function, using a single parameter (K F), which was estimated via model fitting.
Model equations
Mass balance for the lungs, in which topotecan concentration was described by the simple perfusion rate limited distribution model, was described as:
where V Lu represents volume for the lungs, C Lu represents concentration of topotecan in the lungs, C Pl represents concentration of topotecan in the plasma and \( K_{{{\text{p}}_{\text{Lu}} }} \) represents tissue-to-plasma partition coefficient for the lungs. Q He, Q Ki, Q Li, Q Te, Q Mu, Q Sk, Q Ad and Q Br represents blood flow for the heart, kidney, liver, testes, muscle, skin, adipose and brain.
For other tissues where topotecan distribution was perfusion rate-limited (heart, muscle, skin, spleen, gut, brain and adipose), mass balance was described as:
where V T represents total organ volume, Q T represents organ blood flow, C T represents concentration of topotecan in the tissue and \( K_{{{\text{p}}_{\text{T}} }} \) represents tissue-to-plasma partition coefficient for the given tissue.
For the intraperitoneal compartment, mass balance was described as:
where V IP represents volume of IP compartment, C IP represents concentration of topotecan in the IP compartment, \( K_{{{\text{p}}_{\text{IP}} }} \) represents tissue-to-plasma partition coefficient for IP, and CLDIP represents distributional clearance of topotecan between plasma and IP compartments.
Mass balance for the vascular compartment of the testes was described as:
and mass balance for the extravascular compartment of the testes was described as:
where \( V_{{{\text{v}}_{\text{Te}} }} \) and \( V_{{{\text{ev}}_{\text{Te}} }} \) represents the volume of the vascular and extravascular compartments. C Tev and C Teev represents topotecan concentrations in testes vascular and extravascular compartments. Q Te represents blood flow to the testes, PSTe represents permeability-surface area product for the testes, and \( K_{{{\text{p}}_{\text{Te}} }} \) represents tissue-to-plasma partition coefficient for testes.
Mass balance for the vascular compartment of kidney was described as:
and mass balance for the extravascular compartment of kidney was described as:
where \( V_{{{\text{v}}_{\text{Ki}} }} \) and \( V_{{{\text{ev}}_{\text{Ki}} }} \) represents volume of vascular and extravascular compartments for kidney. C Kiv and C Kiev represents topotecan concentrations in kidney vascular and extravascular compartments. Q Ki represents blood flow to the kidney, GFR represents the glomerular filtration rate, CLDKi represents the diffusion clearance of topotecan from the kidney vascular and extravascular compartments, and \( K_{{{\text{p}}_{\text{Ki}} }} \) represents the tissue-to-plasma partition coefficient for kidney. \( V_{{{\text{m}}_{\text{Ki}} }} \) and \( K_{{{\text{m}}_{\text{Ki}} }} \) represent the maximal velocity and affinity parameters for the elimination of topotecan from kidney through the Michaelis-Menten, non-linear elimination pathway. \( V_{{{\text{m2}}_{\text{Ki}} }} \) and \( K_{{{\text{m2}}_{\text{Ki}} }} \) represent parameters for the return of topotecan from the extravascular kidney compartment to vascular compartment.
The mass balance in the liver was described as:
where V Li represents liver volume, C Li represents topotecan concentrations in liver, C Sp represents topotecan concentrations in spleen, and C Gu represents topotecan concentrations in gut. Q Li, Q Gu and Q Sp represent blood flow to the liver, gut and spleen. \( K_{{{\text{p}}_{\text{Sp}} }} \) and \( K_{{{\text{p}}_{\text{Gu}} }} \) represent tissue-to-plasma partition coefficient for spleen and gut. \( V_{{{\text{m}}_{\text{Ki}} }} \) and \( K_{{{\text{m}}_{\text{Ki}} }} \) are the maximal velocity and affinity parameters for the elimination of topotecan from the liver. \( K_{{ 1_{\text{LiKp}} }} \) and \( K_{{ 2_{\text{LiKp}} }} \) were used to describe the tissue-to-plasma partition coefficient for the liver, as a nonlinear function of the concentration of topotecan present in the liver.
