Abstract
Researchers studying mammalian dentitions from functional and adaptive perspectives increasingly have moved towards using dental topography measures that can be estimated from 3D surface scans, which do not require identification of specific homologous landmarks. Here we present molaR, a new R package designed to assist researchers in calculating four commonly used topographic measures: Dirichlet Normal Energy (DNE), Relief Index (RFI), Orientation Patch Count (OPC), and Orientation Patch Count Rotated (OPCR) from surface scans of teeth, enabling a unified application of these informative new metrics. In addition to providing topographic measuring tools, molaR has complimentary plotting functions enabling highly customizable visualization of results. This article gives a detailed description of the DNE measure, walks researchers through installing, operating, and troubleshooting molaR and its functions, and gives an example of a simple comparison that measured teeth of the primates Alouatta and Pithecia in molaR and other available software packages. molaR is a free and open source software extension, which can be found at the doi:10.13140/RG.2.1.3563.4961 (molaR v. 2.0) as well as on the Internet repository CRAN, which stores R packages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Researchers interested in the form-function relationship of mammalian dentitions have begun supplementing traditional morphological measures with new metrics designed to reflect aspects of occlusal surface topography (e.g., Zuccotti et al. 1998; M’Kirera and Ungar 2003; Evans et al. 2007; Boyer 2008; Plyusnin et al. 2008; Evans and Janis 2014; Winchester et al. 2014; Allen et al. 2015). Prominent among these new metrics are: Dirichlet Normal Energy ([DNE] Bunn et al. 2011), Relief Index ([RFI] Boyer 2008), and Orientation Patch Count ([OPC] Evans et al. 2007). These metrics assess different aspects of dental surface topography over entire tooth crowns (or tooth rows) without requiring identification of homologous anatomic features or investigator-defined landmarks. This shift to quantitative, homology-free methods has partly been brought about by advances in computing and scanning technologies, but is also related to the recognition of inter-observer differences when taking traditional measures (see Boyer et al. 2015a). Access to digitized, fine resolution 3D surfaces required for these analyses is rapidly increasing thanks mainly to two important factors: the proliferation and reduced costs of high fidelity scanners and computers needed to produce and process such data; and increased researcher sharing of such data sets through Internet repositories like MorphoSource (Copes et al. 2016).
RFI, OPC, and DNE each bring an informative perspective to dental topography studies. RFI is a ratio of two measures: the surface area of a tooth’s crown and the area of the tooth’s planometric footprint. To date, two different versions of RFI have been developed and have seen common usage in dental topography studies. The first, pioneered by Ungar and M’Kirera (2003), calculates a simple ratio of tooth crown surface area to 2D footprint area with the following formula:
Where A3D equals the three-dimensionally embedded surface area of the tooth, and A2D equals the two-dimensional tooth footprint area in occlusal view. Ungar and M’Kirera (2003) opted to crop the tooth at the lowest point of the occlusal basin, thereby focusing their measure of RFI on the tooth surfaces most likely to be involved in mastication. Boyer (2008) adapted the method to include the entire tooth crown (above the enameled cervix) and incorporated some transformations into the calculation. He proposed the calculation:
where A3D and A2D are equivalent to the values defined by Ungar and M’Kirera’s (2003) RFI calculation. When examining teeth of approximately the same crown height, both of these formulations of RFI will segregate teeth based on total occlusal surface complexity, with more complex occlusal surfaces producing higher RFI values. With Boyer’s (2008) inclusion of the full tooth crown, the latter formulation of RFI will tend to segregate hypsodont (high crowned) teeth apart from brachydont (low crowned) teeth, owing to the longer vertical sidewalls of hypsodont teeth (Boyer et al. 2010; Winchester et al. 2014). The main logic for Boyer’s (2008) formulation is that many non-anthropoid teeth are poorly represented using Ungar and M’Kirera’s (2003) criterion since the functional surfaces of a tooth can extend well below the bottom of the occlusal basin. The decision to use a natural logarithm protects against potential deviations from normality in data sets that represent a large diversity of taxa and a larger range of values than might be expected for analyses restricted to primate clades.
OPC is a measure of the number of separately oriented facets on a tooth surface. It is measured by dividing a tooth surface into contiguous patches sharing an orientation and then summing the number of created patches (Evans et al. 2007). First, each portion of the surface is assigned into one of eight X-Y plane directional categories (similar to North, Northeast, East, Southeast, etc., on a compass), with each category comprising a 45° sector around the central vertical axis (Z-axis) of the tooth. A simple cone-shaped tooth is expected to have an OPC value of eight because a relatively smooth cone-shaped tooth surface will be arbitrarily divided into patches every 45° around the tooth. The surface of a more complex tooth will have more separately oriented contiguous patches, resulting in a higher OPC value.
Dirichlet Normal Energy (DNE) in particular is worth further discussion and explanation. DNE measures the curvature and undulation of a surface (Bunn et al. 2011). DNE over the whole surface is an integral equation calculated with the following formula:
Where e(p) is the Dirichlet Energy Density (different from DNE) at a given point p on a surface, multiplied by the point’s area (although points have infinitesimally small areas, the sum of all the points’ areas on a surface is—in theory—equal to the total area of the surface).
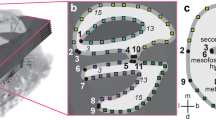
To understand the DNE calculation, consider a unit hemisphere with a radius (r) of 1 (refer to Fig. 1). The Dirichlet Energy Density of a given point is equivalent to the sum of the squared differences between the point’s two principal curvatures and the curvature of the point’s tangent plane (the curvature of a plane—tangent or otherwise—is always zero). Every point (p, Fig. 1a) has an associated tangent plane (t, ghosted above the hemisphere in Fig. 1a), and two principal curvatures, which together describe the shape of the surface about the point (p). In Fig. 1, the two principal curvatures about point p are highlighted as occurring in the planes u (blue) and v (red). The curvature of a surface in any given direction at a point is expressed as the reciprocal of the radius of the largest possible osculating circle (i.e., ‘kissing’ circle that most tightly fits the contour of the surface in a given direction, normal to the surface at the point of interest). The osculating circles associated with the two principal curvatures at p are shown in profile in Fig. 1b. The two principal curvatures about point p in Fig. 1 are both −1 (i.e., \( \frac{-1}{r} \) with r = 1), thus the sum of the squared differences about point p—which is equivalent to the point’s Dirichlet Energy Density—equals 2 (i.e., \( {\left(\frac{-1}{r}\right)}^2+{\left(\frac{-1}{r}\right)}^2 \) where r = 1). Principle curvatures are assigned a positive sign if the osculating circle fits above the surface, and negative if below. Therefore the principal curvatures on the outer surface of a dome (like in this example) would be negative, while the principle curvatures on the inner surface of a bowl would be positive.
