Abstract
Epigenetics refers to alterations in gene expression due to modifications in histone acetylation and DNA methylation at the promoter regions of genes. Unlike genetic mutations, epigenetic alterations are not due to modifications in the gene primary nucleotide sequence. The importance of epigenetics in the initiation and progression of breast cancer has led many investigators to incorporate this novel and exciting field in breast cancer drug development. Several drugs that target epigenetic alterations, including inhibitors of histone deacetylase (HDAC) and DNA methyltransferase (DNMT), are currently approved for treatment of hematological malignancies and are available for clinical investigation in solid tumors. In this manuscript, we review the critical role of epigenetics in breast cancer including the potential for epigenetic alterations to serve as biomarkers determining breast cancer prognosis and response to therapy. We highlight initial promising results to date with use of epigenetic modifiers in patients with breast cancer and the ongoing challenges involved in the successful establishment of these agents for the treatment of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The initiation and progression of breast cancer has been recognized for many years to be secondary to the accumulation of genetic mutations which lead to aberrant cellular function. Genetic mutations, either inherited or sporadic, may result in the activation of oncogenes and the inactivation of tumor suppressor genes. The more recent discovery that reversible alterations in histone proteins and deoxyribonucleic acid (DNA) can also lead to tumorigenesis has introduced a novel term to the field of cancer research known as “epigenetics” [1].
Epigenetics refers to alterations in gene expression that are not a result of changes in the primary nucleotide sequence of a gene, such as is the case with a genetic mutation. Instead, epigenetic alterations may result in changes in chromatin structure leading to a repressive chromatin state and silencing of both gene expression and transcription of DNA into ribonucleic acid (RNA). Epigenetic alterations include both histone hypoacetylation and abnormal methylation of DNA in the promoter region of important genes. Epigenetic regulation is critical for normal growth and development, and alterations may result in a variety of pathological processes including autoimmune diseases and cancer. The deregulation of genes that ensues can impact important cellular functions such as DNA repair and control of the cell cycle and apoptosis.
In this review, we will detail the epigenetic alterations that have been reported in breast cancer and have been associated with disease outcome, as well as the potential to utilize epigenetic modifiers to reverse these alterations. We will also describe the current status of clinical trials which incorporate these agents either alone or as combination strategies in breast cancer, and the challenges that have been identified while transitioning these agents to the solid tumor setting.
Epigenetic Alterations in Cancer
Histone Modification
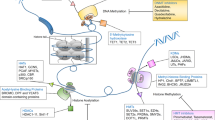
Chromosomal DNA is packaged into chromatin and coils around structural histone proteins. The histones are responsible for maintaining the shape and structure of chromatin. A number of post translational alterations can occur at the amino acid tail of histone proteins which result in a conformational change in the chromatin and therefore in the transcription of important genes such as tumor suppressors. These alterations include acetylation, methylation or phosporylation [2].
Histone acetylation is controlled by a balance in activity between histone acetyltransferase (HAT) and histone deacetylase (HDAC). The former add an acetyl group to histones resulting in uncoiling or “opening” of the chromatin, facilitating gene transcription. The HDACs remove acetyl groups from the histones leading to coiling or “closing” of chromatin which inhibits transcription [3]. The HDACs are critical in the regulation of expression of genes important for cell survival, proliferation, differentiation, and apoptosis [4]. HDACs also act as members of a protein complex responsible for recruitment of transcription factors to the promoter region of genes, including those of tumor suppressors, and regulation of acetylation status of specific cell cycle regulatory proteins [5]. High HDAC expression and histone hypoacetylation have been observed in cancer with associated transcriptional repression of genes, providing a rationale for the investigation of HDAC inhibitors in cancer therapeutics [6].
DNA Methylation
Gene silencing may occur due to methylation of DNA at the promoter region of genes [4]. Adenine, guanine, cytosine and thymine are the four bases which are the building blocks of our genetic make-up. DNA methyltransferase enzymes (DNMTs) add a methyl group (-CH3) to the pyrimidine ring of cytosine to form methyl cytosine (DNA methylation) and play a crucial role in the hypermethylation of tumor suppressor genes. Five DNMT proteins have been discovered in mammals, but only DNMT1, DNMT3a, and DNMT3b have catalytic methyltransferase activity. DNMT1 has a preference for hemi-methylated DNA as a substrate [7], whereas the DNMT3 enzymes are known as de novo methyltransferases and target unmethylated DNA [8].
This process of DNA methylation occurs only to cytosines which precede a guanine in the DNA sequence, known as the CpG dinucleotide. CpG dinucleotides exist throughout the genome and are usually heavily methylated and thus impede transcription of genes at those sites. Where a number of these CpG dinucleotides are found at the promoter regions of genes they are known as CpG islands. In normal tissue CpG islands are more commonly unmethylated, allowing for gene transcription to take place. In cancer, the reverse can be seen with abnormal DNA methylation of CpG islands that impede transcription of important genes such as tumor suppressor genes [1].
Assays
Assays that can identify epigenetic alterations in tumor samples are critical both in the laboratory and in the clinic. When detected, alterations in histone acetylation and gene methylation have several possible clinical utilities. Epigenetic alterations have the potential to identify those at high risk of developing a new cancer, facilitate the early detection of cancer, assist in cancer staging and predict prognosis or response to a particular therapy and present a target for novel therapies [9–12].
HDAC inhibitors target the HDAC enzymes, resulting in hyperacetylation of histone tails. Preclinical investigations have suggested that this histone acetylation may occur approximately 30 min after exposure to HDAC inhibitors, with the effect on chromatin remodeling occurring after more prolonged exposure to these agents (24–48 h minimum) [13]. The hyperacetylation of target histones observed with administration of the HDAC inhibitors is comparable in tumor samples and peripheral blood mononuclear cells (PBMCs) when assessed by standard western blot analysis and other techniques [13, 14]. Analysis of PBMCs represents a useful and non-invasive way to identify the pharmacodynamic effect of HDAC inhibitors, i.e. whether it is “hitting the target.” However, until recently, studies have failed to show a clear correlation between the level of hyperacetylation and response to therapy with HDAC inhibitors [12, 15].
