Abstract
Intellectual disability is a heterogeneous disorder with a wide phenotypic spectrum. Over 1,700 OMIM genes have been associated with this condition, many of which reside on the X-chromosome. The IQSEC2 gene is located on chromosome Xp11.22 and is known to play a significant role in the maintenance and homeostasis of the brain. Mutations in IQSEC2 have been historically associated with nonsyndromic X-linked intellectual disability. Case reports of affected probands show phenotypic overlap with conditions associated with pathogenic MECP2, FOXG1, CDKL5, and MEF2C gene mutations. Affected individuals, however, have also been identified as presenting with additional clinical features including seizures, autistic-behavior, psychiatric problems, and delayed language skills. To our knowledge, only 5 deleterious mutations and 2 intragenic duplications have been previously reported in IQSEC2. Here we report two novel IQSEC2 de novo truncating mutations identified through diagnostic exome sequencing in two severely affected unrelated male probands manifesting developmental delay, seizures, hypotonia, plagiocephaly, and abnormal MRI findings. Overall, diagnostic exome sequencing established a molecular diagnosis for two patients in whom traditional testing methods were uninformative while expanding on the mutational and phenotypic spectrum. In addition, our data suggests that IQSEC2 may be more common than previously appreciated, accounting for approximately 9 % (2/22) of positive findings among patients with seizures referred for diagnostic exome sequencing. Further, these data supports recently published data suggesting that IQSEC2 plays a more significant role in the development of X-linked intellectual disability with seizures than previously anticipated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intellectual disability is defined as intelligence quotient of <70 together with performance lower than age-and-culture-related standards in at least two areas of adaptive function, starting before 18 years of age (American Psychiatric Association 2000). Intellectual disability can be classified in one of four categories including mild, moderate, severe, and profound (American Psychiatric Association 2000). According to researchers, intellectual disability is a very heterogeneous disorder with underlying genetic (5–10 %), chromosomal (22 %), and syndromic (21 %) etiologies (Strømme 2000). In OMIM, over 1,700 gene entries are associated with intellectual disability, many of which reside on the X-chromosome (Chelly et al. 2006). Even though more than 120 X-linked intellectual disability genes are known to be associated with the condition, a specific underlying cause is not identified in over 50 % of families with suspected nonsyndromic X-linked intellectual disability (Shoubridge et al. 2010). X-linked intellectual disability is currently estimated to be responsible for 5–10 % of all cognitive impairment presenting in males (Tran Mau-Them et al. 2013). In addition to major genetic heterogeneity, the variable phenotypic expressivity of X-linked intellectual disability also creates significant challenges for both clinical diagnosis and genetic counseling. Syndromic and non-syndromic forms of X-linked intellectual disability have been extensively reported in the literature, and some genotype–phenotype correlations have been established. One gene in particular, IQSEC2, originally thought to be linked to non-syndromic X-linked intellectual disability, is recently presenting in association with both intellectual disability and additional phenotypic features suggestive of a possible syndrome (Tran Mau-Them et al. 2013 and Shoubridge et al. 2010). The IQSEC2 (IQ motif and Sec7 domain 2) gene is located on chromosome Xp11.22 and encodes guanine nucleotide exchange factors for the ADP-ribosylation factor family of GTP-binding proteins (Nagase et al. 1998 and Shoubridge et al. 2010). This gene is known to play a significant role in the maintenance of homeostasis within the neural environment of the human brain (Shoubridge et al. 2010). A loss of guanine nucleotide exchange factor activity, caused by pathogenic mutations, is likely to be the underlying mechanism in disease (Shoubridge et al. 2010). This is ultimately caused by reduced activation of the ARF6 substrate, which influences the regulation of actin cytoskeleton organization (Shoubridge et al. 2010).