a A hypothetical unit hemisphere (r = 1) for demonstrating the calculation of Dirichlet Normal Energy (DNE). The point p is indicated on top of the hemisphere, and the two principal curvatures about point p occur on the surface of the hemisphere in the planes u (blue) and v (red). The tangent plane (t) associated with p is displayed as semi-transparent above the hemisphere. b Profile views of the two osculating circles associated with the principal curvatures in planes u and v can be seen with colors coordinated respective to the planes in a. Note that each osculating circle has a radius of 1. Because the example put forward here is of a hemisphere, where the curvature of the surface is equal in all directions, the directions on the surface of the principal curvatures about p are arbitrary. Otherwise the two principal curvatures about a point occur in the normal planes capturing the largest and smallest curvatures, respectively, at p. The total surface DNE of this (and all) hemispheres is ~12.566 (or 4 × π, see text for further explanation)
Like p, all other points throughout the hemisphere in Fig. 1 have Dirichlet Energy Density values of 2. Therefore, the Dirichlet Normal Energy of the surface, which is the integral of the Dirichlet Energy Density values across the entire surface, is equivalent to:
resulting in a total DNE value of ~12.566 (or 4 × π), when r = 1 (note that, 2πr 2 is the surface area formula for a hemisphere). By comparison, a flat plane is expected to have a DNE value of 0, regardless of its size. This follows from the Dirichlet Energy Density throughout a plane equaling 0 (the radii of the osculating circles about the principal curvatures of a plane are infinitely large, resulting in principal curvature values of 0). Like the flat plane (or any other surface for that matter), an interesting observation to draw from Eq. (4) is that the size of the hemisphere does not affect the DNE value. The radius of the hemisphere is off-set between the Dirichlet Energy Density and the surface area components of the equation. In this way DNE is purely a measure of the surface’s complexity and curvature, independent of its size and orientation, making it a valuable addition to the dental morphologist’s tool kit (see also Bunn et al. 2011).
These new measures have proven effective for quantitatively distinguishing among the lower molar surfaces of primate species processing differing diets (Boyer 2008; Bunn et al. 2011; Winchester et al. 2014), and at demonstrating adaptive and presumed functional convergences among rodent and carnivoran dentitions in taxa consuming similar ratios of plant/animal matter (Evans et al. 2007). A promising avenue of further development for the above-mentioned measurements is their potential advantage for studying tooth crowns of mammals that lack obviously homologous features—such as rodents, multituberculates, and xenarthrans—whose highly derived crown surface structures bear only remote resemblances to the tribosphenic molar pattern of their ancestors (Bargo et al. 2009; Wilson et al. 2012). Moreover, it is also possible to examine evolutionary series of taxa who resemble one another in occlusal morphology but exhibit evolutionary trends in crown height or surface complexity. The benefits of these homology-free measures have recently been employed in studies involving fossil taxa (e.g., Wilson et al. 2012; Prufrock et al. 2016). However, access to software for performing these nascent analyses has lagged behind the availability of the specimens, scans, and recognition of their usefulness. Although one stand-alone open source software package capable of performing all of these measures has recently become available (MorphoTester http://morphotester.apotropa.com; Winchester 2016); and while there are other packages capable of performing some of these metrics (e.g., Teether; Bunn et al. 2011, and SurferManipulator available at site maintained by A. Evans: http://evomorph.org/surfermanipulator; Evans et al. 2007), there remain significant advantages to implementing these methods in the R scientific computing language (R Core Team 2015).
R provides a flexible and powerful set of tools for data analysis, and offers broad accessibility across computing platforms. R has a wide network of users and contributors, so producing an R package aimed at performing these dental measures may also serve as a seminal platform for extending these metrics and their applications. Other users may develop creative and unanticipated manipulations or extensions of the metrics or their components (or the code). R’s framework allows users to easily build on packages using other preexisting packages or code excerpts (or write their own), an important feature of the R platform and user network helping make it a powerful, versatile, and popular emerging scientific computing language. Additionally, implementing these analyses in R allows for taking advantage of the many powerful R graphics packages and tools. R’s graphics capabilities, utilized here, facilitate a greater understanding and interpretation of DNE, RFI, and OPC. Here we present molaR, a new R package that provides functions for measuring and plotting DNE (Bunn et al. 2011), RFI following Boyer (2008), and a non-rasterized variant of OPC (Evans et al. 2007), on three dimensional surface scans of teeth.
Description
The functions in molaR (outlined in Table 1) are written in the scientific computing language R (R Core Team 2015), and require R version >=3.2.3. The other R packages required to operate molaR include: rgl (Adler et al. 2015), alphahull (Pateiro-Lopez and Rodriguez-Casal 2015), geomorph (Adams and Otárola-Castillo 2013), psych (Revelle 2015), Rvcg (Schlager 2015a), and their dependencies. molaR automatically calls for the installation of these packages upon adding molaR to the user’s library, and also calls them when loading molaR. Importantly, installing and operating rgl requires X11 (XQuartz). Formerly, X11 was a standard component of both the Mac and Windows operating systems. However, since the release of Yosemite (OS X 10.10) for Mac, X11 is no longer included and older versions of X11 are no longer integrated with the Mac operating system. Mac users operating new machines using Yosemite or later, or having upgraded older machines to Yosemite or later, who are trying to install and use rgl or any packages dependent on rgl—such as molaR—will also need to manually install the latest version of X11 (available here: http://www.xquartz.org/). Additionally, alterations in the X11-Mac OS interface have caused some distortions when generating legends in molaR plots (with molaR v. 1.0), which have been repaired with the molaR v. 2.0 release (see molaR Quick Start.R available at the doi:10.13140/RG.2.1.3284.9687 for more details).