The detection of promoter hypermethylation at CpG islands in both preclinical and clinical samples has been facilitated by a technique known as methylation-specific polymerase chain reaction (MSP) [16]. MSP can differentiate between methylated and unmethylated cytosine upon sodium bisulfite treatment of DNA and subsequent amplification of the modified DNA using primer sets specific to the methylated or unmethylated promoters. Quantitative multiplex-methylation specific PCR (QM-MSP) was subsequently found to be a highly sensitive technique which can accurately assess promoter hypermethylation for many genes simultaneously in small samples, termed a “candidate marker approach.” [17] Advances in gene array technologies now also allow for a comprehensive whole-genome methylation array analysis (“methylome analysis”) in cancer samples at low cost [18]. These methods quantify the proportion of methylated cytosines to total cytosines at approximately 27,500 different CpG dinucleotides in 14,500 regions (e.g. the Illumina Infinium HumanMethylation27 array). The areas queried in this method include thousands of well-annotated genes as well as hundreds of methylation hotspots in cancer genes, cancer related targets, and microRNA promoters. As little as 500 ng of input DNA is required by many of these assays [19]. Ongoing research aims to further refine these methods, develop new assays and ultimately assess their clinical utility in well-designed prospective clinical discovery and validation studies.
Epigenetic Modifiers
HDAC Inhibitors
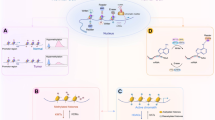
Aberrant HDAC activity has been documented in a variety of tumor types and led to the development of HDAC inhibitors as anticancer therapeutics (Table 1). Currently available HDAC inhibitors target a variety of HDAC isoenzymes with class 1 (HDAC 1, 2, 3 and 8), class 2 (HDAC 4–7 and 9–10), and class 4 (HDAC 11) activity. Modest clinical benefits were previously reported with relatively weak HDAC inhibitors such as valproic acid and phenylbutyrate in advanced solid tumors or hematologic malignancies [20]. More potent HDAC inhibitors including both class-specific inhibitors (entinostat and romidepsin) and pan HDAC inhibitors (vorinostat, belinostat and panobinostat) have been developed recently. Both romidepsin and vorinostat are approved by the Food and Drug Administration (FDA) for treatment of cutaneous T-cell lymphoma (CTCL). In a phase 2b clinical trial in patients with heavily pretreated CTCL, investigators reported a 30 % response rate with use of vorinostat, an impressive median time to progression, and pruritus relief in approximately 30 % of patients. [21] Romidepsin is associated with significant anti-tumor activity following failure of previous treatment in patients with CTCL as well as peripheral T-cell lymphoma (PTCL) [22–24]. Clinical studies in solid tumors are ongoing with HDAC inhibitors, alone or in combination with other agents, with those relevant to breast cancer described later in this review.
DNMT Inhibitors
DNMT inhibitors, also referred to as demethylating agents, have been under preclinical and clinical investigation for over 30 years [25]. The nucleoside analogues, 5-azacitidine (AZA) and decitabine (DAC), have been the most widely studied. Because they are cytidine analogs, both agents are incorporated into DNA after activation to a triphosphate moiety. After formation of an irreversible complex with DNMT1, degradation of the enzyme occurs [26]. This prevents methylation of daughter DNA in CpG islands during DNA replication. In addition, AZA (but not DAC) is converted into a ribonucleoside moiety and is incorporated into RNA, interfering with protein translation. At low concentrations (e.g. 30nM DAC, 300nM AZA), these inhibitors exhibit potent DNA hypomethylation properties, whereas high concentrations (≈3–10 μM) are cytotoxic [27]. The doses of AZA and DAC that were employed in many of the early clinical trials in solid tumors were cytotoxic, reflecting maximum tolerated doses, which likely accounts for the excessive toxicity, and possibly also to lack of overall efficacy, observed in these studies [28]. More recently, clinical trials in hematologic malignancies were designed with a better understanding of the DNA hypomethylating effects of these agents. AZA and DAC have subsequently been approved by the FDA for use in the treatment of myelodysplastic syndrome (MDS). A phase 3 randomized trial in patients with high risk MDS indicated a significant improvement in median overall survival with AZA when compared to conventional care regimens including best supportive care or standard chemotherapy (24.5 versus 15 months) [29]. Impressive disease responses have also been observed with the use of DAC in a similar patient population [30]. Other DNMT inhibitors in an earlier phase of development include DAC analogues such as SGI-110 and zebularine.
Epigenetics and Breast Cancer
Epigenetic alterations are prevalent in breast cancers, prompting much interest in their clinical significance and whether these can be manipulated. Aberrant HDAC activity has been documented in many tumor types. HDAC 1 expression is associated with an advanced stage and aggressive histology in certain cancers [31]. However, in breast cancer core biopsy specimens, HDAC 1 expression is associated with estrogen receptor (ER) and progesterone receptor (PR) expression, earlier stage of disease at diagnosis, and improved disease-free survival [32]. HDAC 6 messenger RNA (mRNA) is more frequently expressed in breast cancer patients with small (less than 2 cm), low grade, ER and PR-positive tumors. However, multivariate analyses failed to confirm that HDAC 6 expression was an independent prognostic factor for survival [33].
When considering aberrant DNA methylation in breast cancer, global DNA hypomethylation is far more prevalent in breast cancer specimens (up to 50 %) compared to that observed in other tumor types. This global hypomethylation has been associated with poor prognostic factors such as tumor size, stage and grade [34]. DNA hypomethylation can also affect individual breast cancer genes [35]. More commonly, breast cancer-related genes are hypermethylated and thus silenced compared to non-cancerous tissue. Methylated genes in breast cancer include those important for growth (e.g. the ER), evasion of apoptosis (e.g. HOXA5 and Twist), invasion and metastasis (e.g. E-cadherin) as well as cell differentiation (e.g. RARbeta) [36]. A panel of seven genes was evaluated using MSP in a variety of breast tissues [37, 38]. In invasive breast carcinomas, almost all specimens contained at least one hypermethylated gene, 80 % contained two, and 60 % contained three or more methylated genes. Only one of eight reduction mammoplasty specimens contained hypermethylated genes. DNA methylation in RASSF1A and CDH1 promoters has also been evaluated by QM-MSP in archival tumor and blood samples from 92 patients with breast cancer and 50 controls. RASSF1A and CDH1 methylation was observed in 82.6 % and 21.7 % of breast cancer tumors respectively, while no methylation was detected in controls. CDH1 methylation levels were significantly associated with lymph node status and breast cancer subtype [39]. In another study, the methylation status of eight genes (DCR1, DAPK1, RASSF1A, DCR2APC, MGMT, GSTP1 and PTEN) was evaluated in snap-frozen primary breast tumors (n = 49). The highest frequencies of promoter methylation was observed for the APC (54 %), DCR1 (40 %), DAPK1 (37 %), RASSF1A (33 %) and MGMT (22 %) genes, with 75 % of specimens associated with aberrant methylation in at least one gene. An association was also identified between MGMT promoter methylation and age, with MGMT methylation more often observed in older patients. Larger tumors showed a higher frequency of RASSF1 and DAPK1 promoter methylation [40]. These studies indicate that promoter methylation of specific genes may be utilized as potential prognostic biomarkers in breast cancer patients if validated in future studies.