The mass balance in the gall bladder compartment and gut lumen compartments were described as:
where A Bile and A GutL represents amount of topotecan present in gall bladder and gut lumen compartments. Tau represents the transit time for the topotecan from gall bladder and gut lumen compartments.
The partial absorption of re-circulated topotecan from the gut lumen compartment to the systemic circulation was described as:
where V Blood represents volume of the blood and K F characterizes the nonlinear topotecan bioavailability.
Parameter estimation
All 13 estimated parameters (CLDIP, \( V_{{{\text{m}}_{\text{Ki}} }} \), \( K_{{{\text{m}}_{\text{Ki}} }} \), \( V_{{{\text{m2}}_{\text{Ki}} }} \), \( K_{{{\text{m2}}_{\text{Ki}} }} \), \( V_{{{\text{m}}_{\text{Li}} }} \), \( K_{{{\text{m}}_{\text{Li}} }} \), Tau, PSTe, CLDKi, \( P_{{ 1_{\text{LiKp}} }} \), \( P_{{ 2_{\text{LiKp}} }} \) and K F) were obtained by fitting all equations, simultaneously, to plasma and tissue data obtained from the 5, 10 and 30 mg/kg doses. Rather than taking the average of the three samples obtained at a given time point, all the data points were naively pooled and included in the data set used for parameter estimation. All the concentration values were defined in nM units. Parameters were fitted using maximum likelihood estimation method of the ADAPT-V program with the variance model:
where Y(t) is the model output at a given time t and Var(t) is the variance associated with the output. σIntercept and σSlope are the variance parameters representing a linear relationship between the standard deviation of the model output and Y(t).
Model evaluation
The main objective for the evaluation of the PBPK model was to test the prediction capability of the model for plasma and tissue concentrations of topotecan in mice. For this purpose, plasma or tissue pharmacokinetics data of topotecan in mice, after IV bolus administration of doses other than the ones used for model development, were either obtained from published reports using Graph-to-Digital Data Converter (Graph Digitizer®, Version 1.9, http://www.geocities.com/graphdigitizer/.htm) or by performing additional experiments. Plasma data from the literature were obtained for IV doses of 1.25 mg/kg [7], 15 mg/kg [22], 20 mg/kg [23] and 80 mg/kg [10]. Additionally, tissue data from literature was obtained following IV administration of 20 mg/kg [23]. The literature derived plasma and tissue concentrations data were then compared with the PBPK model simulated values. The model was also used to simulate the plasma pharmacokinetics of topotecan after IP administration of 20 mg/kg, and for simulation of plasma and tissue profiles for a 1 mg/kg dose. In each case, model predicted data were compared to observed data.
Results
The plasma concentration versus time profiles for topotecan after 5, 10 and 30 mg/kg IV bolus administration in Swiss Webster mice are shown in Fig. 2. Topotecan disposition was non-linear, and AUC values for the 5, 10 and 30 mg/kg doses were 760, 2297 and 47,263 (ng h)/ml, respectively. The apparent systemic clearance decreased dramatically with dose (6.58, 4.35 and 0.635 l/h/kg for 5, 10 and 30 mg/kg).
Plasma pharmacokinetics of topotecan in mice. Symbols represent observed data points for topotecan concentrations in plasma, after IV administration of the drug at different dose levels. Key: (filled circle) 5 mg/kg, (filled diamond) 10 mg/kg and (filled triangle) 30 mg/kg. The lines represent the best fit obtained for the data using the developed PBPK model. Key: (solid line) 5 mg/kg, (dotted line) 10 mg/kg and (dashed line) 30 mg/kg
In Fig. 2, the points represent experimental data and the solid lines depict the model fittings for the PBPK model. Overall, the plasma concentration versus time profiles were captured relatively well by the PBPK model. Compared to the 30 mg/kg dose, the plasma concentration versus time profiles for the 5 and 10 mg/kg doses were slightly overestimated at some time points. Tissue-to-plasma partition coefficients for all tissues, for all three doses, are shown in Table 2. The partition coefficients varied extensively from 0.123 (brain average K p) to 55.3 (kidney average K p). Estimated parameter values for the PBPK model are presented in Table 3.