Installing and Loading molaR
Users interested in installing molaR to their local R library can do so in two ways. First [1], users can download and locally install the appropriate source binary (either Windows or Mac), or tarball from the CRAN web page: http://cran.fhcrc.org/web/packages/molaR/index.html, or download the tarball from the following DOI: 10.13140/RG.2.1.3563.4961. Alternatively [2], users operating a machine connected to the internet can directly install molaR while using R by running the command: install.packages(‘molaR’) directly in the R console. We also provide a quick-start script which walks users through installing molaR and running its functions on an example file, the script contains additional troubleshooting information and is available at the following DOI: 10.13140/RG.2.1.3284.9687.
Examples
The following demonstrations are aimed simultaneously at assisting users in operating and trouble shooting the functions provided in molaR and illustrating the power of the program for visualizing a mammalian tooth crown. The functions in molaR analyze three-dimensional triangular polygon mesh files (PLY files). 3D mesh files must be formatted a specific way to use molaR, and most users can conduct this pre-processing in Avizo or Amira (FEI Visualization Sciences Group, Berlin, Germany). For users desiring a free, open source solution, MeshLab (http://meshlab.sourceforge.net/) has a full complement of mesh surface processing features. Surfaces imported into R for use in molaR must be triangular PLY (Stanford polygon file format) files, of either ASCII or binary format (ASCII and binary files will need to be imported differently, see below). Prior to import, it is recommended that surfaces be simplified to roughly ~10,000 faces (in order to maintain consistency with past projects), and they should be properly oriented (cf. Boyer 2008; Bunn et al. 2011; and Winchester et al. 2014 who did not simplify prior to RFI analysis). A surface is properly oriented for use with molaR if the occlusal plane is parallel with the X- and Y-axes, and perpendicular to the Z-axis (the normal of the occlusal plane can be positive or negative with regards to the Z-axis). Batches of dental surface scans can be uniformly oriented using the R package auto3dgm (Boyer et al. 2015b) and post-hoc batch rotated into the proper orientation using software such as MeshLab or Avizo to ensure consistency among specimens. It is also recommended that scans be smoothed using Avizo’s smoothing functions, although this is not required and should be done at the user’s discretion (see below). Previous work has cropped surface scans to include either [1] the tooth crown surface above the lowest point on an occlusal basin (e.g., Zuccotti et al. 1998; M’Kirera and Ungar 2003) or [2] the entire tooth crown (including the enameled cervix) (e.g., Boyer 2008; Bunn et al. 2011; Winchester et al. 2014). Arguments for choice of cropping can be found in the respective studies. Users interested in manipulating their PLY files in R to accommodate these different cropping approaches should explore the tools included in the Morpho package (Schlager 2015b).
Importing Scans for use in molaR
Once scans are pre-processed, ASCII tooth surface files can be imported either using the read.ply function from the geomorph package (Adams and Otárola-Castillo 2013), or if they were pre-processed using Avizo, with the read.AVIZO.ply function included in molaR. Both of these functions will automatically generate an image of the PLY file in an RGL device window, allowing users to preview the surface prior to analysis. Binary PLY files can be imported using vcgPlyRead contained in the R package Rvcg (Schlager 2015a). The vcgPlyRead function can import scans of any kind, and will automatically repair scans upon import (i.e., by removing any floating vertices); however, vcgPlyRead does not automatically offer a preview of the scan, which may be advantageous in some applications.
Calculating, Plotting, and Troubleshooting DNE
Dirichlet Normal Energy (DNE) can be calculated on imported scans using the DNE function in molaR. DNE is calculated following Bunn et al. (2011) using the formula:
Where the function e estimates the energy density on each individual triangular face of the surface. The energy density for any given face is estimated as:
where \( \mathbf{G}=\left(\begin{array}{ll}{}_{\left\langle u,v\right\rangle}^{\left\langle u,u\right\rangle}\hfill & {}_{\left\langle v,u\right\rangle}^{\left\langle u,v\right\rangle}\hfill \end{array}\right) \), and \( \mathbf{H}=\left(\begin{array}{ll}{}_{\left\langle {n}_u,{n}_v\right\rangle}^{\left\langle {n}_u,{n}_u\right\rangle}\hfill & {}_{\left\langle {n}_v,{n}_v\right\rangle}^{\left\langle {n}_u,{n}_v\right\rangle}\hfill \end{array}\right) \). The variables u and v represent edge vectors on a given triangular face originating from some point p occurring at one vertex of the triangular face being analyzed. The variables n u and n v are derivatives of the normal n in the directions u and v, respectively (Bunn et al. 2011). There are two reasons these calculations appear different from the DNE calculations presented in the introduction. First, due to the limits of computer processing, dental (and other) scans cannot be analyzed as continuous surfaces. That is, the surface must be discretized into measureable components. molaR and other programs like it take advantage of the Stanford PLY file format, which divides a surface up into small triangular faces. The DNE function in molaR treats each of these faces as a stand-in for the surface points described in the DNE discussion in the introduction (description accompanied by Fig. 1). Equation (5) above is therefore calculating the Dirichlet Energy Density, and area for each face. Second, the surface of a biological specimen is irregular and cannot be described with a simple mathematical formula (unlike a hemisphere or plane). So the sum of squared differences between the radii of the osculating circles and the tangent plane at the junctions of each face must be estimated. molaR’s DNE function accomplishes this by comparing each triangular face’s normal with the face’s vertex normals. Each vertex normal is an average of the normals of the adjoining faces. The issues arising from this estimation are overcome with the matrix algebra in Eq. (6) (for more information see Bunn et al. 2011; Winchester 2016).
In addition to the above calculation alterations, the Total Surface DNE calculation excludes two types of faces from the final summation: [1] Boundary faces—defined as triangular faces having an edge (i.e., two vertices) on the boundary of the surface—are excluded from the total surface DNE calculation on the basis that their boundary status makes calculating the vertex normals too unreliable, potentially overestimating Dirichlet Energy Density values. [2] Faces containing the highest 0.1 % (the 99.9th percentile) of Dirichlet Energy Densities are also excluded. Occasionally faces located in deep recesses of the surface or faces erroneously produced during surface reconstruction from scan data return extreme outlier energy density values, which both greatly alter the total energy value of the surface (sometimes doubling or tripling the total value), and are of biologically dubious significance. On a typical ~10,000 face PLY, there tend to be only a small handful of extreme values (three to five faces) worth excluding. Therefore, a threshold of 0.1 % (approximately ten faces) usually captures these faces without excluding too much of the surface, although users can specify the percentile exclusion threshold with the outliers argument. The DNE function provides these faces and their values in a list, and users can re-incorporate these faces into the DNE calculation at their discretion (see below). The DNE function also incorporates a condition-checking step to ensure the calculation of the G identity matrix exhibits sufficient backwards stability (for more details see Winchester 2016). The function DNE initially prints the total surface DNE to the R console, and returns a list object containing several important elements outlined in Table 2. These elements include information on the faces excluded from the total surface energy summation, which users can evaluate and potentially re-incorporate into their analyses.
