Abstract
A novel series of Ni(II)-Schiff base complexes were synthesized from five hydrazide derivatives. All new synthesizes were fully characterized applying analytical and spectral techniques. 1:1 molar-ratio (M:L) was the main ratio isolated that represented by neutral bi-dentate mode of bonding. Square-planer configuration was the only structural formula proposed for all faintly-colored Ni(II) complexes. Such structural forms were supported by UV–Vis data as well as diamagnetic characteristic. All investigated compounds were configured to reach optimum forms by Gaussian09 program under RTD-B3LYP-FC method. Also, HyperChem (8.1) program was implemented towards Ni(II) complexes to extract QSAR parameters after energy-minimization. Fundamental data were obtained after computational execution, among that which offer a good expectation for Ni(II)-5c, Ni(II)-5d and Ni(II)-5e complexes in aimed application field (therapeutic). MOE-docking approach was considered for all Ni(II) complexes versus 3s7s protein, to signify inhibition-characteristic and rank that. The interaction parameters presented Ni(II)-5c, Ni(II)-5d and Ni(II)-5e complexes, as a best anti-tumor inhibitors, which exactly appeared in laboratory screening.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Pyridine derivatives have been widely studied for over a century because of their utilization in many branches of chemistry, such as catalysis and therapeutic applications [1]. Many pyridine-containing compounds display fabulous medicinal properties, including hypnotic and sedative [2], HIV antiviral [3], hepatic endoplasmic reticulum calcium uptake regulator [4], α-glucosidase inhibitory, and anticancer activities [5]. Various research studies are continuing to develop and improve specific target-based anticancer drugs in order to reduce drug toxicity, resistance, dosage, and more [6]. Furthermore, benzohydrazide derivatives are considered serious moieties that display great effective inhibitory activity towards several cancer cells [7]. Increasing interest which focused on structural-based therapeutic reagent, designing approach for structure–activity relationships (SAR) was conducted. This will provide excellent rules for the choosing of novel and high active compounds [8], which can be prepared and examined experimentally. Considerable numbers of clinically effective anticancer drugs, are either naturally existing or designed and prepared from other synthetic analogs. Metal complexes display unique characteristics enhancing their active role as promising therapeutic agents, especially as anticancer drugs [9,10,11]. Great and remarkable characteristics as the ability to produce positively charged ions in aquatic solutions, which may bind with negatively charged biological molecules [12]. The elevated electron affinity of different metal ions can worthy polarize coordinating groups, which droving to propagation of hydrolysis reactions [13]. Recently, significant interest has been oriented to metal-complexes of hydrazides and pyridine based on their biological significant [14]. In recent years, great attention is directed to design and synthesize novel nanosized metal complexes due to their high potential applicability as anticancer and genotoxic agents [15,16,17]. In this work we planned to synthesize benzohydrazide derivatives, to be the nucleus for preparing series of their Ni(II) complexes. Descripting molecular and structural formulae of all synthesizes by implementing all available tools was the following step. Theoretical approach was elaborately handled in this study to establish best atomic skeletons or conducts docking simulation. MOE-docking results will be compared deliberately with that resulted from in-vitro screening against breast-cancer cell line, to assess the conformity-extent.
2 Experimental Work
The implemented chemicals and reagents were Merck and PDH from Sigma-Aldrich, also all solvents used were spectroscopic and used as it is without any treatment or purification.
2.1 Synthesis
Synthesis of Ethyl 4-((3-cyano-4,6-dimethylpyridin-2-yl)amino)-benzoate (3): To a suspension of 2-chloro-4,6-dimethylnicotinonitrile (2) (1.66 g, 0.01 mol) and ethyl p-aminobenzoate (1.65 g, 0.01 mol) in ethyl alcohol (30 ml), HCl (1 ml, conc.) was added. The reaction mixture was suffered heating under reflux, for 8 h followed by cooling at 25 °C. The white precipitate formed was filtered off, dried and recrystallized from ethyl alcohol.
White solid, yield 40%, m.p. = 140–141 °C. IR (KBr); 3309 (N–H), 2223 (C≡N), 1712 cm−1 (C=O). 1H NMR (DMSO-d6) (Fig. 1S): 1.31 (t, J = 7.0 Hz, 3H), 2.39 (s, 3H), 2.41 (s, 3H), 4.28 (q, J = 7.0 Hz, 2H), 6.87 (s, 1H, pyridine H-5), 7.74 (d, J = 8.4 Hz, 2H, Ar–H), 7.87 (d, J = 8.4 Hz, 2H, Ar–H), 9.34 ppm (s, 1H, NH). Analysis for C17H17N3O2; Calcd (Found): C, 69.14 (69.14); H, 5.80 (5.80); N, 14.23 (14.23)%.
Synthesis of 4-((3-Cyano-4,6-dimethylpyridin-2-yl)amino)-benzohydrazide (4): To a suspension of ethyl benzoate derivative (3) (2.95 g, 0.01 mol) in ethyl alcohol (30 ml), hydrazine hydrate (1 ml, 0.01 mol) was added. The reaction components were heated under reflux for 12 h and then cooled to 25 °C. The precipitate that formed was collected by filtration and then recrystallized from ethyl alcohol.
White crystals, yield 51%, m.p. = 240–242 °C. IR (KBr): 3340, 3287 (NH and NH2), 2217 (C≡N), 1611 cm−1 (C=O). 1H NMR (DMSO-d6) (Fig. 1S); 2.35 (s, 3H), 2.37 (s, 3H), 4.44 (s, 2H, NH2), 6.80 (s, 1H, pyridine H-5), 7.65 (d, J = 9.0 Hz, 2H), 7.74 (d, J = 8.0 Hz, 2H), 9.14 (s, 1H, NH), 9.60 ppm (s, 1H, NH). Analysis for C15H15N5O; Calcd. (Found): C, 64.04 (64.15); H, 5.37 (5.34); N, 24.90 (24.96)%.