Previously Reported Mutations in IQSEC2
Mutations in IQSEC2 have been linked to mechanisms causing neurocognitive and neuropsychiatric phenotypes. Case reports of affected probands show phenotypic overlap with conditions associated with deleterious MECP2, FOXG1, CDKL5, and MEF2C gene alterations (Tran Mau-Them et al. 2013). Historically, only five deleterious IQSEC2 mutations were reported in association with X-linked intellectual disability. In 1988, a family (called MXR1) was identified as having a hereditary, non-syndromic form of X-linked mental retardation (Suthers et al. 1988). This family, later analyzed in a multi-family research project, was found to carry a c.2587C>T change in exon 8 of IQSEC2, leading to a p.R863W substitution (Shoubridge et al. 2010). An unrelated second family in the same study (called MRX18) was identified as having a c.2402C>A (p.Q801P) mutation in exon 6 of IQSEC2 (Shoubridge et al. 2010). A third unrelated family (called US166) with the same phenotype was identified as having a c.2273G>A change in exon 5 leading to a p.R758Q substitution (Shoubridge et al. 2010). Finally, a fourth unrelated Australian family (called AU128) was identified as having a c.1075C>T change in exon 4 leading to a p.R359C substitution in the IQ-like domain of IQSEC2 (Shoubridge et al. 2010). The fifth mutation, a de novo nonsense mutation (ARG855*1), was identified in a male proband with no other affected family members (Rauch et al. 2012).
Based on additional studies of 2 affected female probands, one with an X-chromosome translocation, it was also discovered that the IQSEC2 gene uniquely escapes X-inactivation and can also lead to intellectual disability in carrier females (Li and Carrel 2008; Nagase et al. 1998, and Shoubridge et al. 2010). It has been well-established for some time that various genes on the X-chromosome can “escape” the inactivation phenomenon (Li and Carrel 2008). The underlying mechanism for this genetic mechanism is still not completely understood; however, from a clinical standpoint, female patients presenting with relative X-linked phenotypic features has been unmistakably observed (Li and Carrel 2008). The female proband reported by Morleo et al. (2008) was found to carry a balanced de novo t(X;20)(p11.2;q11.2) karyotype rearrangement involving a disruption in IQSEC2. She presented clinically with severe infantile seizures, global developmental arrest, hypsarrhythmia, and severe mental retardation. This female proband also had left talipes equinovarus, congenital hip dislocation, esotropia requiring surgical correction, no crawling or walking, myocolonic seizures involving both arms (refractory to therapy), severe hypotonia, stopped speaking, and cannot eat solid foods. MRI studies showed non-specific volume loss and periventricular white matter changes in the brain (Morleo et al. 2008). X-inactivation studies revealed an extremely skewed X-inactivation pattern with dominant use of the translocated X-chromosome (Morleo et al. 2008).
In 2013, two de novo intragenic duplications on the X-chromosome involving IQSEC2, and one nonsense mutation were described in three additional male patients presenting with a severe intellectual disability plus additional clinical features including neonatal hypotonia, delayed motor skills, seizures, strabismus, autistic-like behavior, stereotypic midline hand movements, microcephaly, little-to-no walking, little-to-no language skills, significant behavioral issues, and mildly abnormal facial features. Only one of the three probands presented with abnormal MRI findings (Tran Mau-Them et al. 2013).
Two Novel Mutations in IQSEC2 Identified on Exome Sequencing
In this report, we present two unrelated probands carrying novel de novo mutations in the IQSEC2 gene identified through diagnostic exome sequencing. These two patients clearly show phenotypic overlap and expansion to cases previously reported in the literature. Both probands have significant developmental delay, seizures, hypotonia, vision impairments, plagiocephaly, autistic-like features, absent language skills, and abnormal MRI findings. The first patient additionally has mild facial hypoplasia, bilateral coxa valga, inability to track objects, and showed no interest in pleasure activities. The second patient also has mild beaking of the nasal tip, a round face, mild prognathia with slight maxillary hypoplasia, widely spaced nipples, hyperextensibility, absent deep tendon reflexes, and “doughy” skin. Family histories were unremarkable for intellectual disability and seizure disorders. Despite the limited number of cases with known IQSEC2-associated disease reported in the literature, our data clearly suggest that this gene plays a larger role in the cause of X-linked cognitive impairment than previously thought. IQSEC2 should also be considered a candidate gene for sequencing in the patients presenting with intellectual disability plus seizures, and additional consideration is warranted with regards to the syndromic nature of its phenotypic association.