Perfusion rate limited, non-eliminating organs
Topotecan concentration versus time data for the heart, lungs, gut, spleen, adipose, skin, brain and muscle, along with the model predicted profiles, are presented in Fig. 3. Concentration versus time curves for the organs declined in parallel with the plasma concentration of topotecan and, in general, the model was able to capture the overall behavior of the data very well. For the skin, the model-predicted topotecan tissue concentrations for the 30 mg/kg dose were over-predicted at later time points. Given the nature of the skin, it is possible that concentrations in skin tissue are under-measured due to poor tissue homogenization (which would not be well-appreciated through recovery experiments that assess drug extraction from skin tissue homogenates). Except for the brain and skin, tissue-to-plasma partition coefficients were more than unity, suggesting significant tissue binding. No parameters were estimated for these tissues, and the characterization of all concentrations in the tissues were driven by standard physiological parameters for mice.
The pharmacokinetics of topotecan in heart, lungs, gut, spleen, adipose, skin, brain and muscle (tissues that are described by the perfusion rate-limited sub-models). Symbols represent observed topotecan concentrations in tissues after IV administration of the drug at different dose levels. Key: (filled circle) 5 mg/kg, (filled diamond) 10 mg/kg and (filled triangle) 30 mg/kg. The lines represent the best fit obtained for the data using the developed PBPK model. Key: (solid line) 5 mg/kg, (dotted line) 10 mg/kg and (dashed line) 30 mg/kg
Permeability-limited tissues and elimination organs
Topotecan concentration versus time data for the testes, peritoneum, kidney, and liver, along with the model predicted profiles, are presented in Fig. 4. The model was able to capture the overall behavior of the data very well with the exception of initial time points for the kidney after 5 mg/kg, where an over-prediction of tissue concentrations was observed. Testes concentration versus time curves declined in a manner dissimilar to the plasma concentration of topotecan, clearly indicating permeability-limited drug disposition. The permeability-surface area product for testes (PSTe) was estimated to be 1.90E−04 l/h, which was ~2% of the value of blood flow to the testes. The PSTe was also estimated with a good precision and CV% was 6.13. The peritoneal compartment was connected to the plasma compartment via a distributional clearance (CLDIP), which was estimated with good precision (CV% of 27.5, final estimate: 1.52E−03 l/h). Parameters describing the nonlinear elimination of topotecan from kidney and liver were also estimated with good precision (Table 3). Parameters responsible for describing the rate of topotecan transfer from vascular to extravascular compartments (CLDKi), and from extravascular to vascular compartment of kidney (\( V_{{{\text{m2}}_{\text{Ki}} }} \)) were estimated with less precision (Table 3). The lack of availability of separate data for these two sub-compartments may explain the uncertainty in the estimated values. Parameters describing tissue-to-plasma partition coefficients for the liver (\( K_{{ 1_{\text{LiKp}} }} \), \( K_{{ 2_{\text{LiKp}} }} \)) were also estimated with good precision.
Time course of topotecan concentration in kidney, liver, testes and peritoneum (elimination organs and tissues described by permeability rate-limited sub-models). Symbols represent observed topotecan concentrations in tissues after IV administration of the drug at different dose levels. Key: (filled circle) 5 mg/kg, (filled diamond) 10 mg/kg and (filled triangle) 30 mg/kg. The lines represent the best fit obtained for the data using the developed PBPK model. Key: (solid line) 5 mg/kg, (dotted line) 10 mg/kg and (dashed line) 30 mg/kg
Enterohepatic circulation (EHC)
After a steep decline in plasma concentrations immediately after IV dosing (steep alpha phase), topotecan exhibited relatively prolonged exposure (shallow beta phase, Fig. 2). At low doses, there was virtually no elimination of drug from the body for hours, suggesting recycling of drug. This recycling was described as an enterohepatic cycling (EHC) process, and was characterized with a 2-compartment transit model. The transit time of topotecan through these two compartments was estimated to be 5.32 h (CV% 3.17). The partial absorption of recycled topotecan from gut lumen compartment was described by a nonlinear bioavailability function, and the parameter that relates topotecan bioavailability nonlinearly with the amount of drug present in gut lumen (K F) was estimated with a good precision (Table 3).