2.1.1 Synthesis of N′-arylidene-4-((3-cyano-4,6-dimethylpyridin-2-yl)amino)-benzohydrazide 5a–e
A mixture of benzohydrazide derivative (4) (0.56 g, 0.002 mol) and 0.002 mol from appropriate para substituted benzaldehyde (namely; benzaldehyde, 4-methylbenzaldehyde, 4-methoxybenzaldehyde, 4-nitro-benzaldehyde and 4-chlorobenzaldehyde) in 20 ml ethyl alcohol, was refluxed for 2 h. The crystalline solid that formed upon cooling was filtered off to afford N′-arylidene derivatives (5a–e).
N′-Benzylidene-4-((3-cyano-4,6-dimethylpyridin-2-yl)amino)-benzohydrazide (5a): White solid (C22H19N5O), yield 60%, m.p. = 260–261 °C. IR (KBr); 3412, 3257 (N–H), 2208 (C≡N), 1648 cm−1 (C=O).
4-((3-Cyano-4,6-dimethylpyridin-2-yl)amino)-N′-(4-methyl-benzylidene)benzohydrazide (5b): White solid (C23H21N5O), yield 56%, m.p. = 270–271 °C. IR (KBr); 3337, 3220 (N–H), 2219 (C≡N), 1640 cm−1(C=O).1H NMR (DMSO-d6) (Fig. 1S): 2.33 (s, 3H),2.37 (s, 3H), 2.39 (s, 3H), 6.83 (s, 1H, pyridine H-5), 7.26 (d, J = 8.5 Hz, 2H), 7.60 (d, J = 8.0 Hz, 2H), 7.73 (d, J = 8.5 Hz, 2H), 7.85 (d, J = 8.5 Hz, 2H), 8.39 (s, 1H, CH = N), 9.27 (s, 1H, NH), 11.67 ppm (s, 1H, NH).
4-((3-Cyano-4,6-dimethylpyridin-2-yl)amino)- N′-(4-methoxy-benzylidene)benzohydrazide (5c): White solid(C23H21N5O2), yield 55%, m.p. > 300 °C.IR (KBr): 3331, 3252 (N–H), 2219 (C≡N), 1638 cm−1(C=O).1H NMR (DMSO-d6) (Fig. 1S); 2.39 (s, 3H), 2.41 (s, 3H), 3.81 (s, 1H, OCH3), 6.85 (s, 1H, pyridine H-5), 7.02 (d, J = 8.4 Hz, 2H), 7.67 (d, J = 8.4 Hz, 2H), 7.75 (d, J = 8.4 Hz, 2H), 7.86 (d, J = 8.4 Hz, 2H), 8.39 (s, 1H, CH = N), 9.25 (s, 1H, NH), 11.60 ppm (s, 1H, NH).
4-((3-Cyano-4,6-dimethylpyridin-2-yl)amino)-N′-(4-nitrobenzylidene)-benzohydrazide (5d): Yellow solid (C22H18N6O3), yield 78%, m.p. = 295–298 °C. IR (KBr): 3292, 3206 (N–H), 2228 (C≡N), 1666 cm−1(C=O). 1H NMR (DMSO-d6) (Fig. 1S); 2.38 (s, 3H), 2.40 (s, 3H), 6.86 (s, 1H, pyridine H-5), 7.75 (d, J = 9.0 Hz, 2H), 7.87 (d, J = 8.0 Hz, 2H), 7.98 (d, J = 8.0 Hz, 2H), 8.29 (d, J = 9.0 Hz, 2H), 8.52 (s, 1H, CH = N), 9.30 (s, 1H, NH), 12.03 ppm (s, 1H, NH).
N′-(4-Chlorobenzylidene)-4-((3-cyano-4,6-dimethylpyridin-2-yl)-amino)benzohydrazide (5e): White solid (C22H18ClN5O), yield 71%, m.p. = 269–270 °C. IR (KBr); 3302, 3214 (N–H), 2209 (C≡N), 1660 cm−1(C=O). MS m/z (%) (Fig. 2S); 405 (M+ + 2, 10.26), 404 (M+ + 1, 43.47), 403 (M+, 100.00), 251 (16.44), 250 (32.45), 90 (17.43), 77 (10.27).
2.1.2 Synthesis of Ni(II)-Schiff Base Complexes
Ethanolic solution of 2 mmol from each N′-arylidene ligand 5a–e (0.739, 0.767, 0.799, 0.829 and 0.808 g, sequentially) was added to ethanolic NiCl2.6H2O (0.334, 2 mmol) solution. The reaction mixtures were heated under reflux for 2–3 h. 0.5 g of sodium acetate were added to each reaction-mixture to precipitate the aimed Ni(II) complexes. The faint colored complexes (yellow or orange) were isolated by filtration and washed by EtOH and ether, then kept to dray in desiccator under CaCl2. All isolated complexes are stable, non-hygroscopic and have high m.p. (˃ 300 °C).