The prevalence of ER methylation has specifically been examined after establishing that the ER indeed has a CpG island in its A and B promoters and first exon [41]. The ER gene is unmethylated at the CpG island in normal tissues and in several ER-positive human breast cancer cell lines. Only 36 % of human breast cancers that express both the ER and PR proteins are methylated at the ER promoter, compared to 72 % of tumors that are ER-positive but PR-negative, and 100 % of tumors that are ER and PR-negative [41]. These observations prompted further investigation as to whether reversal of methylation at the ER promoter would sensitize the tumors to hormone treatment and is described below.
Whether methylation of specific breast cancer genes can predict clinical outcomes has also been investigated using a genome-wide methylation array technology (methylome analysis) [42]. The Illumina Infinium HumanMethylation27 array was used to analyze both primary invasive breast cancers (n = 103) and normal breast samples (n = 21). A higher frequency of methylation is observed in ER-positive tumors compared to ER-negative tumors, specifically, higher methylation at 5,264 loci in ER-positive versus at 3,112 loci in ER-negative tumors. The hypermethylated loci in ER-negative tumors, however, cluster closer to the transcriptional start sites than in the ER-positive tumors; perhaps indicating a tighter control of transcriptional repression. The methylation patterns of ER-positive and ER-negative tumors are also distinct with 27 (ER-positive) and 13 (ER-negative) loci showing the highest subtype specificity in individual tumor samples. This information, if validated, may help identify molecular pathways best targeted in the individual breast cancer subtypes. Finally, an attempt was made to develop an epigenomic signature that would aid prediction of outcome in breast cancer patients. Investigators identified a 100 CpG loci signature that was significantly associated with disease progression in patients newly diagnosed with breast cancer treated with either no adjuvant therapy, hormonal or chemotherapy. Approximately 20 % of the loci included in this signature were from homeobox-containing genes including HOX suggesting a key role in tumor progression. Others have also reported associations between aberrant DNA methylation and breast cancer outcomes [43]. However, additional studies are needed to define the exact role of gene methylation signatures in predicting clinical outcome and response to therapy. Based on the higher frequency of methylation observed in ER-positive tumors in these studies, it is possible that ER-positive breast cancer may represent a better target for epigenetic therapy than other tumor subtypes.
DNA methylation profiling has also identified specific breast cancer subtypes that are distinct from the “intrinsic subtypes” classified by gene-expression profiling in recent years (luminal A, luminal B, HER2 and basal) [44]. Methylome analysis performed on frozen primary tumor samples, led to the identification of six different methylation clusters [45]. It was shown for the first time that DNA methylation profiles can reflect the cell-type composition of the tumor microenvironment, with a T lymphocyte infiltration of these tumors in particular in HER2-enriched and basal-like tumors. Interestingly, high expression of certain immune-related genes were found to be associated with improved relapse-free survival providing further insight into the importance of the immune system and tumor microenvironment in certain breast cancer subtypes.
An area of great interest is whether methylation of specific breast cancer genes can predict benefit from breast cancer therapies. For example the antitumor activity of poly(adenosine diphosphate)-ribose polymerase (PARP) inhibitors in BRCA1/BRCA2-associated cancers has been recently, of intense interest to the oncology community [46]. Interestingly, the BRCA1 gene has been shown to be inactivated in sporadic breast and ovarian cancers by DNA methylation [10]. Whether this epigenetic modification confers sensitivity to PARP inhibitors was investigated by a group of international collaborators [47]. As with BRCA mutation, hypermethylation of the gene in a breast cancer cell line was associated with equal sensitivity to PARP inhibitors as did the BRCA1 mutation. Treatment of this cell line with AZA restored expression of the gene. In addition, 36.7 % (25 of 68 tumors) of “triple-negative” (ER/PR/HER2-negative) breast tumors were found to exhibit BRCA1 methylation, indicating that this may be a population that may benefit from PARP inhibitors in the clinic.
Another area of intense research at this time is the association between epigenetics and microRNAs (miRNAs), which are small, non-coding RNA molecules with the ability to regulate gene expression. miR are downregulated in many tumor types including breast cancer and have the potential to be used as biomarkers for early breast cancer detection or prognosis [48, 49]. Hypermethylation of miRNAs may also lead to their silencing and inability to function as tumor suppressors [50]. Whether miRNAs reflect a novel target in breast cancer requires further evaluation.
Preclinical Activity of the HDAC Inhibitors
Laboratory research conducted to date supports the investigation of HDAC inhibitors for the treatment of breast cancer. Vorinostat, for example, induces differentiation or arrests growth of a wide variety of human carcinoma cells including breast cancer cells [51, 52]. Vorinostat also reduced tumor incidence in NMU-induced rat mammary tumorigenesis by 40 % [53]. In vitro studies demonstrated that vorinostat inhibits clonogenic growth of both ER-positive and ER-negative breast cancer cell lines by inducing G1 and G2/M cell cycle arrest and subsequent apoptosis [54]. Exposure to low concentrations of vorinostat is also associated with accumulation of cells mainly in G1, while higher vorinostat concentrations cause cell cycle arrest predominantly in G2/M [52].
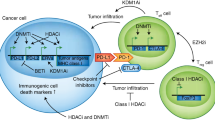
The ability of the HDAC inhibitors to relieve transcriptional repression in preclinical breast cancer models has also been investigated. The accumulation of acetylated H3 and H4 histone tails in conjunction with re-expression of a functional ER in ER-negative breast cancer cell lines has been observed with a novel HDAC inhibitor, scriptaid [55]. Treatment of ER-negative breast cancer cell lines with vorinostat is associated with reactivation of silenced ER, as well as downregulation of DNMT1 and EGFR protein expression [56]. The significance of an epigenetically reactivated ER was demonstrated when tamoxifen sensitivity was restored in the ER-negative MDA-MB-231 breast cancer cells following treatment with both HDAC (trichostatin A) and DNMT inhibitors (DAC) [57]. Entinostat has been shown to induce not only re-expression of ERα, but also the androgen receptor and the aromatase enzyme (CYP19) both in vitro and in triple-negative breast cancer xenografts [58]. In addition, the combination of entinostat and letrozole resulted in a significant and durable reduction in the xenograft tumor volume when compared to treatment with either agent alone. These experiments have provided the strong rationale for combining epigenetic modifiers with hormonal therapy in breast cancer clinical trials [58]. Interestingly, many of these studies also indicate that a strategy which combines HDAC and DNMT inhibitors is more efficacious than either agent alone with respect to both re-expression of silenced genes and restoration of response to tamoxifen and aromatase inhibitors [55, 59, 60].