Some aspects of the total number of faces included in the PLY are worth considering in the broader context of the DNE calculation. As is demonstrated in the introduction, DNE is (in theory) unaffected by the size of the surface, therefore intuition suggests that the number of faces on a PLY surface should have no affect on the calculation of DNE. This is only partly true however. The DNE calculation could be significantly altered if the surface is overly simplified resulting in a PLY file that does not accurately represent the contours of the original specimen. That is, if a surface is represented by too few polygon faces to accurately capture all of the relevant curvature on the surface, then DNE may be underestimated as simplification obliterates surface details. In this regard, the ~10,000 face standard has been demonstrated to provide sufficient and biologically meaningful accuracy in measuring dental specimens (Bunn et al. 2011; Winchester 2016). However, if two PLY files represent a surface equally accurately, yet one of them has significantly fewer faces, the DNE calculation on the file containing fewer faces will tend to be lower. This is expected due to the exclusion of boundary faces from the summation in the molaR DNE function. As a PLY file contains more and more faces, the faces on the boundary will come to represent a smaller and smaller percentage of the total DNE surface calculation (see Winchester 2016). Although a larger number of faces may more accurately represent the DNE value of a surface, this will come at a cost of processing speed, and there appear to be rapidly diminishing returns beyond the ~10,000 face standard. Furthermore, users should consider the role the Dirichlet Energy Density outliers play when surfaces contain far more than the recommend ~10,000 faces. Implicit in the removal of these outlier faces is the acknowledgement that a perfect representation of the surface—and therefore exact calculation of DNE—is not possible. Thus, the ~10,000 face recommendation is a compromise between biologically meaningful accuracy and processing speed, and is consistent with previously published work (e.g., Bunn et al. 2011; Winchester et al. 2014; Prufrock et al. 2016; Winchester 2016).
The DNE3d function allows users to project Dirichlet Energy Values (Dirichlet Energy Density × Face Area) onto a three-dimensional surface image (Fig. 2). DNE3d requires an object created by the function DNE, and has several arguments that allow users to customize their plots (see Table 3). In cases where two teeth display substantially different total surface DNE values, coloration in plots obtained from DNE3d may not provide a direct comparison between the two teeth, because DNE3d defaults to setting a relative color range for each surface. The setRange argument allows users to manually set the range of energy values displayed on the surface. By default, the DNE3d graphics function represents the surface energy on a log-scale. Users will find that there is a significant right-tail skew associated with energy values, and plotting these values on a linear scale obscures differences in the lower values making surfaces appear more homogenous (see Fig. 2a and b). This default can be overridden with the logColors logical argument.
Surface views of two lower first molar teeth from mantled howler monkeys (Alouatta palliata) from the Duke Evolutionary Anthropology Department Comparative Collection (a & b: DU-LP 07; and c & d: DU-LP 09) with variable levels of wear and total DNE. a: DNE values per PLY face of a worn A. palliata tooth in linear scale. b: DNE values per PLY face for the same tooth pictured in a but in log-scale. Due to the significant right tail skew of DNE face values, distinguishing DNE surface complexity is difficult without log scale, as can be seen in comparing a & b. c: Automatically set log-scaled view of DNE values per PLY face for a relatively unworn A. palliata tooth. d: Manually set log-scaled view of DNE values per PLY face for the same tooth pictured in c. Tooth in d is scaled to match DNE face values range for tooth pictured in b. Direct comparisons can be made between b & d unlike between b & c. Total surface DNE value for tooth pictured in a & b is: 310.651, (specimen number: DU-LP 07). Total surface DNE value for tooth pictured in c & d is: 174.556, (specimen number: DU-LP 09)
Two types of errors can occur when attempting to run the DNE function. First, an error stating: Error in ‘$< −.data.frame’(‘*tmp*’, “Dirichlet_Energy_Densities”, value = 0): replacement has 1 row, data has 0 will occur when the surface has no boundaries (i.e., is an enclosed object). In this case, it is recommended that the file be cropped to only include the enamel cap (or a hole be opened somewhere on the surface). Another potential error states: Error in if (which(pts == i) == 1) { : argument is of length zero. This error occurs when the PLY file contains a vertex that has no faces associated with it (i.e., a floating point not associated with the rest of the surface). This can sometimes occur when cropping and processing CT-scans into PLY files. In this case, it is recommended that the file be re-cropped, being sure to eliminate all points not contained on the surface of interest. Alternatively, the user can read the PLY file into R with the function vcgPlyRead from the Rvcg package. This function will automatically eliminate ‘floating’ vertices.
Calculating, Plotting, and Troubleshooting RFI
Relief Index (RFI) can be calculated on imported scans using the RFI function. RFI in molaR is calculated following Boyer (2008) using the formula from Eq. (2) above. The RFI function automatically prints the RFI value, three-dimensionally and two-dimensionally embedded areas of the surface to the console. RFI also returns a list object containing the seven subsidiary objects outlined in Table 4. Researchers desiring to use Ungar and M’Kirera’s (2003) original formulation of RFI can extract the un-transformed 3D and 2D area measures from the RFI output.
The plotting function RFI3d enables users to visualize the RFI calculation by displaying the three-dimensional surface juxtaposed to its two-dimensional footprint (Fig. 3). RFI3d requires an object created by the RFI function. It also contains several arguments that allow users to customize their plots (see Table 5). Users interested in only displaying the tooth’s footprint can make the tooth surface completely transparent by setting Opacity = 0.