2.2 CT-DNA Binding Methodology
The binding-grade of Ni(II)-Schiff base complexes toward CT-DNA (calf thymus), was screened by applying spectrophotometric titration technique. Amount of CT-DNA (50 mg) was dissolved in bi-distilled water at pH = 7 by a constant stirring and kept overnight at 4 °C. Mixing of 5.0 mM of tris(hydroxymethyl)-aminomethane with NaCl (50 mM) dissolved in bi-distilled water at pH = 7, yields Tris–HCl buffer. A stock solution of DNA was prepared in Tris–HCl buffer, the absorbance of this solution was measured at 260 and 280 nm, the ratio of A260/A280 recorded at 1.8–1.9 range. That points to pure DNA solution, which free from protein [18]. A concentration of DNA-stock solution (5.25 × 10−4 M), was determined by knowing molar absopititvity-coefficient value (ε = 6600 M−1 cm−1) at 260 nm. 2.5 × 10−5 M was the concentration used from each complex, which faced a gradual additive from CT-DNA amount that covering concentration range upon 0.00 to 1.58 × 10−4 mol l−1 . The blank solution includes all quantitative additives (buffer and DNA) except the tested compound (complex), which be used as a reference, to cancel the absorption of DNA. Using 1 cm quartz cuvette, the absorption of each concentration was measured over 200–900 nm range at 25 °C versus blank solution cell. The binding constant (Kb) for each tested complex towards DNA, was estimated by the following equation; [DNA]/(εa − εf) = [DNA]/(εb − εf) + 1/Kb (εa − εf) [19], through relation between slope/intercept, from the plots of [DNA]/(εa − εf) vs.[DNA]. Where; the molar concentration of DNA is [DNA], the extinction coefficient observed is εa = Aobs/[compound] at definite [DNA]. Also, εf is the extinction-coefficient for each free Ni(II) complex, εb is the extinction-coefficient of the complex at full-bonding towards DNA.
2.3 Computational Studies
2.3.1 Conformational Strategy
The optimized structural forms for all new compounds were demonstrated over Gaussian 09 program-screen [20] after choosing the appropriate method. According to polarizable model, a default ethanol solvent was used for configuration process upon DFT/B3LYP method. 3-21G was the diffusion function basis set suitable for Schiff base derivatives and their Ni(II) complexes [16, 17]. The yielded files (log and chk) facilitate estimating significant indexes, which lead to broad visualization for their discriminated characteristics. All optimized structures were visualized over Gauss-View program [21], for schematic numbering and other significances. Such significances were obtained based on frontier energy-gaps (∆E) by utilizing standard equations [22, 23].
2.3.2 Molecular Operating Environmental-Docking (MOE) Approach
MOE docking (vs. 2015) approach is the most recent software concerning with docking simulation technique that aims to evaluate the inhibition extent towards pathogen-proteins. Elected protein was 3s7s, as the PDB file of crystal structure for human placental aromatase complexed to breast cancer drug [24]. This simulation technique was implemented for all new synthesizes starting with the orientation till optimization for tested compound. The pre-optimization configuration was achieved by adding hydrogen atoms, atomic charges and potential energies. In addition to, important parameters were adjusted by MMFF94x force field during energy minimization [16, 17, 24]. The configured compounds were saved as MDB format that know ready for docking process. The orientation of 3s7s protein was the following interest which started by built hydrogen atoms over receptors after removing water molecules. Automatically connecting the receptor types followed by fixing the potential energies, were the second stage. Site-finder was done over line helix of protein amino acids [25]. The dummies were adjusted over alpha-sites in protein and then the docking process can be started. The docking simulation process consumed variable times covering all tested compounds but all processes were the average of 30 poses. Such poses were adjusted by London dG scoring function that improved twice by triangle Matcher methods. The binding rank was discriminated among the tested compounds through scoring energy values. Meanwhile, ligand type, receptors, interaction type, H-bond length and energy content were the other features recorded for docked complexes. The influential effect of H-bond length (≤ 3.5 Å) was extended to be the most significant judger on the magnitude of interaction validity at all. Moreover, the interaction patterns as well as the surface maps were also extracted to confirm comparative features.
2.4 Anticancer Screening
In National Research Centre of Egypt, this screening was executed at Bioassay-Cell Culture Laboratory. Using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay method, the cytotoxic efficiency of new Ni(II)-Schiff base complexes, were evaluated [26]. From ATCC (American Type Culture Collection, USA) Tissue culture Unite, MCF-7 cell line was purchased. In sterile area of laminar flow-cabinet safety-class (Baker, SG403INT, Sanford, ME, USA), this investigation procedure was done. At 37 °C under 5% CO2, MCF-7 cell, was suspended in medium of DMEM from 1% mixture of antibiotic–antimycotic (10,000 U ml−1 Potassium Penicillin, 10,000 µg ml−1 Streptomycin Sulfate and 25 µg ml−1 Amphotericin B) and 1% l-glutamine. Using water jacketed carbon dioxide incubator (Sheldon, TC2323, Cornelius, OR, USA)at37 ºC, the cell batch was cultured for 10 days, then seeded by10 × 103 cells/well in fresh medium for complete growth (96-well microtiter plastic plates) under 5% CO2. After aspirating media cultivated, fresh medium was added (without serum) and cells were incubated alone (negative control) or by variable concentrations from tested samples (in DMSO) till reaching, 100, 50, 25, 12.5, 6.25, 3.125, 0.78 and 1.56 μg ml−1 (final concentrations). Then and after incubation for 48 h the medium was aspirated and 40 μl MTT salt (2.5 μg ml−1) were added. Such addition was carried out on each well which still incubated along further 4 h under 5% CO2 at 37 °C. 200 μl of 10% Sodium dodecyl sulphate (SDS) prepared in deionized water, were added to each well to stop the reaction and dissolving the crystals formed, then incubated overnight at 37 °C. Cytotoxic natural agent (positive control) composes of 100 µg ml−1, gives 100% lethality under the same conditions [26,27,28]. The absorbance was measured at 595 nm (reference λ = 620 nm) using microplate multi-well reader (Bio-Rad Laboratories Inc., model 3350, Hercules, California, USA). Implementing independent t-test by SPSS 11 program, the statistics were tested for samples versus negative control (cells with complexes). The viability percentage was calculated using the following relation; (Reading of extract/Reading of negative control) − 1) × 100.