The RARβ gene, well known for its’ tumor suppressive effects in epithelial cells, can also be subject to gene silencing by epigenetic modification in breast cancer models. Reactivation of the gene has been demonstrated with use of both HDAC and DNMT inhibitors [9]. Clinical studies investigating the retinoids in various breast cancer populations to date have yielded disappointing results, but perhaps the lack of efficacy observed relates to the fact that RARβ expression was not evaluated in the majority of these studies [61, 62]. It is possible that more careful patient selection based on RARβ expression in the primary tumor and the use of epigenetic modifiers to reactivate the pathway if silenced will be necessary to produce clinically meaningful results.
Pretreatment of various tumor cell lines with HDAC inhibitors increases the cytotoxicity of chemotherapy. Administering the HDAC inhibitor after chemotherapy did not achieve the same results, suggesting that pretreatment with these agents may open the chromatin structure and thus facilitate an enhanced anti-cancer effect of chemotherapy drugs that target DNA [63]. In breast cancer cell lines with amplification and overexpression of HER2, HDAC inhibitor use depleted HER2 by attenuation of its mRNA levels and promotion of proteosomal degradation. HDAC inhibition also enhanced apoptosis induced by trastuzumab, docetaxel, epothilone B, and gemcitabine [64]. HDAC inhibitors also significantly enhance trastuzumab-induced growth inhibition in trastuzumab-sensitive, HER2-overexpressing breast cancer cells, providing a strong rationale for clinical studies with this combination in patients with HER2-positive disease [65, 66].
Preclinical Activity of the DNMT Inhibitors
Scientists evaluated the administration of low doses of AZA and DAC in breast cancer cell lines and in xenograft models in an attempt to harness the tumor “reprogramming” effect of these agents. Nanomolar (nM) doses of these agents (e.g. 100nM DAC and 500nM AZA) resulted in the development of an anti-tumor “memory” response which inhibited the growth of cancer cells, including traditionally resistant stem-like cells [67]. These effects were observed without evidence of immediate cytotoxicity. Sustained, genome-wide alterations in promoter methylation and gene expression which affected major cell signaling pathways were also observed. The alteration of promoter methylation of specific tumor suppressor genes including ER, BRCA-1, E-cadherin, PTEN and MASPIN have been demonstrated previously in breast cancer cell lines exposed to these agents in a number of other studies [68–70]. In addition, DNMT inhibitors have been shown to sensitize breast cancer cell lines to the chemotherapeutic agent doxorubicin by inducing tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) [71]. Preclinical studies combining a DNMT inhibitor with an HDAC inhibitor have also yielded promising results as described above [55, 59, 60]. Ongoing studies aim to elucidate novel combination strategies relevant to breast cancer treatment paradigms including combinations of epigenetic modifiers with tamoxifen, aromatase inhibitors, trastuzumab, and cytotoxic agents, amongst others.
Breast Cancer: Clinical Experience
Based on the approval of both DNMT inhibitors and the HDAC inhibitors for various hematologic malignancies [21, 23], clinical investigators extended studies of epigenetic modifiers to the solid tumor arena either as single agents or in combination with both standard and investigational therapies. A number of early phase solid tumor clinical trials have been completed or are ongoing (Table 2).
HDAC Inhibitors
Vorinostat is a potent HDAC inhibitor which targets class 1 and 2 HDACs. This agent has been shown to inhibit proliferation of both ER-positive and ER-negative breast cancer cell lines and induce cell cycle arrest [52]. It was the first HDAC inhibitor available for investigator-initiated trials. Antitumor activity was observed in a phase 1 trial investigating oral vorinostat in patients with advanced cancer [72]. Based on the results from this phase 1 study, alongside strong preclinical rationale, a phase 2 trial of single agent vorinostat was designed in the advanced breast cancer setting. Fourteen patients (8 ER/PR-positive, 4 HER2-positive) were treated with single agent oral vorinostat at a dose of 200 mg oral twice daily for 14 days of a 21 day cycle [73]. Median number of prior chemotherapies was only 1.5 (range 0-2). The study failed to reach its primary endpoint (response rate by RECIST criteria), however, stable disease was observed in almost 30 % of patients (n = 4) with time to progression of 4, 8, 9 and 14 months in these patients. The therapy was well tolerated with the most common adverse events including fatigue, nausea and diarrhea. In addition, a presurgical or “window” biomarker study has evaluated surrogate markers of response with short term administration of vorinostat. Vorinostat 300 mg given twice daily for six doses to women awaiting definitive breast surgery was associated with a reduction in proliferation-related gene expression by RT-PCR (reverse transcriptase polymerase chain reaction) using the Oncotype Dx assay. However, significant changes in cell death, candidate gene methylation or expression of candidate genes such as the ER were not observed. The results support preclinical findings that while the agent may inhibit proliferation, ER and other gene expression modulation is more likely to be seen following the combination of a HDAC inhibitor with another anti-cancer agent or epigenetic modifier [74].
Whether the addition of a HDAC inhibitor to hormonal strategies for breast cancer can reverse resistance to hormonal therapy and therefore improve breast cancer outcomes has been investigated. Preclinical models which suggested that the efficacy of tamoxifen can be enhanced by vorinostat [14] prompted the development of a phase 2 trial in advanced breast cancer patients. Women with hormone-resistant breast cancer (n = 43) received oral vorinostat 400 mg daily (21 days of a 28 day cycle) and tamoxifen 20 mg daily, with the investigators reporting an objective response rate of 19 % and a clinical benefit rate of 40 % [15]. The combination of agents was well tolerated with no unexpected adverse events. Approximately 60 % of patients received prior tamoxifen in the adjuvant setting, and 54 % had received two prior lines of therapy with aromatase inhibitors. Histone hyperacetylation and high baseline HDAC 2 levels were predictive of response. These findings suggest that vorinostat may indeed restore responsiveness to tamoxifen in patients with hormone-resistant disease, and support the development of a randomized trial for further delineation of the clinical relevance of the combination.