Surface view of mantled howler monkey lower first molar (specimen number: DU-LP 09) with surface footprint. a Opacity of tooth surface set to 1 (completely opaque). b Same tooth with opacity of tooth surface set to 0.5 (semi-transparent). The footprint is projected below and colored blue in both plots
The RFI function (like OPC, see below) is highly sensitive to the orientation of the imported surface. To accurately calculate the projected two-dimensional area it is critical that the occlusal plane of the tooth be perpendicular to the Z-axis (positive–negative orientation is irrelevant). Deviations from this condition will cause errors in the footprint area calculation. When analyzing teeth without the occlusal surface normal to the Z-axis (or nearly so) or when surfaces have not been properly simplified, the RFI function may display this error: Error in tri.mesh(X) : duplicate data points. The RFI function relies on an alpha-convex hull algorithm contained in the alphahull package (Pateiro-Lopez and Rodriguez-Casal 2015). This alpha hull algorithm requires unique (no duplicate) points when calculating the two-dimensional surface footprint. The above error may occur during the execution of the alpha hull algorithm when either the projection of a misaligned surface or the failure to simplify to ~10,000 faces cause more than one point to occupy the same X-Y coordinates on the footprint boundary. Proper alignment and surface simplification will usually correct this error.
Calculating, Plotting, and Troubleshooting OPC and OPCR
Orientation Patch Count (OPC), originally developed by Evans et al. (2007), is a count of the number of separately oriented ‘facets’ or ‘patches’ on a tooth surface. Each triangular face on the mesh surface is binned into one of eight directions, corresponding to the direction on the X-Y plane in which the face is oriented (Evans et al. 2007). After each face is binned, contiguous faces that share an orientation are grouped into a common patch. The sum of the patches yields the OPC value. Imported surfaces can be analyzed for OPC using the OPC function and associated arguments in molaR (Table 6). The OPC function allows users to specify patch count inclusion criteria using two up-front arguments: [1] minimum_faces which sets the minimum number of surface triangles a patch must have in order to be counted (defaults to three or more to maintain consistency with MorphoTester; Winchester 2016), and [2] minimum_area which specifies a minimum percentage of total surface area a patch must occupy in order to be counted. Engaging minimum_area automatically disables minimum_faces, and so the two cannot be used simultaneously. Upon completion, the OPC function prints the total patch count, and the total number of patches per orientation to the console; it also returns a large and highly structured object whose contents are worth exploring and are explained in Table 7.
Orientation Patch Count Rotated (OPCR) is a modification of OPC that accounts for variations in OPC values arbitrarily arising from initial specimen orientation during analysis. OPCR as originally defined is the mean of eight OPC values iterated step-wise between 0° (initial orientation of the specimen) and 45° (Evans and Janis 2014). OPCR can be calculated using the function OPCr, which passes inclusion criteria on to the OPC function. OPCr defaults to measuring OPC in eight evenly-spaced iterative steps however, users can specify an alternative number of steps and step sizes using the arguments in OPCr (Table 8). OPCr is an executive function, iteratively calling OPC and storing the patch count for each iteration before averaging them. In order to conserve memory and speed during processing, only the final patch count of each iteration is stored during the execution of OPCr. The object produced by OPCr is a list including the OPCR value, and the OPC values and degrees rotated for each iterative call of OPC (Table 9).
As with the calculation of RFI, using the molaR function RFI, OPC as calculated by molaR is susceptible to imprecise specimen orientation. The OPC function expects enamel cap surfaces oriented with the occlusal plane perpendicular to the Z-axis. Improper alignment will result in a skewed patch count, which can be quickly detected by examining the distribution of patches sorted into each directional bin. Improperly oriented surfaces will typically have directional bins containing few or zero patches.
An error sometimes encountered when using OPC states: Error: evaluation nested too deeply: infinite recursion / options(expressions=)?. This error occurs when the recursive patch-clumping function (part of the larger OPC function) encounters an exceptionally large patch (i.e., a patch containing a large number of faces). The base R framework has a default limit of 5000 nested expressions so as to safeguard against infinite recursion. Running the command: options(expressions = 10,000) will override the default limit and reset it to 10,000. R has an upper limit of 500,000 nested recursions, which cannot be overridden. Resetting the limit to 10,000 is almost always high enough to complete any call of the OPC function when using a surface which has been properly down-sampled to ~10,000 faces. Yet, if necessary, the upper limit can be set higher. Users should be aware however, that the command options(expressions = x) will reset the upper recursion limit throughout their entire R framework. This alteration to the R framework can only be undone by resetting the number of expressions, or restarting R. Approaching the upper limit of nested expressions increases the risk of segmentation fault, which can cause R to crash.
Also contained in molaR is the plotting function OPC3d. OPC3d colors the surface mesh according to the directional bins, allowing users to visualize the orientation patches on a surface (Fig. 4). OPC3d requires an object created by the OPC function, not the OPCr function. Like the other plotting functions provided by molaR, OPC3d has several arguments that allow users to customize their plots (see Table 10).
Surface views of mantled howler monkey lower first molar (specimen number: DU-LP 09) with mesh surface triangles colored by directional bins to display OPC. The orientation patches are outlined in black in addition to being colored by orientation. Six perspectives of the same OPC analysis are displayed with consistent bin coloration: a) three-quarters view, b) mesial view, c) distal view, d) occlusal view, e) lingual view, and f) buccal view
molaR Output Cross-Compatibility
In order to check the compatibility of the results from molaR with other packages performing some or all of these measures, 13 digital, cropped, lower second molar specimens (six Pithecia pithecia, and seven Alouatta spp.) were taken from the Winchester et al. (2014) data set (fully available on MorphoSource; Copes et al. 2016) for re-analysis. The file options for each specimen include a tiff stack of microCT images, a ‘raw’ PLY surface file that contains an unaltered surface generated from the microCT data, and a ‘cropped’ PLY surface file that contains the same specimen cropped at the enamel cervix and oriented so the occlusal plane is approximately parallel with the X-Y plane. We collected DNE, RFI, and OPCR from the ‘cropped’ PLY files for each specimen (Supp. 1) using the default package settings in both MorphoTester (Winchester 2016) and molaR (see Table 11). An additional measure of OPCR was collected using SurferManipulator (Evans et al. 2007; Table 11).