2.5 Analysis Techniques
The analytical techniques implemented in this study to analyze all new synthesizes (ligands and Ni(II) complexes) were schematically depicted (Scheme 1S). Evaluation of nickel and chloride content was carried out by complexometric and gravimetric analysis, respectively using standard procedures [29].
3 Results and Discussion
Analytical results proposed equi-molar ratio, for all Ni(II) complexes except of Ni(II)-5d complex, which has 1:2 ratio (M:L). Analytical and physical characteristic were summarized in Table 1. The molar conductivity values (Λm, 10−3 M) were measured for four Ni(II) complexes that displayed excellent solubility in solvent (DMSO). The non-conducting feature was efficiently prevailed with all measured complexes except Ni(II)-5e [30]. The conducting appearance of Ni(II)-5e complex was suggested based on Λm value (48.93 Ω−1cm2 mol−1). This value matches with presence of ionizable chloride ion, which expelled outer coordination sphere after the presence of water molecule. The inductive effect of p-substituents play significant role on coordination behavior which led to different molecular formula (will be discussed in detail later).
3.1 Synthesis mechanism of Schiff bases
This study started by synthesis of 4,6-dimethyl-3-cyanopyridin-5-one (1) from acetylacetone and cyanoacetamide in ethyl alcohol and potassium carbonate [31], (Scheme 1). Heating pyridine compound (1) with phosphorus oxychloride, furnished by 2-chloro-4,6-dimethylnicotinonitrile (2) [31].
The chlorine atom of 2-chloronicotinonitrile derivative 2 proved to be reactive towards nucleophilic-substitution by nitrogen-nucleophile such as; ethyl 4-aminobenzoate to afford ethyl 4-(3-cyano-pyridin-2-ylamino)benzoate scaffold (3) (Scheme 2). The compatible elemental and spectral analysis supported the proposed structure of benzoate compound (3). Its IR spectrum displayed absorptions at 3309, 2223 and 1712 cm−1 to indicate the presence of N–H, nitrile (C≡N) and carbonyl of ester (COOEt) groups. The 1H NMR signals were identified as triplet at 1.31 ppm and quartet at 4.28 ppm for the protons of ethoxy group (–O–CH2–CH3). The singlet-signals at 2.39 and 2.41 ppm assign for methyl groups-protons. The proton of pyridine C-5 resonated as singlet at 6.87 ppm, while the aromatic protons appeared as two doublet signals at 6.74 and 7.87 ppm. The proton of N–H function resonated as singlet at 9.34 ppm. The hydrazide derivative (4) was achieved by refluxing ethyl 4-(3-cyano-pyridin-2-ylamino) benzoate compound (3) with hydrazine hydrate in EtOH for 2 h. The structure of compound (4) was confirmed by spectroscopic techniques including IR and mass spectral analysis. Its IR spectrum exhibited absorption bands at 3340 and 3287 cm−1 for N–H stretching of NH and NH2 groups. The absorptions at 2217 and 1611 cm−1are referring to nitrile (C≡N) and carbonyl (N–C=O) groups, respectively. 1H NMR spectrum indicated the absence of any signal related to ethoxy group and displayed singlet signals of hydrazide moiety (CONHNH2) at 4.44 ppm (NH2) and 9.14 ppm (NH). Condensation of hydrazide (4) with different aromatic benzaldehydes (benzaldehyde, 4-methylbenzal-dehyde, 4-methoxybenzaldehyde, 4-nitrobenzaldehyde and 4-chlorobenzaldehyde) proceeded by boiling in EtOH to afford Schiff’s bases (5). The structure of 5 was confirmed because of their spectral data and satisfactory elemental analysis. Their IR spectra showed absorption band at wave number ranged from 1638 to 1666 cm−1corresponding to carbonyl group. The proton of azomethine group (N–N = CH–) resonated as singlet signal in 1H NMR spectra in the range of 8.39–8.52 ppm.
3.2 Comparative Vibrational Study
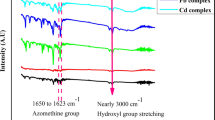
The synthesized Schiff base ligands appeared favorable for neutral bi-dentate binding-mode in most Ni(II) complexes. This appearance was supported by the state of functional-group bands (Table 2), which attributing to, υ(NH), υ(C=O), υ(C=N)pr, υ(C=N21–N) and υ(CN) [32]. More or less un-shifted vibrational-bands that assign for nitrile and pyridine (C=N) groups, asserts on their ruling-out from coordination (Fig. 3S). Mostly, lower shifted appearance for υ(C=O) and υ(C=N21) was recorded, except with Ni(II)-5d spectrum that favor the excluding of N21 from contribution. Neutral mono-dentate mode of bonding was proposed for (5d)derivative, which sounds preferable in presence of highly inductive p-substituted group (NO2). Such group caused electron deficient suffered by close functional-groups especially that contributing in resonance. The exclusion of C=N21 group, was expected and supported by un-changeable band at 1604 cm−1[33]. Ni(II)-5d complex was appeared by unique molar-ratio (1 M:2L) although high steric hindrance, which was not obstacle for this ratio. This proposal was confirmed firstly by analytical data (elemental and mass spectrum) and asserted by IR spectrum, which exhibited the bands by noticeable raise in intensity. The high covalent feature of conjugated anion (chloride) devotes the covalent attachment of the two atoms in all complexes, except for Ni(II)-5e. This is due to the competition of chloride atom with one water molecule that always overcome and contributing. This feature was supported electrically, through the conductivity measurements for 10−3 M, which displayed value agrees with conducting feature for mono-anion. Water molecules were proposed attached physically or chemically with coordination sphere, through the appearance of new bands assign for δr(H2O)and δw(H2O) [34]. Such was supported by TGA analysis, which will be discussed later. υ(Ni–N) and υ(Ni–O) vibrational-bands were recorded at lower wavenumber region, to confirm new bond types between ligand and nickel atom [35].