Model evaluation
Figure 5 shows digitalized data along with model simulations of topotecan plasma concentrations observed following IV bolus administration of 1.25, 15, 20 and 80 mg/kg doses in mice. The PBPK model was able to capture the experimental data for doses that fall within the dose range studied for model development (i.e., 15 and 20 mg/kg) very well. Even experimental data for the doses outside of the range of doses studied for model development (i.e., 1.25 and 80 mg/kg) were captured reasonably well. Overall, the model was able to provide reasonably accurate predictions of topotecan plasma concentrations for a wide range of different doses. Unfortunately, it is difficult to assess the significance of the differences between model predictions and the observed data, as the variability associated with the mean, observed data is not available. Figure 5 also shows comparison between plasma data obtained following IP administration of 20 mg/kg dose and simulations performed with the PBPK model. The model was found to predict the plasma pharmacokinetics of topotecan after IP administration of the drug very well.
Comparison of PBPK model-predicted and observed concentrations of topotecan in mouse plasma after IV or IP administration of the drug at different dose levels. For IV administered doses the solid circles represent mean concentrations of topotecan obtained from digitalization of literature data and solid lines represent the PBPK model predictions. For IP administered dose the solid circles represent observed mean experimental value with error bar representing standard deviation and the solid line represent the PBPK model prediction
The PBPK model was also evaluated for its accuracy in predicting topotecan tissue concentrations in mice following IV bolus administration of the drug. To the best of our knowledge there is only one study [23] that extensively describes data for changes in topotecan tissue concentrations with time, which was selected as our evaluation dataset. Figure 6 shows digitalized data, along with model simulations of topotecan tissue concentrations, following IV bolus administration of 20 mg/kg to mice. In general, the tissue concentration versus time profile was captured quite well by the PBPK model, with the exception of initial time points for the liver and the last time point for the kidney.
The model was further evaluated for its ability to predict the plasma and tissue concentration of topotecan at doses outside of the range used for model development. Figure 7 shows the experimental data following IV bolus administration of 1 mg/kg in mice along with model simulations. The plasma and tissue concentration versus time profiles were captured reasonably well by the PBPK model, with the exception of a few tissues like the adipose and kidney.
Discussion
The present work describes the development and evaluation of a PBPK model for topotecan disposition in plasma and in tissues after IV or IP administration to mice. The model will be used to facilitate the development and testing of an inverse targeting strategy [24, 25], where anti-topotecan antibodies will be administered to alter the time-course of topotecan disposition in systemic tissues.
In many respects, topotecan is a complex drug, and several simplifying assumptions have been made to limit the intricacy of the PBPK model structure. Topotecan is known to undergo reversible hydrolysis from the biologically active lactone form to an inactive carboxylate form. However, due to the rapidity of this process, and due to reports from several studies suggesting that effects correlate best with the total concentrations of topotecan (i.e., the sum of the lactone and carboxylate concentrations) [26, 27], we have only analyzed and characterized the pharmacokinetics of total topotecan in plasma and in tissues. Insignificant plasma protein binding [19] and rapid distribution of topotecan in red blood cells were assumed; hence, no effort was made to separate free and bound drug in plasma or blood. Topotecan is well-known to bind to its pharmacological target (topoisomerase-I) and make a triple complex with DNA [28], but no attempts were made to incorporate specific, saturable drug binding in tissues. Topotecan is also a substrate for several transporters (BCRP, P-glycoprotein, and MRP-4); however, due to the lack of literature data on the individual expression of these proteins, nonlinear topotecan efflux was modeled as though it was associated with a single transporter.