In ENCORE 301, investigators evaluated the role of entinostat, a class 1 selective HDAC inhibitor, in combination with exemestane (steroidal aromatase inhibitor, AI) in a randomized phase 2 study in the advanced breast cancer setting. Postmenopausal women who had received 0–1 prior chemotherapy and had progressed on a non-steroidal AI were randomized to exemestane 25 mg daily plus entinostat 5 mg or placebo weekly [75]. A significant improvement in progression-free survival (PFS) was noted in the entinostat arm compared to placebo (median 4.28 versus 2.27 months, respectively). Preliminary results suggested that overall survival (OS), a secondary endpoint, was also significantly longer in the entinostat arm versus the placebo arm (26.94 versus 20.33 months, respectively). These promising results met the pre-specified study statistical plan and support the need for a phase 3 study. Interestingly, in a subset analysis examining protein acetylation in patients receiving entinostat (n = 27), the median PFS was 8.5 months in those exhibiting protein lysine hyperacetylation versus those who did not, and this was apparent after just 2 weeks of therapy [12]. It is not known whether the addition of entinostat in this setting resulted in the re-expression of the ER, a question which will be answered in ongoing studies [76]. It is also not clear whether the observed benefit reflects activity of entinostat or its combination with exemestane. A phase 3 study is in the planning stages to further delineate the role of entinostat and the potential role of lysine acetylation as a biomarker predicting efficacy.
To date, few studies have combined HDAC inhibitors with standard cytotoxic agents specifically in breast cancer populations. Phase 1 studies in advanced solid tumors have combined vorinostat either with doxorubicin [13], or with a paclitaxel and carboplatin combination [77] and suggested enhanced antitumor activity. In the study which combined vorinostat with doxorubicin, histone hyperacetylation correlated with pre-treatment HDAC 2 expression. Vorinostat has also been investigated in the 1st line metastatic setting in combination with paclitaxel and bevacizumab [78]. In a single arm study, an overall response rate of 55 % was observed which is relatively consistent with that seen in studies evaluating the paclitaxel/bevacizumab combination alone [79]. Median progression-free survival was 11.9 months and median overall survival was 29.4 months. Diarrhea was the only toxicity observed in addition to that expected with paclitaxel and bevacizumab. The investigators demonstrated increased acetylation of Hsp90 and α-tubulin in tumor biopsies obtained from a subset of patients (n = 7). Finally, a neoadjuvant study randomizing patients with primary operable breast cancer to 12 weeks of nab-paclitaxel and carboplatin plus vorinostat or placebo has recently been completed [80]. Patients with both triple-negative and intermediate or high grade hormone receptor-positive breast cancer were included and the primary objective was to determine pathologic complete response rates to neoadjuvant chemotherapy with or without vorinostat.
Accumulating evidence has demonstrated the effectiveness of HDAC inhibitors in combination with several other agents in vitro leading to new clinical trials in which HDAC inhibitors are combined with retinoids [81], antiHER2 strategies [82, 83], hypomethylating agents [84, 85], and many other novel agents.
DNMT Inhibitors
Early phase 1 and 2 trials investigating the role of demethylating agents in solid tumors, including breast cancer, yielded a number of anecdotal responses [86, 87]. A review of previously published work indicated that DNMT inhibitors were associated with response rates as high as 18 % in breast cancer [28]. The doses of AZA that were employed in these studies, however, were far higher than doses used in clinical trials today (as high as 188 mg/m2/day) and likely exerted cytotoxic activity as opposed to relief of transcriptional repression as an anti-cancer strategy [28]. Interestingly, the highest rates of response were reported in patients treated at a dose which approximates the 75 mg/m2/day dosing employed in hematologic malignancies.
Ongoing clinical studies with administration of DNMT inhibitors at the presumed optimal epigenetic dose aim to elucidate the biological effects of these agents, and to assess clinical efficacy, alone or in combination with other anti-cancer agents. The ability of single agent AZA to induce expression of the ER and PR genes in patients with triple-negative breast cancer who are awaiting definitive breast cancer surgery is under investigation using a 75 mg/m2/day dosing schedule [88]. Based on the preclinical evidence previously described which suggests that a combination of epigenetic modifiers may be more successful in re-expression of silenced genes and restoration of hormonal therapy responsiveness, patients with advanced triple-negative and hormone-resistant breast cancer are being enrolled in an ongoing multicenter phase 2 clinical trial and receive the combination of low dose AZA (40 mg/m2) on days 1–5 and 8–10, and entinostat 7 mg on days 3 and 10 of a 28 day cycle. Tumor biopsies prior to and after therapy are collected to assess modulation of candidate gene methylation and expression, such as the ER. Patients may transition to an optional continuation phase at the time of disease progression in which the same epigenetic therapy is administered with the addition of hormonal therapy [89]. Indeed, in a recently published trial exploring the combination of AZA and entinostat in advanced non-small cell lung cancer patients, investigators observed that the regimen was well tolerated and associated with a number of objective responses [85]. These included a complete response as well as a partial response in a patient without progression of disease for 2 years after completing the clinical trial. Interestingly, a number of patients were found to have unexpected major objective responses to subsequent anti-cancer strategies, raising the question as to whether these agents may prime tumor cells to respond to subsequent therapies. A phase 1/2 Canadian trial investigating the combination of decitabine and vorinostat in patients with advanced solid tumors or hematologic malignancies has also indicated clinical activity. Stabilization of disease for 4 or more cycles was observed in 29 % evaluable patients; two of these patients had metastatic breast cancer [90].
The combination of DNMT inhibitors with standard chemotherapy has not been extensively evaluated in the breast cancer setting. Based on strong preclinical evidence that the addition of AZA could overcome platinum resistance through DNA hypomethylation, patients with both platinum resistant and refractory ovarian cancer received the combination of AZA and carboplatin after being enrolled into a phase 1b/2 study. An overall response rate of 22 % was observed in the platinum-resistant patients (disease progression within 6 months of platinum, n = 18) suggesting that further evaluation of the combination was warranted [91]. A phase 1/2 clinical trial of AZA and nab-paclitaxel is ongoing in patients with advanced solid tumors and breast cancer [92]. Whether combining DNMT inhibitors with standard therapies or novel agents will result in clinical benefit for patients with breast cancer remains to be seen. In the meantime, robust preclinical data should support the development of new concepts in order to maximize the chance of success with these agents in the solid tumor arena.