3.3 UV–Vis Spectral-Data
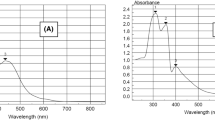
Electronic spectral analysis as well as magnetic moment measurement (Table 3) were considered the most significant to recognize and discriminate structural formulae of transition metal complexes. This study was achieved in DMSO solvent over broad UV–Vis spectral-range (200–1200 nm), which covering allowed and forbidden transitions [36]. The diamagnetic feature (μeff = 0) of all investigated complexes beside their faint colors (yellow or orange), likely emphasizes square-planer configuration. Electronic transition spectra of all Ni(II)-Schiff base complexes (Fig. 4S), exhibited bands according to intra-ligand, charge transfer and ligand field transitions. The π → π* and n → π*were the intra-ligand transitions, which appeared at 33,259, − 30,303 and 31,250–26,315 cm−1 ranges, respectively [37]. While, the bands appeared at 25,000–23,256 cm−1 range, signify Ni–O and Ni–N charge transfer [38]. Regarding ligand-field transitions of d8-systems in square-planer configuration, which represented dz2 as a ground orbital, were clearly remarked and assigned. Bands recorded at 19,230–20,000 and 12,500–16,380 cm−1 ranges are assigning to 1A1g → 1A2g and 1A1g → 1B1g transitions, respectively in square-planer geometry (Fig. 1). While, the band recorded at 11,000 cm−1 in Ni(II)-5b complex-spectrum, refers to spin-forbidden transition [39].
3.4 Mass Spectral-Analysis
The further molecular conformation was conducted by mass spectral analysis under 40 °C min−1 heating rate at 70 eV over 50–1000 m/z range. Such-identification tool was carried out by using electron-spray for collision-induced ionization followed by fragmentation. For instance, Ni(II)-5d and Ni(II)-5e complexes, were the investigated examples (Figs. 2 and 3S), which accordingly confirm the rest. Whereas, the molecular ion peaks were appeared at m/z = 958.94 (I = 14%) and 534.34 (I = 12%) and match with M+ and (M+ + 1)-H2O ions, respectively. The reduced intensity of molecular ion peaks is a normal feature for instability of molecular ion that leads to fragmentation [40]. The base peak recorded with the Ni(II)-5d and Ni(II)-5e complexes spectra, were m/z = 575.87 and 334.83, respectively that coincided with bulky molecular ions inside the coordination sphere. The most powerful peaks that corresponding to metal ion isotopes, which signify and confirm the complexation process. Such peaks undoubted appeared close to m/z ≈ 60 that verifying the presence of nickel atom.
3.5 TGA Analysis
This analytical tool interested in details for quality of binding inside the coordination sphere, particularly with solvent molecules. This analysis was conducted for all complexes at 10 °C min−1, as a heating rate under nitrogenous atmosphere covering 20–800 °C range. All endothermic degradation stages, were appeared extended to cover four to five stages (Fig. 6S). The thermal stability of investigated complexes appeared varied but instability prevailed, except for Ni(II)-5d and Ni(II)-5e complexes. The water molecules proposed attached with most complexes, were strictly verified over the first degradation stage by matched mass-losses (found and calculated). Regarding thermal-degradation behavior over all investigated complexes was relatively-fixed in degradation behavior. So, the decomposition pathway was easily established over all stages and displayed (Table 4). The residual part recorded at ≈ 600 °C was NiO molecule, which sometimes polluted with carbon atoms.
3.6 XRD Analysis
Such analysis which dealing with the topography and crystal-lattice dynamic [41] evaluates the magnitude of crystallinity over three complexes (as example). Extracted patterns that displayed in Figs. 3 and 5S over 10° < 2θ < 80° range, appeared having extent of nano-crystallinity. So, crystallite parameters were estimated by using known equations through FWHM method [42] (Table 5). FWHM, particulate size, 2θ, d spacing, relative intensity, crystal strain (ε) and dislocation density (δ), were the parameters calculated. The estimated sizes were appeared in 5.79–11.53 nm range, which signify their distinguish nanometer characteristics. The reduced crystal strain and dislocation density, enhanced the extent of regularity in crystal building and atomic packing. Miller indices (hkl) were obtained from VESTA software [43] based on known equations [44]. Such indices were 100, 103 and 130. This axial indices close to tetragonal feature with threefold symmetry that remains unchanged by rotation.
3.7 CT-DNA Binding Study
Applying spectrophotometric titration method, the binding quality of tested Ni(II) complexes against CT-DNA was tested (Fig. 8S). This investigation reflects a shadow on the degree of blocking for DNA-active sites by influence of tested complexes, which lead to its control inside infected cell. Intersect-binding constant (Kb) was extracted for four complexes based on an observable hyper-chromic shift in charge transfer band (≈ 350 nm) by 2–3 nm. This shift was observed clearly with increasing DNA amount over constant complex-concentration. The binding feature may be of electrostatic attraction or occlusion inside grooves appeared from helix-deterioration. The magnitude of hyper-chromic shift, as a result for extended complex structure, which primates electrostatic attraction with DNA-active sites. Also, the occlusion of binding-complex inside minor or major grooves that appeared through re-organizing DNA helix or fractional damage for double helix-surface. The binding constants (Kb, M−1) for treated Ni(II) complexes were estimated utilizing known equations [45]. The values calculated were as follow; 2.6 × 105, 6.48 × 105, 6.02 × 105 and 1.6 × 106 For the complexes Ni(II)-5a, Ni(II)-5b, Ni(II)-5c and Ni(II)-5e complexes, respectively. This values matched perfectly with the magnitude of inductive effect of p-substituents, which attached such effect with binding efficiency. Hammett's equation [46], clarifies the direct relation between σR and binding efficiency (Kb) (Fig. 4). The high inductive effect of substituted groups that conducts to lack of electron-density over fictional-groups and led to intensive binding towards DNA-helix.