Few publications have investigated topotecan distribution in tissues; however, due to the promising cytotoxic effects of topotecan for brain tumors, coupled with poor brain penetration, the brain disposition of topotecan has been an active area of research. Attempts to characterize the brain exposure of topotecan have been limited to the application of simple compartmental models [29]. To our knowledge, no attempts have been made to model the disposition of topotecan in other tissues, and no prior attempts have been made to develop a PBPK model for topotecan.
The model described in the present work was used to characterize the tissue disposition of topotecan in several tissues (heart, lungs, gut, spleen, adipose, skin, brain and muscle), for three different doses (5, 10 and 30 mg/kg), simply based on physiological parameters obtained from literature and with the use of experimentally determined tissue-to-plasma concentration ratios. In general, topotecan disposition was well described for these tissues with the use of assumptions of perfusion-limited distribution. Of note, the low value brain to plasma K p observed in our experiments is consistent with reported values [15], and is believed to be related to BCRP1 and P-gp, two dominant transporters at the blood brain barrier that work in concert to efflux topotecan from the brain.
Because we are developing the PBPK model to predict and characterize the effect of the “inverse targeting” strategy, which uses IP route for drug administration, we have also attempted to characterize the disposition of topotecan in the peritoneum. The volume of the peritoneal compartment is thought to be dynamic and, unfortunately, a steady-state or time-averaged estimate of the peritoneal fluid volume has not been reported in the literature (to our knowledge). For the purpose of modeling, the volume of peritoneal cavity was fixed arbitrarily to 0.5 ml, and the distribution of topotecan between the peritoneal and plasma compartments was described with a simple, linear distribution clearance (CLDIP). Drug loss in the peritoneal cavity was considered by fixing the bioavailability of drug following IP dosing to 62%, and binding of drug in the peritoneal compartment was accounted for by using the experimentally determined average K p value. As shown in Fig. 4, the model was able to capture well the peritoneal disposition of topotecan. The model was also able to predict the plasma pharmacokinetics of topotecan after IP administration 20 mg/kg very well (Fig. 5), which provides some support for the assumptions made for the volume of peritoneal compartment and the fixed value of bioavailability.
As blood concentrations are the driving force for the tissue concentrations, it is extremely important for any PBPK model to capture well the plasma pharmacokinetics of drug. One of the major issues encountered while developing the present model was the substantial, dose-dependent nonlinearity of topotecan plasma disposition. Topotecan is a well-known substrate of transporters and the nonlinear behavior of drug was expected. However, apart from the apparent renal and total clearance values of topotecan, no information is available on the intrinsic clearance of topotecan from the liver or kidney. The renal clearance of topotecan is reported to exceed GFR, and it is known that the drug is subject to active renal secretion [7]. Hence, a nonlinear clearance pathway from kidney was included within the model, along with a linear pathway relating to GFR. Although not metabolized considerably in liver, topotecan is reported to be significantly eliminated through active, biliary secretion [11, 16]. Owing to an expectation for capacity-limited biliary secretion, the model includes a nonlinear elimination pathway from the liver. Despite taking into account all of the above mentioned possible governing factors for the plasma pharmacokinetics of topotecan, the preliminary model was not able to characterize the plasma pharmacokinetics of topotecan for all three doses simultaneously. Model-predictions for topotecan disposition could not replicate the observed slow, terminal decline of drug concentrations in plasma and in tissues. This problem was solved by incorporation entero-hepatic cycling, using a classic transit compartment approach [30]. Drug eliminated from the liver was collected in a gallbladder compartment and then transported to the gut lumen, as defined with a two-step transit process. Some of the drug secreted in bile is eliminated in the feces [11], and the remaining drug is absorbed into the systemic circulation in a concentration dependent manner [31]. Hence, the model incorporates an input function of topotecan from gut lumen to plasma, which includes a nonlinear function for bioavailability (Eq. 12). With inclusion of enterohepatic cycling kinetics, the model was able to characterize the plasma concentrations of topotecan reasonably well for all three doses, simultaneously (Fig. 2).