Challenges and Future Directions
Because epigenetic alterations are frequently detected in breast tumors, agents that target these alterations are of great interest and are under ongoing investigation. Some promise has been observed clinically in studies in which a hormonal agent was added to HDAC inhibitors in the phase 2 setting [15, 75]. Phase 3 trials will confirm or refute this success. However, several challenges must be overcome to allow for optimal incorporation of epigenetic modifiers into the solid tumor and specifically breast cancer clinical treatment paradigm.
The dose and schedule of the DNMT inhibitors in breast cancer clinical trials remains to be defined. Laboratory studies have shown that AZA and DAC optimally inhibit DNA methylation when used at lower than cytotoxic doses with prolonged exposures [27, 67]. This reflects what is observed clinically in patients with hematologic malignancies treated with these agents [29, 84]. The exact impact of using epigenetic modifiers at an optimally epigenetic dose instead of a cytotoxic dose is yet unknown in solid tumors, despite the supposition that anti-cancer activity will be enhanced. Ongoing clinical trials in breast cancer patients aim to elucidate this question [89].
Rational combinations of epigenetic modifiers with other anti-cancer agents such as hormonal therapy, standard chemotherapy, anti-HER2 therapy, retinoids and novel agents such as Hsp90 inhibitors or other small molecule inhibitors may yield greater results than with either agent alone in the clinic. Again, what schedule of administration is utilized may be key and should ideally be modeled preclinically prior to the initiation of clinical trials. For example, an attempt to enhance efficacy to standard chemotherapy by “priming” cells with DNMT inhibitors prior to the administration of chemotherapy is under investigation [93].
Optimizing the use of the clinically available epigenetic modifiers is clearly important. An oral form of AZA is currently in development which may be far more convenient for patients than the intravenous and subcutaneous routes employed at this time. A number of new agents are also in development which may circumvent some of the limitations of the currently available drugs such as their in vivo deamination by cytidine deaminase (CDA), and tendency to be subject to drug resistance. For example, SGI-110 is a DAC analogue in phase 1 development that is largely resistant to CDA such that resistance to therapy may be avoided with this agent [94]. The development of DNMT inhibitors that do not require incorporation into DNA for activity may also circumvent drug resistance by targeting DNMT directly. In addition, the development of DNMT inhibitors selective for the various DNMT enzymes may be relevant if a disease is found to be driven by an individual enzyme, minimizing unwanted off-target effects [95]. Finally, novel agents that target histone methyltransferase (e.g. DOTL1) [96] or histone demethylase (e.g. LSD1) [97] regulate DNMT1 stability and may be utilized in the clinic in the future, most likely in combination with other agents including epigenetic modifiers.
Robust biomarkers of response to epigenetic modifiers in the breast cancer setting remain elusive. Protein lysine acetylation may predict response to entinostat in women receiving the combination of entinostat plus exemestane [12]. Histone hyperacetylation and higher baseline HDAC 2 levels were found to correlate with response in a single arm phase 2 trial of vorinostat and tamoxifen in women with hormone-resistant breast cancer [15]. Validation of these findings is necessary before these potential biomarkers can be used clinically. Potential biomarkers of response to epigenetic agents in other disease settings include DNMT3a mutations in acute myeloid leukemia [98], and demethylation of a set of four lung cancer-associated epigenetically silenced genes in patients with lung cancer [85]. The prospective collection of both blood and tissue samples as a component of clinical trials will be essential to define the population of patients who will benefit from these therapies in the future.
Another important issue for clinical investigators is the design of clinical trials which incorporate epigenetic modifiers. Traditionally, response rate and time to tumor progression have been utilized as primary endpoints for therapeutic trials, as a surrogate for survival from cancer. However, a high response rate with AZA and DAC did not translate to a survival benefit in hematologic malignancy studies. In breast cancer, specifically, single agent vorinostat resulted in clinical benefit as evidenced by stable disease in 30 % of patients, but did not yield objective responses by RECIST criteria [99]. Whether a modification of the standard response assessment criteria is necessary for trials investigating epigenetic agents is a matter for discussion, as has been proposed by those investigating immune therapies as anti-cancer agents [100].
Conclusion
The field of epigenetics is an exciting and ever evolving area of investigation in cancer research and therapeutics. Despite the approval of the HDAC and DNMT inhibitors for varying indications in hematologic malignancies, significant challenges remain regarding the addition of these agents to the armamentarium of drugs used to treat solid tumors including breast cancer. The potential for the HDAC inhibitors to overcome resistance to hormonal therapy in advanced breast cancer patients has been highlighted in recent times, but requires confirmatory trials before this information can be used in the clinic. Importantly, we are moving closer to identifying a robust biomarker to predict response to the HDAC inhibitors, but again validation is required and will hopefully be achieved in future clinical trials. Important questions remain regarding the optimal dose, schedule and combination of epigenetic modifiers in the breast cancer setting. In this era of personalized medicine, investigators continue to design preclinical experiments and clinical trials in order to answer these questions such that the right patients may benefit from these agents in the not too distant future.
Abbreviations
- AZA:
-
5-azacitidine
- CDA:
-
cytidine deaminase
- CTCL:
-
cutaneous T-cell lymphoma
- DAC:
-
decitabine
- DNA:
-
deoxyribonucleic acid
- DNMT:
-
DNA methyltransferase
- DNMTs:
-
DNA methyltransferase enzymes
- ER:
-
estrogen receptor
- FDA:
-
Food and Drug Administration
- HAT:
-
histone acetyltranserase
- HDAC:
-
histone deacetylase
- MDS:
-
myelodysplastic syndrome
- miRNAs:
-
micro RNAs
- mRNA:
-
messenger RNA
- MSP:
-
methylation-specific polymerase chain reaction
- uM:
-
micromolar
- nM:
-
nanomolar
- PARP:
-
poly(adenosine diphosphate)-ribose polymerase
- PBMCs:
-
peripheral blood mononuclear cells
- PTCL:
-
peripheral T-cell lymphoma
- PR:
-
progesterone receptor
- QM-MSP:
-
quantitative multiplex methylation-specific PCR
- RNA:
-
ribonucleic acid
- RT-PCR:
-
reverse transcriptase polymerase chain reaction
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
References
Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54.
Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36:75–88.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80.
Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28.
Arts J, de Schepper S, Van Emelen K. Histone deacetylase inhibitors: from chromatin remodeling to experimental cancer therapeutics. Curr Med Chem. 2003;10:2343–50.
Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–69.
Pedrali-Noy G, Weissbach A. Mammalian DNA methyltransferases prefer poly(dI-dC) as substrate. J Biol Chem. 1986;261:7600–2.