3.8 Computational Studies
3.8.1 Conformational Study
This computational study has a great impact on verifying the mode of bonding within the complexes. This based on the distribution of functional groups upon the optimum structures of the ligands. After geometry optimization process, the position of donor sites may facilitate or hinder their contribution in coordination. Such may considered the further asserting tool for the bonding inside the complexes, which really achieved (see Sect. 3.8.1.2). Accordingly, Gaussian09 software under DFT/B3LYP method was applied upon the ligands and their Ni(II) complexes. All synthesizes were faced configuration under economical basis set (3-21G) until executing optimization process that yielded the structural-forms displayed (Figs. 1 and 9S). In addition to, important files (log and chk) were excreted that feed us with essential characteristics for treated compounds (Tables 6, 7). Regarding chk files and implementing HOMO–LUMO energy band splitting (ΔE), functional physical-indexes were calculated (Table 6) and frontier-orbital images were also extracted (Figs. 5 and 10–11S). According to HOMO and LUMO images and with respect to Schiff base derivatives (Fig. 10S), intensive appearance for such molecular orbitals around hydrazide-moiety that responsible for coordination, was recorded. This signifies the extent of facility in electronic transitions among the coordinating groups. Moreover and according to HOMO and LUMO images of Ni(II) complexes (Figs. 5 and 11S), appeared fairly localized in metal atom and surrounding. This points to the influence of metal atom on electronic distribution over the whole compound, which lead to easiest electronic transition at all.
3.8.1.1 Functional Physical Parameters
Electronegativity (x), chemical potential (μ), global-hardness (η), global-softness (s), global-electrophilicity (ω) and absolute-softness (ϭ), were the physical parameters calculated for all synthesizes to build their functional vision. Based on HOMO–LUMO energy band splitting (ELUMO-EHOMO) such parameters were estimated using known relations [22, 23]. From comparative point of view, regarding such parameters and with respect to Schiff base derivatives (5a–e), we notice the following; (i) the p-substituent impact was remarked clearly through the significant difference among the whole parameters. (ii) The favored characteristics (x, s, ω and ϭ) that orient to excellent properties, displayed the priority of (5d) derivative, which already has the powerful p-substituted group (nitro). While and with respect to Ni(II) complexes, we noticed the following; (i) general enhancement was recorded for all characteristics based on reduced ΔE values, in comparing with free ligands. (ii) Preferred electronegativity and softness properties were progressed with Ni(II)-5d upon the rest complexes. This demonstrates the positive influence of inductive-effect for p-substituent on functional-characteristics. Such priority points to promising biological feature, which coincides directly with the softness and electrostatic properties [47, 48]. (iii) Electrophilicity index (ω) was appeared varied significantly among Ni(II)-complexes, which also confirms the impact of substituents on that. Ni(II)-5d exhibited high electrophilicity, which already expected based on shortage in electron-density, which accompanied by high electron-acquiring ability from surrounding [49]. Dipole moment, excitation-energy, oscillator-strength and formation energy, were the other physical parameters extracted from log files after optimization process (Table 7). According to dipole moment values, were agreed successfully with p-substituent-behavior, which already varied covering donating (Me and OMe) and withdrawing (Cl and NO2) groups. The reduced dipole moment value was recorded with Ni(II)-5d complex in compatible with electrophilicity feature. The values of excitation energy and oscillator strength, exhibited enhanced superiority of complexes in stability and bond-strength towards excitation process. Finally and according to formation energy values, appeared very compatible and weighted for advanced stability of complexes.
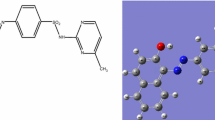
3.8.1.2 Molecular Electrostatic Potential (MEP)
Electron-cloud maps (Figs. 6, 10S and 12S) were demonstrated over the atoms after building new cube over new surface-maps, to illustrate the magnitude of electron density over the molecule. The maps discriminate nucleophilic and electrophilic sites, by blue and red colors, respectively, while the middle-feature appeared green. The electrostatic maps of Schiff base derivatives (5a–e) (Fig. 10S), appeared by extensive range (≈ ± 9.8 × 10−2) with favorable focus on amide and hydrazide centers. Such promotes their nucleophilic contribution towards Ni(II) ion [25]. While, the electrostatic maps of Ni(II) complexes, display extended range (≈ ± 0.16 × 102), exceeded free derivatives especially with Ni(II)-5b and Ni(II)-5c complexes. This exceeding may refer to the contribution of covalent-chloride as electron-dense atoms beside the role of p-substituents, which cannot ignored.
3.8.2 Quantitative Structural Characteristic Relationships Study
This computational study (QSAR) is a key player in judging on reactivity-magnitude of treated compound, towards the intended application. Such study was executed after energy minimization upon HyperChem (8.1) program, for all Ni(II) complexes. Under semi-empirical (AM1) setup that followed by Molecular Mechanics force field (MM+) [14,15,16,17], the treatment was done without fixation for any parameter till equilibrium. The use of Polake-Ribiere conjugated gradient algorithm method [50], conducts to energy minimization characteristic. Surface area, hydration energy, reactivity, polarizability and log p were the QSAR parameters obtained (Table 8). Generally, promising feature can be expected easily for such complexes based on distinguish QSAR values, particularly the reactivity and Log p values. As known, a strong revers relation between partition-coefficient (log P) with that of biological activity [16, 17, 51]. So finally, Ni(II)-5d, Ni(II)-5e and Ni(II)-5c may offer vigorous results in biological application.