Despite good characterization of plasma topotecan disposition, initial versions of the model were unable to capture the tissue profiles of drug in the elimination organs (i.e., kidney and liver), using assumptions of flow-limited and linear tissue distribution. These two tissues were found to exhibit high tissue-to-plasma K p values, which demonstrated substantial dose-dependencies (Tables 1, 2). The dose-dependencies in tissue distribution may relate to saturable binding of the drug in the tissue or, perhaps, saturable uptake or efflux of drug due to capacity-limited drug transport [15]. Although several tissues showed a decrease in K p with dose, fixed values for K p were found to allow good characterization of tissue data for all tissues except the liver and the kidney. To improve the fit of liver data, a non-linear function (Eq. 9) was employed (Fig. 3). The kidney data were quite complex, showing a pattern similar to a downward parabola for changes in K p with increasing dose. We hypothesize that this pattern may be due to combination of nonlinear binding of drug in the tissue and involvement of an efflux transporter. Hence, to better characterize topotecan concentration in kidney, the organ was divided into vascular and extra-vascular sub-compartments, which were connected via linear and nonlinear transport pathways. The modified model was able to capture the kidney data of topotecan for 10 and 30 mg/kg doses well, but the parameters connecting sub-compartments were not estimated with good precision (Table 3). This likely relates to a lack of sufficient data to characterize the complexities associated with the transport processes.
The final model was evaluated for utility in predicting plasma and tissue concentrations of topotecan after IV or IP administration. Figure 5 shows the evaluation result for plasma pharmacokinetics of topotecan after 1.25, 15, 20 and 80 mg/kg IV bolus administration and 20 mg/kg IP bolus administration. As evident from Fig. 5, the model was able to predict the plasma pharmacokinetics of topotecan after IV or IP drug administration of different doses very well. Figure 6 shows the evaluation results for tissue pharmacokinetics of topotecan after a 20 mg/kg IV dose. As shown in Fig. 6, the model also satisfactorily predicted the tissue pharmacokinetics of topotecan from the literature, at a different dose level than those used for model development. Additional external validation, using data well below the range used in model development (i.e., 1 mg/kg), was also demonstrated (Fig. 7). In comparing observed data and model simulations, it is important to note that the 1 mg/kg dose used for evaluation is 5 times lower than the lowest dose used for model development, and hence nonlinear processes are expected to be much more prominent determinants of plasma and tissue exposure.
The PBPK model presented within this work, which has been structured to be consistent with known physiology and cellular processes impacting topotecan disposition, has been shown to be quite robust. It is anticipated that this model will help to facilitate the prediction of topotecan disposition within several drug targeting strategies that are currently under development. Additionally, due to its physiological basis, there is promise that the model may be quickly scaled to facilitate cross-species predictions.