Subjects and Methods
Patient 1 Clinical History
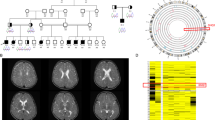
The 4-year old male proband was seen at the Mayo Clinic Department of Genetics for clinical evaluation. The proband has one unaffected brother and one unaffected sister (Fig. 1). His prenatal history was unremarkable, and no pregnancy concerns were reported. His sister was diagnosed with congenital hip dysplasia, but family history was otherwise unremarkable and consanguinity was denied. The patient was initially noted to be delayed at 6 months of age, when he was not meeting expected milestones and developed plagiocephaly. Parents reported that this child “never cried”. Hypotonia was noted at that time. At 2 years of age, he developed intractable seizures. At 4 years of age, the patient remains non-verbal and non-ambulatory, with limited ability to maintain a sitting position. He has minimal eye-tracking, and a history of strabismus. A skeletal survey revealed brachiocephaly with mild face hypoplasia and bilateral coxa vaga. He is non-dysmorphic in appearance. Clinicians also noted that upon physical exam, the proband showed no interest in relationships or pleasure activities, and did not respond to his name. He has frequent hand flapping activity, suggestive of an autism spectrum disorder. MRI studies showed a diffusely small brain relative to the cranium, likely due to generalized cerebral and cerebellar volume loss.
Clinicians in this case searched for this proband’s underlying etiology through a variety of biochemical, genetic, and physiological tests. With the inability to produce a diagnostic explanation in this case after a lengthy investigation process, diagnostic exome sequencing was pursued.
Patient 2 Clinical History
The 3-year old male proband was seen at the Children’s Hospital & Clinics of Minnesota Department of Genetics for clinical evaluation. He was born full term to healthy, nonconsanguineous parents and has one unaffected brother and one unaffected sister not tested through diagnostic exome sequencing (Fig. 2). His delivery was uncomplicated. He first rolled over at 6 months of age. His parents first became concerned around 6 months of age when he was not sitting up and noted unusual eye movements. He was diagnosed with hypotonia, strabismus, astigmatism, and cortical vision impairment at 9 months. A brain MRI showed hypoplastic corpus callosum. The patient was referred to the Genetics Clinic and was seen at 13 months of age. Several studies were recommended, including carbohydrate-deficient-transferrin, array comparative genomic hybridization, and repeat MRI. At a return visit at 16 months of age, he was rolling from back to front, babbling and laughing and had one word. Repeat brain MRI at 17 months revealed increased T2 flair signaling in the white matter, overall reduced volume, and hypoplastic corpus callosum. At around 2 years of age, he presented with myoclonic seizures, which increased in frequency and became intractable. He continued to make slow developmental progress and could sit when placed in a sitting position and began to play with toys. He had positional plagiocephaly with mild relative microcephaly (OFC 5th percentile, weight 8th percentile, length 13th percentile), mild beaked nasal tip, round face, mild prognathia with slight maxillary hypoplasia, high-arched palate, widely-spaced nipples, and mild hyperextensibility. Deep tendon reflexes could not be obtained. Stereotypic hand movements including bringing the fists to midline were noted. He also has some purposeful use of his hands. At age 4 he continued to make slow developmental progress and was attempting to sit independently. He continued to have frequent myoclonic seizures. A ketogenic diet was ineffective. He bangs his head occasionally, but does not seriously injure himself. He had a gastrostomy tube placed for failure to thrive. Extensive biochemical, genetic, and physiological testing was performed to try to identify the underlying etiology of this child’s condition. Diagnostic exome sequencing was pursued once it became clinically available (Fig. 5).
IRB Exemption, Informed Consent and Sample Collection
Based on the definition of research as outlined in The Federal Policy for the Protection of Human Subjects (45 CFR 46.102(d)), case reports of 3 or fewer patients do not meet the criteria for human-subject research and do not need IRB review and approval since these reports do not involve the formulation of a research hypothesis, implementation of research investigation, or systematic and prospective data collection. Therefore, it was determined that our 2 cases reported here met the criteria for exemption from IRB review and approval under these Federal regulations. Proper documented informed consent meeting Federal criteria was obtained from all patients and their family members involved in this case report, in compliance with the requirements outlined in The Federal Policy for the Protection of Human Subjects (Protection of Human Subjects 2009).