3.8.3 MOE-Docking Approach Study
This study is the exclusive technical approach in drug designing industry, which offer virtual view simulate the interaction between tested compound (proposed drug) and cell pathogen protein [15]. Molecular Operating Environmental module (vs. 2015) was used to accomplish such simulation for all investigated Ni(II) complexes against crystal structure of breast-cancer protein(3s7s). The ligand interaction parameters (Table 9), interaction validity patterns and surface-mapping (Figs. 7 and 13S) were extracted for each complex. Ligand types, receptors, interaction feature, H-bonding length, energy content in addition to scoring energy, were the interaction parameters used to evaluate the extent of inhibition. Comparative insight upon Schiff base-3s7s complexes (5a–e), offers the following remarks; (i) according to ligand binding sites, were focused on N9, 11, 20 and O19 atoms. (ii) According to receptor types, Asparagine, Glycine, Lysine, Glutamate, Cysteine, Valine and Methionine were the contributing amino acids in interaction patterns. (iii) According to interaction types, H-donor and H-acceptor were the only exhibited by true binding approach with length ≤ 3.5 Å [16, 17]. (iv) Finally and according to scoring energy values (kcal mol−1), derivatives 5d (− 7.1906) and 5e (− 8.3743) displayed superior inhibition score. With respect to Ni(II)-Schiff bases-3s7s complexes, the following remarks were aggregated; (i) the interacting sites were, O32, 52; N9,19, 21 and 6-ring. (ii) Regarding receptor types contributing inside interaction patterns were, Glycine, Lysine, Glutamate, Glutamine, Aspartate and Methionine. (iii) Regarding the interaction types actually considered, were H-donor, H-acceptor and pi-H that having real lengths. (iv) Finally and regarding scoring energy values, Ni(II)-5d, Ni(II)-5c and Ni(II)-5e complex exhibited superior inhibition score against breast cancer protein (3s7s) [25]. With respect to interaction validity patterns as well as surface-mapping (Figs. 7 and 13S), we noticed that, the receptor exposure has prevalence over ligand-exposure. Also, the interaction types were covered all polar, acidic, basic and greasy beside a broad proximity-contour surrounding the tested compound.
3.9 In-Vitro Anticancer Screening
As we know that, the scientific research faces a great challenge towards overcoming cancer daises particularly, breast cancer which annually leads to death of many women. Consequently, we concerned with in-vitro screening for new synthesized Ni(II) complexes, against breast cancer cell line (MCF-7). The inhibition efficiency was tested for series of concentrations from each tested complex as; 12.5, 25, 50 and 100 μM, to estimate the IC50. Doxorubicin, which is known by its distinct anti-tumor efficiency, was the reference drug used for comparison. This comparison facilitates ranking the inhibition-attitude among the whole tested compounds and then throw a light on that promising. Demonstrating a relationship between the concentration (µg ml−1) and cell-viability (%), leads to estimatingIC50 (µg ml−1) values as mentioned in Table 10. The values displayed superiority of Ni(II)-5e and Ni(II)-5c complexes, which appeared having very strong inhibition towards breast-cancer cell line. Also, Ni(II)-5d has a significant inhibition but the rest displayed moderate activity.
4 Conclusions
Novel Ni(II)-Schiff base complexes were prepared and characterized. The mode of bonding was strongly affected by p-substituents features. The structural formula appeared steady with all Ni(II) complexes by square-planer form. The conformational study was implemented to establish the best atomic distribution inside the structures. The docking simulation was applied for all complexes against breast-cancer protein. After simulation process, expected distinguish characteristic was recorded with most treated complexes. In-vitro screening was tested for all Ni(II) complexes, against MCF-7 cell line. Promising inhibition activity was recorded with Ni(II)-5c and Ni(II)-5e complexes.
References
F. Xu, W. Li, W. Shuai, L. Yang, Y. Bi, C. Ma, H. Yao, S. Xu, Z. Zhu, J. Xu, Eur J. Med. Chem. 173, 1–14 (2019)
C.S. McCrae, A. Ross, A. Stripling, N.D. Dautovich, Clin. Interv. Aging 2, 313–326 (2007)
T.S. Harrison, L.J. Scott, Drugs 65, 2309–2336 (2005)
X. Wang, G. Mick, K. McCormick, Physiol. Rep. 7, e14151 (2019)
A.M. Al-Majid, M.S. Islam, S. Atef, F.F. El-Senduny, F.A. Badria, Y.A.M.M. Elshaier, M. Ali, A. Barakat, A.F.M. Motiur Rahman, Molecules 24, 1332 (2019)