References
Markman M (1997) Topotecan: an important new drug in the management of ovarian cancer. Semin Oncol 24(1 Suppl 5):S1–S5
Anand A, Chan SY (2004) The use of topotecan for relapsed ovarian cancer in accordance with the National Institute for Clinical Excellence guidance 2001: the Nottingham experience. Clin Oncol (R Coll Radiol) 16(8):543–548
Herben VM, ten Bokkel Huinink WW, Beijnen JH (1996) Clinical pharmacokinetics of topotecan. Clin Pharmacokinet 31(2):85–102
Takimoto CH, Arbuck SG (1997) Clinical status and optimal use of topotecan. Oncology (Williston Park) 11(11):1635–1646
Brogden RN, Wiseman LR (1998) Topotecan. A review of its potential in advanced ovarian cancer. Drugs 56(4):709–723
Kollmannsberger C, Mross K, Jakob A, Kanz L, Bokemeyer C (1999) Topotecan—a novel topoisomerase I inhibitor: pharmacology and clinical experience. Oncology 56(1):1–12
Zamboni WC, Houghton PJ, Johnson RK, Hulstein JL, Crom WR, Cheshire PJ, Hanna SK, Richmond LB, Luo X, Stewart CF (1998) Probenecid alters topotecan systemic and renal disposition by inhibiting renal tubular secretion. J Pharmacol Exp Ther 284(1):89–94
Guichard S, Montazeri A, Chatelut E, Hennebelle I, Bugat R, Canal P (2001) Schedule-dependent activity of topotecan in ovcar-3 ovarian carcinoma xenograft: pharmacokinetic and pharmacodynamic evaluation. Clin Cancer Res 7(10):3222–3228
van Warmerdam LJ, ten Bokkel Huinink WW, Rodenhuis S, Koier I, Davies BE, Rosing H, Maes RA, Beijnen JH (1995) Phase I clinical and pharmacokinetic study of topotecan administered by a 24-hour continuous infusion. J Clin Oncol 13(7):1768–1776
Chen J, Lu Q, Balthasar JP (2007) Mathematical modeling of topotecan pharmacokinetics and toxicodynamics in mice. J Pharmacokinet Pharmacodyn 34(6):829–847
Herben VM, Schoemaker E, Rosing H, van Zomeren DM, ten Bokkel Huinink WW, Dubbelman R, Hearn S, Schellens JH, Beijnen JH (2002) Urinary and fecal excretion of topotecan in patients with malignant solid tumours. Cancer Chemother Pharmacol 50(1):59–64
Rosing H, Herben VM, van Gortel-van Zomeren DM, Hop E, Kettenes-van den Bosch JJ, ten Bokkel Huinink WW, Beijnen JH (1997) Isolation and structural confirmation of n-desmethyl topotecan, a metabolite of topotecan. Cancer Chemother Pharmacol 39(6):498–504
Rosing H, van Zomeren DM, Doyle E, Bult A, Beijnen JH (1998) O-glucuronidation, a newly identified metabolic pathway for topotecan and n-desmethyl topotecan. Anticancer Drugs 9(7):587–592
Leggas M, Adachi M, Scheffer GL, Sun D, Wielinga P, Du G, Mercer KE, Zhuang Y, Panetta JC, Johnston B, Scheper RJ, Stewart CF, Schuetz JD (2004) Mrp4 confers resistance to topotecan and protects the brain from chemotherapy. Mol Cell Biol 24(17):7612–7621
de Vries NA, Zhao J, Kroon E, Buckle T, Beijnen JH, van Tellingen O (2007) P-glycoprotein and breast cancer resistance protein: two dominant transporters working together in limiting the brain penetration of topotecan. Clin Cancer Res 13(21):6440–6449
Jonker JW, Smit JW, Brinkhuis RF, Maliepaard M, Beijnen JH, Schellens JH, Schinkel AH (2000) Role of breast cancer resistance protein in the bioavailability and fetal penetration of topotecan. J Natl Cancer Inst 92(20):1651–1656
Hendricks CB, Rowinsky EK, Grochow LB, Donehower RC, Kaufmann SH (1992) Effect of P-glycoprotein expression on the accumulation and cytotoxicity of topotecan (SK&F 104864), a new camptothecin analogue. Cancer Res 52(8):2268–2278
Leger F, Loos WJ, Fourcade J, Bugat R, Goffinet M, Mathijssen RH, Verweij J, Sparreboom A, Chatelut E (2004) Factors affecting pharmacokinetic variability of oral topotecan: a population analysis. Br J Cancer 90(2):343–347
Chen J, Balthasar JP (2005) High-performance liquid chromatographic assay for the determination of total and free topotecan in the presence and absence of anti-topotecan antibodies in mouse plasma. J Chromatogr B 816(1–2):183–192