Prior to analyzing surfaces, the PLY files were processed in Avizo (for workflow, see Fig. 5). The orientation of each surface was standardized such that the occlusal plane was normal to the positive Z-axis to allow for consistent processing in SurferManipulator. For some surfaces, this required rotating the mesh by 180° about the X-axis, while for others no reorientation was required. The set of oriented PLYs was saved as STL files for analysis by SurferManipulator, then further processed prior to importing into MorphoTester or molaR. For the analyses (i.e., calculation of DNE, RFI, and OPCR) with MorphoTester and molaR, the surfaces were simplified to ~10,000 faces using Avizo’s Simplification Editor, smoothed using Avizo’s Smooth Surface function at 100 iterations with a lambda value of 0.6, and saved as PLY files. This smoothing step was aimed at removing signal noise that can originate during scanning or reconstruction of CT data and manifests as errors in the orientation of PLY faces. Unsmoothed surfaces are much more rugged and can lead to considerable inconsistency in DNE values (see Winchester 2016). Smoothing is an important step and is recommended when measuring DNE in particular in order to maintain consistency with methods employed in past studies (Bunn et al. 2011; Winchester et al. 2014; Fig. 5). Unlike versions of the PLY files measured in MorphoTester and molaR, the STL files analyzed in SurferManipulator were neither simplified, nor smoothed. STL files were converted to Surfer (Golden Software version 8) grids using SurferManipulator’s File Format Conversion dialog on default settings. The resolution of resulting grid files were then standardized by regridding using the Surfer Functions dialog with an exact number of 50 rows. OPCR was then calculated in SurferManipulator’s CSV Viewer dialog, with a ‘Min. patch’ value of 2. Note that SurferManipulator excludes patches with total number of cells equal to or less than the ‘Min. patch’ value, thus the smallest patches considered in the SurferManipulator analyses contained three cells, analogous to the three face minimum utilized in MorphoTester and molaR calculations of OPCR.
The DNE measurement workflows for the values presented in Table 11, and other works, which previously presented DNE. Box colorations indicate different software platforms. Workflow proceeds from left to right. Note that all tooth surfaces for Bunn et al. (2011), Winchester et al. (2014), and the ‘raw,’ ‘smoothed,’ and ‘Mesh-faired’ tooth surfaces associated with this publication are available on MorphoSource for download and re-analysis, in addition to surfaces from earlier works. Values of RFI and DNE published in these earlier studies should be indistinguishable (excepting for rounding errors) to values produced by molaR on identical surface meshes subjected to the same processing
The results for DNE, RFI, and OPCR are very similar when comparing molaR and MorphoTester when the surface scans are subject to identical processing and when implicit fairing is left on its default (disabled) setting in MorphoTester (see Fig. 5 and Table 11). The minor differences in DNE between the two software programs stem from variation in the identification of outlier faces. The MorphoTester and molaR default settings identify the outliers slightly differently. molaR excludes faces on the basis of the estimated Dirichlet Energy Density value for each triangular face on the mesh, not each face’s Dirichlet Energy Quantity (which is density × area). MorphoTester’s default settings, on the other hand, exclude faces based on the Dirichlet Energy Quantity. molaR therefore biases towards removing very small triangular faces, and more frequently removes faces found in surface recesses. molaR tends to exclude a lower percentage of the total surface, and thus regularly calculates a slightly higher DNE value for surfaces than does MorphoTester (Table 11). Users can explore the energy density values and face areas for each trianglar face in the molaR DNE output and opt to alter outlier exclusion criteria via adjusting the exclusion threshold with the outlier argument in the DNE function. MorphoTester has options that allow users to identify outliers using either Dirichlet Energy Density or Dirichlet Energy Quantity. Ultimately, while there are differences in DNE values returned from MorphoTester versus molaR, these differences are negligible and far smaller than the standard error produced in the samples using either software package (Table 11).
Table 11 also contains the original results for the 13 specimens analyzed here as reported in Winchester et al. (2014), whose reported DNE values differ substantially from the values produced using molaR and MorphoTester (reported raw data available in supplementary materials). Winchester et al. (2014) used a different software package, Teether (Bunn et al. 2011) to calculate DNE. The differences in DNE values stem from a previously undescribed implicit fairing step that was built into the Teether DNE software (and is also included among the DNE options in MorphoTester). Thus, the DNE values calculated by Bunn et al. (2011) and Winchester et al. (2014) include an additional smoothing step (implicit fairing) in addition to the cropping, simplifying, and smoothing operations performed in Avizo and Amira reported in their methods (Fig. 5). Being the first researchers to experiment with DNE calculations on molars, Bunn et al. (2011) and Winchester et al. (2014) were primarily focused on presenting the conceptual application rather than the technical implementation of DNE to tooth crowns. Therefore, they opted to simplify the description of their methods in order to maximize their readership’s appreciation of the larger concepts. However, without the inclusion of this additional smoothing step, replication of their results in other software is currently impossible. We therefore sought to mimic their implicit fairing step on the 13 PLY files that had been simplified and smoothed in Avizo as described above using a Laplacian Smooth operation in MeshLab with three steps, 1D boundary smoothing, and cotangent weighting enabled. The calculated values are not identical, as this smoothing algorithm does not perfectly replicate the implicit fairing smooth used in Teether, however they are remarkably close (Table 11 molaR ‘Mesh-faired’ vs Winchester et al. 2014; Fig. 5). Further refinement of the smoothing procedures and parameters will likely enable backwards and horizontal compatibility of data sets.
Reflecting on the above comments, an obvious issue of concern in the application of DNE to dental studies is the considerable variation in surface pre-processing across projects already in the literature (Fig. 5), and therefore the general reproducibility and compatibility of results. To address this we make four recommendations. First [1], researchers should explicitly state in their methods (or supplemental materials), all steps included in their PLY surface processing workflow, or reference a publication whose explicit procedures they followed without deviation. Second [2], researchers should make publicly available all raw and processed PLY surfaces analyzed in their work, using data sharing repositories such as MorphoSource (Copes et al. 2016). Third [3], we tentatively suggest that researchers should avoid the additional implicit fairing smoothing operation, since it is a redundant step for surfaces already smoothed in Avizo, MeshLab, or Amira, and occasionally causes DNE calculation errors in MorphoTester (though these bugs are known and being addressed). However, this suggestion does make combining previously published data sets with new analyses inappropriate. Therefore, fourth [4], researchers aiming to combine previously published DNE values with new values must verify their compatibility, which can be done with access to the raw and processed PLY files.