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57.
Fackler MJ, Rivers A, Teo WW, et al. Hypermethylated genes as biomarkers of cancer in women with pathologic nipple discharge. Clin Cancer Res. 2009;15:3802–11.
Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4.
Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28.
Ordentlich P. Pharmacodynamic analysis of ENCORE 301, a placebo-controlled, randomized phase 2 study of exemestane with and without entinostat in postmenopausal ER + breast cancer patients demonstrates an association of lysine hyperacetylation with clinical outcome. Presented at the AACR Molecular Targets Conference 2011 (Abstract PR-6).
Munster PN, Marchion D, Thomas S, et al. Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br J Cancer. 2009;101:1044–50.
Hodges-Gallagher L, Valentine CD, Bader SE, Kushner PJ. Inhibition of histone deacetylase enhances the anti-proliferative action of antiestrogens on breast cancer cells and blocks tamoxifen-induced proliferation of uterine cells. Breast Cancer Res Treat. 2007;105:297–309.
Munster PN, Thurn KT, Thomas S, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–35.
Licchesi JD, Herman JG. Methylation-specific PCR. Methods Mol Biol. 2009;507:305–23.
Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–52.
Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98.
Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47(3):142–50.
Gore SD, Weng LJ, Zhai S, et al. Impact of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res. 2001;7:2330–9.
Olsen EA, Kim YH, Kuzel TM, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–15.
Piekarz RL, Frye R, Turner M, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–7.
Piekarz RL, Frye R, Prince HM, et al. Phase II trial of romidepsin in patients with peripheral T-cell lymphoma. Blood. 2011;117(22):5827–34.
Whittaker SJ, Demierre MF, Kim EJ, et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28:4485–91.
Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93.
Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41.
Qin T, Youssef EM, Jelinek J, et al. Effect of cytarabine and decitabine in combination in human leukemic cell lines. Clin Cancer Res. 2007;13:4225–32.
Cowan LA, Talwar S, Yang AS. Will DNA methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics. 2010;2:71–86.
Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–32.
Steensma DP, Baer MR, Slack JL, et al. Multicenter study of decitabine administered daily for 5 days every 4 weeks to adults with myelodysplastic syndromes: the alternative dosing for outpatient treatment (ADOPT) trial. J Clin Oncol. 2009;27:3842–8.
Miyake K, Yoshizumi T, Imura S, et al. Expression of hypoxia-inducible factor-1alpha, histone deacetylase 1, and metastasis-associated protein 1 in pancreatic carcinoma: correlation with poor prognosis with possible regulation. Pancreas. 2008;36:e1–9.
Krusche CA, Wulfing P, Kersting C, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat. 2005;90:15–23.
Zhang Z, Yamashita H, Toyama T, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10:6962–8.
Soares J, Pinto AE, Cunha CV, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–8.
Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–73.
Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–82.
Pu RT, Laitala LE, Alli PM, Fackler MJ, Sukumar S, Clark DP. Methylation profiling of benign and malignant breast lesions and its application to cytopathology. Mod Pathol. 2003;16:1095–101.
Fackler MJ, McVeigh M, Evron E, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–5.
Sebova K, Zmetakova I, Bella V, et al. RASSF1A and CDH1 hypermethylation as potential epimarkers in breast cancer. Cancer Biomark. 2011;10:13–26.
Tserga A, Michalopoulos NV, Levidou G, et al. Association of aberrant DNA methylation with clinicopathological features. Oncol Rep. 2012;27(5):1630–8.
Lapidus RG, Nass SJ, Butash KA, et al. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer research. 1998;58:2515–9.
Fackler MJ, Umbricht CB, Williams D, et al. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71:6195–207.
Hill VK, Ricketts C, Bieche I, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011;71:2988–99.
Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3:726–41.
Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34.
Veeck J, Ropero S, Setien F, et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol. 2010;28:e563–4. author reply e5-6.
Schrauder MG, Strick R, Schulz-Wendtland R, et al. Circulating Micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2011;7:e29770.
Ferracin M, Querzoli P, Calin GA, Negrini M. MicroRNAs: toward the clinic for breast cancer patients. Semin Oncol. 2011;38:764–75.
Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–9.
Said TK, Moraes RC, Sinha R, Medina D. Mechanisms of suberoylanilide hydroxamic acid inhibition of mammary cell growth. Breast Cancer Res. 2001;3:122–33.
Munster PN, Troso-Sandoval T, Rosen N, Rifkind R, Marks PA, Richon VM. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–7.
Cohen LA, Amin S, Marks PA, Rifkind RA, Desai D, Richon VM. Chemoprevention of carcinogen-induced mammary tumorigenesis by the hybrid polar cytodifferentiation agent, suberanilohydroxamic acid (SAHA). Anticancer Res. 1999;19:4999–5005.
Huang L, Pardee AB. Suberoylanilide hydroxamic acid as a potential therapeutic agent for human breast cancer treatment. Mol Med. 2000;6:849–66.
Keen JC, Yan L, Mack KM, et al. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2′-deoxycytidine. Breast Cancer Res Treat. 2003;81:177–86.
Zhou Q, Shaw PG, Davidson NE. Inhibition of histone deacetylase suppresses EGF signaling pathways by destabilizing EGFR mRNA in ER-negative human breast cancer cells. Breast Cancer Res Treat. 2009;117(2):443–51.
Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66:6370–8.
Sabnis GJ, Goloubeva O, Chumsri S, Nguyen N, Sukumar S, Brodie AM. Functional activation of the estrogen receptor-alpha and aromatase by the HDAC inhibitor entinostat sensitizes ER-negative tumors to letrozole. Cancer Res. 2011;71:1893–903.
Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 2001;61:7025–9.
Fan J, Yin WJ, Lu JS, et al. ER alpha negative breast cancer cells restore response to endocrine therapy by combination treatment with both HDAC inhibitor and DNMT inhibitor. J Cancer Res Clin Oncol. 2008;134:883–90.
Cassidy J, Lippman M, Lacroix A, Peck G. Phase II trial of 13-cis-retinoic acid in metastatic breast cancer. Eur J Cancer Clin Oncol. 1982;18:925–8.
Veronesi U, Mariani L, Decensi A, et al. Fifteen-year results of a randomized phase III trial of fenretinide to prevent second breast cancer. Ann Oncol. 2006;17:1065–71.
Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–300.
Fuino L, Bali P, Wittmann S, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther. 2003;2:971–84.