Nedelman JR, Gibiansky E, Lau DT (1995) Applying bailer’s method for AUC confidence intervals to sparse sampling. Pharm Res 12(1):124–128
Holder DJ (2001) Comments on nedelman and jia’s extension of satterthwaite’s approximation applied to pharmacokinetics. J Biopharm Stat 11(1–2):75–79
De Cesare M, Zunino F, Pace S, Pisano C, Pratesi G (2000) Efficacy and toxicity profile of oral topotecan in a panel of human tumour xenografts. Eur J Cancer 36(12):1558–1564
Fujita H, Okamoto M, Takao A, Abe H, Ishii R, Arase H (1995) Pharmacokinetics of SK & F 104864 in experimental animals. II. Tissue distribution. Gan To Kagaku Ryoho 22(12):1789–1792
Balthasar J, Fung HL (1994) Utilization of antidrug antibody fragments for the optimization of intraperitoneal drug therapy: studies using digoxin as a model drug. J Pharmacol Exp Ther 268(2):734–739
Balthasar JP, Fung HL (1996) Inverse targeting of peritoneal tumors: selective alteration of the disposition of methotrexate through the use of anti-methotrexate antibodies and antibody fragments. J Pharm Sci 85(10):1035–1043
O’Dwyer PJ, LaCreta FP, Haas NB, Halbherr T, Frucht H, Goosenberg E, Yao KS (1994) Clinical, pharmacokinetic and biological studies of topotecan. Cancer Chemother Pharmacol 34(Suppl):S46–S52
Grochow LB, Rowinsky EK, Johnson R, Ludeman S, Kaufmann SH, McCabe FL, Smith BR, Hurowitz L, DeLisa A, Donehower RC et al (1992) Pharmacokinetics and pharmacodynamics of topotecan in patients with advanced cancer. Drug Metab Dispos 20(5):706–713
Strel’tsov SA, Mikheikin AL, Grokhovskii SL, Oleinikov VA, Kudelina IA, Zhuze AL (2002) Interaction of topotecan, DNA topoisomerase I inhibitor, with double-stranded polydeoxyribonucleotides. 4. Topotecan binds preferably to the GC base pairs of DNA. Mol Biol (Mosk) 36(5):912–930
Zhuang Y, Fraga CH, Hubbard KE, Hagedorn N, Panetta JC, Waters CM, Stewart CF (2006) Topotecan central nervous system penetration is altered by a tyrosine kinase inhibitor. Cancer Res 66(23):11305–11313
Bischoff KB, Dedrick RL, Zaharko DS, Longstreth JA (1971) Methotrexate pharmacokinetics. J Pharm Sci 60(8):1128–1133
Yamagata T, Kusuhara H, Morishita M, Takayama K, Benameur H, Sugiyama Y (2007) Improvement of the oral drug absorption of topotecan through the inhibition of intestinal xenobiotic efflux transporter, breast cancer resistance protein, by excipients. Drug Metab Dispos 35(7):1142–1148
Fenneteau F, Turgeon J, Couture L, Michaud V, Li J, Nekka F (2009) Assessing drug distribution in tissues expressing P-glycoprotein through physiologically based pharmacokinetic modeling: model structure and parameters determination. Theor Biol Med Model 6(2):1–13
Baxter LT, Zhu H, Mackensen DG, Butler WF, Jain RK (1995) Biodistribution of monoclonal antibodies: scale-up from mouse to human using a physiologically based pharmacokinetic model. Cancer Res 55(20):4611–4622
Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP (1997) Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health 13(4):407–484
Davies B, Morris T (1993) Physiological parameters in laboratory animals and humans. Pharm Res 10(7):1093–1095
Fennell TR, Brown CD (2001) A physiologically based pharmacokinetic model for ethylene oxide in mouse, rat, and human. Toxicol Appl Pharmacol 173(3):161–175
Gearhart JM, Jepson GW, Clewell HJ, Andersen ME, Conolly RB (1994) Physiologically based pharmacokinetic model for the inhibition of acetylcholinesterase by organophosphate esters. Environ Health Perspect 102(Suppl 11):51–60
Acknowledgment
This work was supported by grant CA118213 from the National Cancer Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shah, D.K., Balthasar, J.P. Physiologically based pharmacokinetic model for topotecan in mice. J Pharmacokinet Pharmacodyn 38, 121–142 (2011). https://doi.org/10.1007/s10928-010-9181-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-010-9181-1