Y.-L. Zhang, F.-Q. Zhao, J. Inorg. Organomet. Polym. 28, 2714–2720 (2018)
H.C. Wang, X.Q. Yan, T.L. Yan, H.X. Li, Z.C. Wang, Molecules 21, 1012 (2016)
F.A. Saad, M.G. Elghalban, N. El-Metwaly, H. El-Ghamry, A.M. Khedr, Appl. Organometal. Chem 31, e3721 (2017)
D.C. Culita, L. Dyakova, G. Marinescu, T. Zhivkova, R. Spasov, L. Patron, R. Alexandrova, O. Oprea, J. Inorg. Organomet. Polym. 29, 580–591 (2019)
H.A. El-Boraey, M.A. El-Salamony, J. Inorg. Organomet. Polym. 29, 684–700 (2019)
X.-L. Wan, L. Dong, J.-Z. Wang, H.-T. Shi, L.-B. Yang, J. Inorg. Organomet. Polym. 29, 1184–1191 (2019)
R.W.-Y. Sun, D.-L. Ma, E.L.-M. Wong, C.-M. Che, Dalton Trans. 43, 4884–4892 (2007)
K.L. Haas, K.J. Franz, Chem. Rev. 109, 4921–4960 (2009)
H.A. Katouah, J.H. Al-Fahemi, M.G. Elghalban, F.A. Saad, I.A. Althagafi, N.M. El-Metwaly, A.M. Khedr, Mater. Sci. Eng. C 96, 740–756 (2019)
F.A. Saad, N.M. El-Metwaly, A.M. Khedr, J. Inorg. Organomet. Polym. 29, 1606–1624 (2019)
N. El-Metwaly, I. Althagafi, H.A. Katouah, J.H. Al-Fahemi, T.M. Bawazeer, A.M. Khedr, Appl. Organomet. Chem. 33, e5095 (2019)
I. Althagafi, M.G. Elghalban, N.M. El-Metwaly, J. Inorg. Organomet. Polym. (2019). https://doi.org/10.1007/s10904-018-01062-3
J. Marmur, J. Mol. Biol. 3, 208–2018 (1961)
A. Wolfe, G.H. Shimer, T. Meehan, Biochemistry 26, 6392–6396 (1987)
M.J. Frisch et al., Gaussian 09, Revision D (Gaussian Inc., Wallingford, CT, 2010)
R. Dennington II, T. Keith, J. Millam, GaussView, Version 4.1.2 (Semichem Inc., Shawnee Mission, KS, 2007)
R.C. Chikate, S.B. Padhye, Polyhedron 24, 1689 (2005)
R.F. George, Eur. J. Med. Chem. 47, 377 (2012)
M.H. Abdellattif, M.A. Hussien, E. Alzahrani, Int. J. Pharm. Sci. Res. 9, 1000–1019 (2018)
I. Althagafi, N. El-Metwaly, T.A. Farghaly, Molecules 24, 1741 (2019)
T. Mosmann, J. Immunol. Methods 65, 55–63 (1983)
F. Denizot, R. Lang, J. Immunol. Methods 89, 271–277 (1986)
R.R. Weichselbaum, H.J. Mauceri, N.N. Hanna, M.A. Beckett, D.H. Gorski, M.-J. Staba, K.A. Stellato, K. Bigelow, R. Heimann, S. Gately, M. Dhanabal, G.A. Soff, V.P. Sukhatme, D.W. Kufe, Nature 394, 287–291 (1998)
A.I. Vogel, Text Book of Quantitative Inorganic Analysis (Longman, London, 1986)
W.J. Geary, J. Coord. Chem. Rev. 7, 81–122 (1971)
A. Yassin, Chem. Heterocycl. Compd. 45, 35–41 (2009)
M.S. Morales-Ríos, P.Y. López-Camacho, O.R. Suárez-Castillo, P. Joseph-Nathan, Tetrahedron Lett. 48, 2245–2249 (2007)
J. Song, H. Yamataka, Z. Rappoport, J. Fluor. Chem. 124, 119–122 (2003)
K.S. Abu-Melha, N.M. El-Metwally, Spectrochim. Acta A 70, 277–283 (2008)
N.M. El-Metwaly, R.M. El-shazly, I.M. Gabr, A.A. El-Asmy, Spectrochim. Acta A 61, 1113–1119 (2005)
A.B.P. Lever, Inorganic Electronic Spectroscopy (Elsevier, Amsterdam, 1986)
N.M. El-Metwally, G.A.A. Al-Hazmi, Spectrochim. Acta A 107, 289–295 (2013)
N.M. El-Metwally, K.S. Abu-Melha, Transit. Met. Chem. 32, 828–834 (2007)
A.A. Abou-Hussen, N.M. El-Metwaly, E.M. Saad, A.A. El-Asmy, J. Coord. Chem. 58, 1735–1749 (2005)
M.S. Refat, N.M. El-Metwaly, Spectrochim. Acta A 81, 215–256 (2011)
B.D. Cullity, Elements of X-Ray Diffraction, 2nd edn. (Addison-Wesley, Reading, 1993)
S. Velumani, X. Mathew, P.J. Sebastian, S.K. Narayandass, D. Mangalaraj, Sol. Cells 76, 347–358 (2003)
K. Momma, F. Izumi, J. Appl. Crystallogr. 44, 1272–1276 (2011)
B.D. Cullity, S.R. Stock, Elements of X-Ray Diffraction, 3rd edn. (Prentice Hall, New Jersey, 2001)
A.Z. El-Sonbati, A.F. Shoair, A.A. El-Bindary, M.A. Diab, A.S. Mohamed, J. Mol. Liq. 209, 635–647 (2015)
L.H. Abdel-Rahman, A.M. Abu-Dief, N.M. Ismail, M. Ismael, J. Inorg. Nano-Met. Chem. 47, 467–480 (2017)
G.A. Al-Hazmi, K.S. Abou-Melha, N.M. El-Metwaly, K.A. Saleh, Bioinorg. Chem. Appl. 2018, 7176040 (2018)
H. Allal, Y. Belhocine, E. Zouaoui, J. Mol. Liq. 265, 668–678 (2018)
I. Fleming, Frontier Orbital's and Organic Chemical Reactions (Wiley, London, 1976)
M.M. Al-Iede, J. Karpelowsky, D.A. Fitzgerald, Pediatr. Pulmonol. 51, 394–401 (2016)
N. El-Metwaly, J.H. Al-Fahemi, I. Althagafi, A.M. Khedr, H.A. Katouah, J. Inorg. Organomet. Polym. (2019). https://doi.org/10.1007/s10904-019-01233-w
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abumelha, H.M., Al-Fahemi, J.H., Althagafi, I. et al. Deliberate-Characterization for Ni(II)-Schiff Base Complexes: Promising In-Vitro Anticancer Feature that Matched MOE Docking-Approach. J Inorg Organomet Polym 30, 3277–3293 (2020). https://doi.org/10.1007/s10904-020-01503-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10904-020-01503-y