Huang X, Wang S, Lee CK, Yang X, Liu B. HDAC inhibitor SNDX-275 enhances efficacy of trastuzumab in erbB2-overexpressing breast cancer cells and exhibits potential to overcome trastuzumab resistance. Cancer Lett. 2011;307:72–9.
Bali P, Pranpat M, Swaby R, et al. Activity of suberoylanilide hydroxamic Acid against human breast cancer cells with amplification of her-2. Clin Cancer Res. 2005;11:6382–9.
Tsai HC, Li H, Van Neste L, et al. Transient Low Doses of DNA Demethylating Agents Exert Durable Anti-tumor Effects on Hematological and Epithelial Tumor Cells. Cancer Cell. 2012;21(3):430–46.
Krawczyk B, Fabianowska-Majewska K. Alteration of DNA methylation status in K562 and MCF-7 cancer cell lines by nucleoside analogues. Nucleosides Nucleotides Nucleic Acids. 2006;25:1029–32.
Krawczyk B, Rudnicka K, Fabianowska-Majewska K. The effects of nucleoside analogues on promoter methylation of selected tumor suppressor genes in MCF-7 and MDA-MB-231 breast cancer cell lines. Nucleosides Nucleotides Nucleic Acids. 2007;26:1043–6.
Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90.
Xu J, Zhou JY, Tainsky MA, Wu GS. Evidence that tumor necrosis factor-related apoptosis-inducing ligand induction by 5-Aza-2′-deoxycytidine sensitizes human breast cancer cells to adriamycin. Cancer Res. 2007;67:1203–11.
Kelly WK, O’Connor OA, Krug LM, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–31.
Luu TH, Morgan RJ, Leong L, et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–42.
Stearns V, Jacobs LK, Tsangaris TN, et al. Association of vorinostat with decrease in gene expression of proliferation-related genes in tumors from women with newly diagnosed breast cancer. Abs 3097. Journal of Clinical Oncology (suppl). 2010;28.
Yardley DA, Ismail-Khan RR, Klein PM. Entinostat, a Novel histone deacetylase inhibitor, added to exemestane improves PFS in advanced breast cancer in a randomized, phase II, Double-blind study. Presented at the San Antonio Breast Cancer Symposium 2011 (Abstract PD01-04).
http://clinicaltrials.gov/ct2/show/NCT01234532?term=chumsri&rank=3. Accessed online February 13th 2012.
Ramalingam SS, Parise RA, Ramanathan RK, et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13:3605–10.
Ramaswamy B, Fiskus W, Cohen B, et al. Phase I-II study of vorinostat plus paclitaxel and bevacizumab in metastatic breast cancer: evidence for vorinostat-induced tubulin acetylation and Hsp90 inhibition in vivo. Breast Cancer Res Treat. 2012; 132(3):1063–72.
Gray R, Bhattacharya S, Bowden C, Miller K, Comis RL. Independent review of E2100: a phase III trial of bevacizumab plus paclitaxel versus paclitaxel in women with metastatic breast cancer. J Clin Oncol. 2009;27:4966–72.
Connolly RM, Leal JP, et al. Early change in 18-fluorodeoxyglucose (FDG) uptake on positron emission tomography (PET) predicts response to preoperative systemic therapy (PST) in HER2-negative primary operable breast cancer: translational breast cancer research consortium (TBCRC008). Presented at the American Society of Clinical Oncology (ASCO) meeting 2012. J Clin Oncol. 2012;30(suppl; abstr 10509).
Raffoux E, Cras A, Recher C, et al. Phase 2 clinical trial of 5-azacitidine, valproic acid, and all-trans retinoic acid in patients with high-risk acute myeloid leukemia or myelodysplastic syndrome. Oncotarget. 2010;1:34–42.
http://clinicaltrials.gov/ct2/show/NCT00258349?term=swaby&rank=2. Accessed online February 13th 2012.
http://clinicaltrials.gov/ct2/show/NCT01118975?term=vorinostat+and+breast+cancer&rank=2. Accessed online February 13th 2012.
Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9.
Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non–small cell lung cancer. Cancer Discovery. 2011;1(7):598–607.
Weiss AJ, Metter GE, Nealon TF, et al. Phase II study of 5-azacytidine in solid tumors. Cancer Treat Rep. 1977;61:55–8.
Weiss AJ, Stambaugh JE, Mastrangelo MJ, Laucius JF, Bellet RE. Phase I study of 5-azacytidine (NSC-102816). Cancer Chemother Rep. 1972;56:413–9.
http://clinicaltrials.gov/ct2/show/NCT01292083?term=NCT01292083&rank=1. Accessed online January 7th 2012.
Connolly RM, Jankowitz RC, Andreopoulou E, et al. A Phase 2 study investigating the safety, efficacy and surrogate biomarkers of response of 5-Azacitidine (5-AZA) and Entinostat (MS-275) in Patients with Advanced Breast Cancer. SABCS 2011 Ongoing Trials Session (Abs OT3 − 01 − 06).
Stathis A, Hotte SJ, Chen EX, et al. Phase I study of decitabine in combination with vorinostat in patients with advanced solid tumors and non-Hodgkin’s lymphomas. Clin Cancer Res. 2011;17:1582–90.
Fu S, Hu W, Iyer R, et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer. 2011;117:1661–9.
http://clinicaltrials.gov/ct2/show/NCT00748553?term=NCT00748553&rank=1. Accessed online January 8th 2012.
Scandura JM, Roboz GJ, Moh M, et al. Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood. 2011;118:1472–80.
Chuang JC, Warner SL, Vollmer D, et al. S110, a 5-Aza-2′-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther. 2010;9:1443–50.
Foulks JM, Parnell KM, Nix RN, et al. Epigenetic drug discovery: targeting DNA methyltransferases. J Biomol Screen. 2011;17:2–17.
Yao Y, Chen P, Diao J, et al. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J Am Chem Soc. 2011;133:16746–9.
Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2012;131(3):777–89.
Metzeler KH, Walker A, Geyer S, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26(5):1106–7.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
Wolchok JD, Hoos A, O’Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–20.
Acknowledgements
Supported by Specialized Program Of Research Excellence in Breast Cancer (P50 CA88843), Stand Up To Cancer, and QVC and Fashion Footwear Association of New York (FFANY).
Disclosure
Dr. Stearns has received investigator-initiated grants from Abraxis, Merck, Novartis, and Pfizer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Connolly, R., Stearns, V. Epigenetics as a Therapeutic Target in Breast Cancer. J Mammary Gland Biol Neoplasia 17, 191–204 (2012). https://doi.org/10.1007/s10911-012-9263-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-012-9263-3