RFI also shows slight variation among: MorphoTester, molaR, and the Winchester et al. (2014) results. First, the differences between Winchester et al. (2014) and molaR arise from several variations in the calculation procedures. Winchester et al. (2014) computed A3D from smoothed versions of the original files, which were not down-sampled to 10,000 faces, and did so directly in Avizo. Winchester et al. (2014) then calculated A2D using scaled planometric footprint images exported out of Avizo and then measured in ImageJ (Abramoff et al. 2004). Though the differences in RFI values are small, they are in the predicted direction given this difference in protocol (the values from Winchester et al. [2014] are higher). That is, down-sampling the surface of a tooth from the typical 100,000–200,000 faces initially produced from a μCT scan to 10,000 faces tends to reduce A3D. Additionally, the pixel-based counting system used by ImageJ to assess area tends to produce lower values for A2D than the approach taken by molaR. Like Winchester et al. (2014), Boyer (2008), and Bunn et al. (2011) followed the same procedure. Thus on balance, the approach taken by Winchester et al. (2014) make slightly higher estimates for A3D and slightly lower for A2D resulting in a slightly larger RFI as compared to the analyses performed here using molaR. Second, the variation between molaR and MorphoTester (smaller than that when comparing molaR to earlier works) arises from a small difference in the calculation of the two-dimensional footprint areas. MorphoTester and molaR use two different methods to calculate the projected footprint area. Both software packages initially project the 3D surface onto a plane parallel with the occlusal plane. molaR then uses the alpha hull algorithm contained in the R package alphahull (Pateiro-Lopez and Rodriguez-Casal 2015), which identifies and then traces the points on the outer boundary of the 2D projection. Once these points are identified, the enclosed area is calculated. MorphoTester relies on a pixel-counting method (very similar to ImageJ). The program renders the 2D projection as a rasterized image and associates a pixel-to-unit scale-bar with it, then counts the weighted pixels to get the 2D area. Like the DNE calculation, the RFI calculation differences between molaR and MorphoTester are negligible (Table 11).
The OPCR values (Table 11) produced by molaR and MorphoTester under the stated protocols, and using the software’s default settings are identical. However, the OPCR values produced on the same set of teeth are dramatically different when comparing molaR and MorphoTester to SurferManipulator, which was used by Winchester et al. (2014) and whose results we have attempted to reproduce with SurferManipulator again here (Table 11). The variant of OPC employed by molaR and MorphoTester accounts for face X-Y plane orientation on fully three-dimensional surfaces, rather than on the 2.5 dimensional raster surfaces (meaning flat surfaces with a unique Z-value for every X-Y cell) originally employed by Evans et al. (2007). This distinction in approach results in different OPCR values when using OPCr in molaR and MorphoTester as compared to employing SurferManipulator. As a result, until further refinement of the OPCR calculation employed by molaR (or MorphoTester) can be developed, aimed at producing identical results with those of SurferManipulator, researchers should not combine data produced from molaR (or MorphoTester) with those generated from SurferManipulator.
The OPCR values reported by Winchester et al. (2014) were calculated using SurferManipulator, which prior to molaR and MorphoTester was the only software package available to perform the patch-clumping functions intrinsic to the measurement. Our attempt to replicate those values using SurferManipulator failed, though the results were overall more consistent with the findings of Winchester et al. (2014) than the three-dimensional analyses performed by molaR and MorphoTester (Table 11). The reasons for differences in OPCR values calculated in this study compared to those calculated for the same specimens by Winchester et al. (2014) remain mysterious, as care was taken to replicate methods as closely as possible; however, there are several potential causes of the discrepancies. The orientations of PLY files obtained from MorphoSource were not altered, except to rotate some surfaces by 180° such that the occlusal plane was normal with respect to the positive Z-axis. OPC is highly sensitive to tooth orientation (as emphasized above), and if any additional changes to surface orientations were made by Winchester et al. (2014) that were not reflected in the PLY files downloaded from MorphoSource, these changes alone could easily explain the differences in our calculations (unlike the differences in calculated DNE values, which are immune to orientation). Additionally, SurferManipulator offers a wide variety of user options, which can be introduced at many steps, and thorough documentation of every step in the protocol is rarely reported, despite the many studies that have employed it. Differences in file format conversion procedures, data standardization, or OPCR parameters will result in different OPCR values, and are a potential source of influence for the reported differences between this study and Winchester et al. (2014). The only notable difference we detected in our protocol from that of Winchester et al. (2014) was the use of STL files rather than PLY files as the initial 3D mesh. Researchers wishing to duplicate previously published results using SurferManipulator should directly contact the authors of those works for their workflow.
Finally, SurferManipulator was designed in 2008 using the Microsoft Visual Basic language and development environment to run with Windows 2000 and Windows XP operating systems (OS). While Visual Basic is still compatible with modern Windows OS, its official support ended in 2008 and it has not received updates to accommodate more recent, or 64-bit OS, and the ActiveX objects it requires to function are no longer included in or supported by Windows. The program Surfer, which SurferManipulator uses to rasterize tooth scans and perform calculations of slope and aspect, has been updated many times since the version available in 2008 (version 8) and the one available now (version 13). Changes to the gridding algorithms used in Surfer could potentially change OPCR calculations of identical tooth surfaces and may explain the differences between the values obtained in this study versus those calculated by Winchester et al. (2014). While SurferManipulator is still an effective tool at calculating OPC and OPCR and distinguishing teeth based on their surface complexity (Table 11), these considerations underscore the need for software packages that are compatible with current OS, take full advantage of modern computing power, and utilize the full extent of three-dimensional data such as molaR and MorphoTester.
Plotting Extensions Using rgl
The plots produced from the molaR package can be printed and enhanced through functions available in the rgl package (Adler et al. 2015). Users can employ rgl.snapshot to export high quality PNG files out of R for publications after custom orienting their surfaces. Additionally, rgl contains several options to make short videos that enable rotating views of the surfaces in RGL windows. Users may find these options enable maximum visualization of their results. At the time of publication, issues arising from the rgl package’s interaction with X11 in Mac OS >=10.10 are causing distortions in the legends printed by DNE3d and OPC3d in molaR v. 1.0. These problems have been resolved with the release of molaR v. 2.0, (more information can be found in the molaR Quick Start.R file located at DOI: 10.13140/RG.2.1.3284.9687).
Running molaR Functions in a Batch
Included in the package is a batch operation function entitled molaR_Batch. This function automates the collection of DNE, RFI, OPC, and OPCR from multiple PLY files. Users should set their working directory to a folder containing the PLY files they wish to analyze for DNE, RFI, OPC, or OPCR. Users operating the molaR_Batch function can select the analyses they wish to run with the logical arguments DNE, RFI, OPCr, and OPC. The details argument, if enabled, will include information such as 3D area, 2D area, and OPC values of different rotations during OPCR in the function output. Additionally, the parameters used to run DNE, OPC or OPCr functions can be adjusted to user preference in a variety of arguments that parallel those found in their respective functions. After analyses are complete, the molaR_Batch function will produce a CSV file containing all of the requested calculations, located in the same working directory folder as the original PLY files.
Citing molaR
Authors publishing work using the analysis or visualization tools contained in molaR should cite this paper, and the appropriate progenitor works. We ask users not to cite the molaR package manual, which can be found on the CRAN webpage: https://cran.r-project.org/web/packages/molaR/molaR.pdf.
References
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Internatl 11:36–42
Adams DC, Otárola-Castillo E (2013) geomorph: an R package for the collection and analysis of geometric morphometric shape data. Method Ecol Evol 4:393–399
Adler D, Murdoch D, Nenadic O, Urbanek S, Chen M, Gebhardt A, Bolker B, Csardi G, Strzelecki A, Senger A (2015) rgl: 3D visualization using OpenGL. R package version 0.95.1247
Allen KL, Cooke SB, Gonzales LA, Kay RF (2015) Dietary inference from upper and lower molar morphology in platyrrhine primates. PLoS ONE 10:e0118732. doi:0118710.0111371/journal.pone.0118732
Bargo MS, Vizcaíno SF, Kay RF (2009) Predominance of orthal masticatory movements in the early Miocene Eucholaeops (Mammalia, Xenarthra, Tardigrada, Megalonychidae) and other megatherioid sloths. J Vertebr Paleontol 29:870–880
Boyer DM (2008) Relief index of second mandibular molars is a correlate of diet among prosimian primates and other euarchontan mammals. J Hum Evol 55:1118–1137
Boyer DM, Evans AR, Jernvall J (2010) Evidence of dietary differentiation among late Paleocene-early Eocene plesiadapids (Mammalia, Primates). Am J Phys Anthropol 142:194–210
Boyer DM, Puente J, Gladman JT, Glynn C, Mukherjee S, Yapuncich GS, Daubechies I (2015b) A new fully automated approach for aligning and comparing shapes. Anat Rec 298:249–276
Boyer DM, Winchester J, Kay RF (2015a) Technical Note: The effects of differences in methodology among some recent applications of shearing quotients. Am J Phys Anthropol 156:166–178
Bunn JM, Boyer DM, Lipman Y, St. Clair EM, Jernvall J, Daubechies I (2011) Comparing Dirichlet normal surface energy of tooth crowns, a new technique of molar shape quantification for dietary inference, with previous methods in isolation and in combination. Am J Phys Anthropol 145:247–261
Copes LE, Lucas LM, Boyer DM (2016) A collection of non-human primate CT scans house in MorphoSource, a repository for 3D data. Sci Data
Evans AR, Janis CM (2014) The evolution of high dental complexity in the horse lineage. Ann Zool Fenn 51:73–79
Evans AR, Wilson GP, Fortelius M, Jernvall J (2007) High-level of similarity of dentitions in carnivorans and rodents. Nature 445:78–81
M’Kirera F, Ungar PS (2003) Occlusal relief changes with molar wear in Pan troglodytes troglodytes and Gorilla gorilla gorilla. Am J Primatol 60:31–41
Pateiro-Lopez B, Rodriguez-Casal A (2015) alphahull: generalization of the convex hull of a sample of points in the plane. R package version 2.0
Plyusnin I, Evans AR, Karme A, Glonis A, Jernvall J (2008) Automated 3D phenotype analysis using data mining. PLoS ONE 3:e1742. doi:1710.1137/journal.pone.0001742
Prufrock KA, Boyer DM, Silcox MT (2016) The first major primate extinction: an evaluation of paleoecological dynamics of North American stem primates using a homology free measure of tooth shape. Am J Phys Anthropol
R Core Team (2015) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing
Revelle W (2015) psych: Procedures for Personality and Psychological Research, Northwestern Univeristy, Evanston, IL http://cran.r-project.org/package=psych Version=1.5.8
Schlager S (2015a) Package ‘Rvcg’ Manipulations of Triangular Meshes Based on the ‘VCGLIB’ API https://cran.r-project.org/web/packages/Rvcg/index.html Version=0.13.1.2
Schlager S (2015b) Morpho: Calculations and Visualisations Related to Geometric Morphometrics https://cran.r.project.org/web/packages/Morpho/index.html Version=2.3.1.1
Ungar PS, M’Kirera F (2003) A solution to the worn tooth conundrum in primate functional anatomy. Proc Natl Acad Sci USA 100:3874–3877
Wilson GP, Evans AR, Corfe IJ, Smits PD, Fortelius M, Jernvall J (2012) Adaptive radiation of multituberculate mammals before the extinction of the dinosaurs. Nature 483:457–460
Winchester JM (2016) MorphoTester: an open source application for morphological topographic analysis. PLoS ONE 11:e0147649. doi:10.1371/journal.pone.0147649
Winchester JM, Boyer DM, St. Clair EM, Gosselin-Ildari AD, Cooke SB, Ledogar JA (2014) Dental topography of platyrrhines and prosimians: convergence and contrasts. Am J Phys Anthropol 153:29–44
Zuccotti LF, Williamson MD, Limp WF, Ungar PS (1998) Technical Note: Modeling primate occlusal topography using geographic information systems technology. Am J Phys Anthropol 107:137–142
Acknowledgments
The authors thank redditor /u/Kiss_My_Wookiee for help in naming the package, and Roxanne Larsen for assistance with deciphering MATLAB code. The authors declare no known conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 72 kb)
Rights and permissions
About this article
Cite this article
Pampush, J.D., Winchester, J.M., Morse, P.E. et al. Introducing molaR: a New R Package for Quantitative Topographic Analysis of Teeth (and Other Topographic Surfaces). J Mammal Evol 23, 397–412 (2016). https://doi.org/10.1007/s10914-016-9326-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10914-016-9326-0