A blood sample was collected from each proband at their respective medical institutions and sent to Ambry Genetics (Aliso Viejo, CA) for diagnostic exome sequencing. Genomic deoxyribonucleic acid (gDNA) was isolated from whole blood from both patients. Samples were prepared using the SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA) (Gnirke et al. 2009). The enriched exome libraries were then applied to the solid surface flow cell for clonal amplification and sequencing using paired-end, 100-cycle chemistry on the Illumina HiSeq 2000 (Illumina, San Diego, CA).
Methodology
Initial data processing and base calling, including extraction of cluster intensities, was done using RTA 1.12.4 (HiSeq Control Software 1.4.5). Sequence quality filtering script was executed with the Illumina CASAVA software (ver 1.8.2, Illumina, Hayward, CA). Data yield (Mbases), %PF (pass-filter), # of reads, % of raw clusters per lane, and quality scores were examined in Demultiplex_Stats.htm file. The sequence data were aligned to the reference human genome (GRCh37) and variant calls were generated using CASAVA and Pindel. Exons plus at least 2 bases into the 5′ and 3′ ends of all the introns were analyzed. Data analysis focused on nonsense variants, small insertions and deletions, canonical splice site alterations or non-synonymous missense alterations. The Human Gene Mutation Database (HGMD), the Single Nucleotide Polymorphism database (dbSNP), 1,000 genomes, HapMap data and online search engines (e.g., PubMed) were used to search for previously described gene mutations and polymorphisms (HapMap 2003; Sherry et al. 2001, and Stenson et al. 2009). The filtering pipeline protects all variants annotated within the Human Gene Mutation Database (HGMD) and/or the Online Mendelian Inheritance in Man (OMIM) databases. Stepwise filtering included the removal of common SNPs, intergenic and 3′/5′ UTR variants, non-splice-related intronic variants, and lastly synonymous variants. Variants were then filtered further based on family history and possible inheritance models. Data are annotated with the Ambry Variant Analyzer tool (AVA), including nucleotide and amino acid conservation, biochemical nature of amino acid substitutions, population frequency (ESP and 1,000 genomes), and predicted functional impact (including PolyPhen and SIFT in silico prediction tools) (Gitiaux et al. 2013; HapMap 2003, and Grohmann et al. 2004). Each candidate mutation was assessed by a molecular geneticist to identify the most likely causative mutation(s). Multiple sequence alignments were viewed using IGV (integrative genomics Viewer software) (Robinson et al. 2011).
Identified candidate alterations were confirmed using automated fluorescence dideoxy sequencing. Co-segregation analysis was performed using each available family member (Table 1). Amplification primers were designed using PrimerZ. PCR primers were tagged with established sequencing primers on the 5′ end. Sequencing was performed on an ABI3730 (Life Technologies, Carlsbad, CA) using standard procedures.
Results
Exome sequencing of the two probands resulted in an average of 15.6 Gb of sequence per sample Mean coverage of captured regions was an average of 120× per sample, with >91 % covered with at least 10× coverage, an average of >91 % base call quality of Q30 or greater, and an overall average mean quality score of <Q35. Stepwise filtering removal of common SNPs, intergenic and 3′/5′ UTR variants, non-splice-related intronic variants, and synonymous variants resulted in ~11,000 variants per patient (Table 2).
Bioinformatics and manual filtering for patient #1 revealed 56 genes (68 unique alterations) post-family history inheritance model filtering based on autosomal and X-linked dominant and recessive and Y-linked inheritance models. Manual review of each alteration to rule out sequencing artifacts and polymorphisms along with medical interpretation to rule out genes lacking significant clinical overlap with the patient’s evaluated phenotype resulted in 5 genes (5 unique alterations). Among these, one notable gene (one unique alteration) with potential clinical relevance was detected: IQSEC2 c.2582G>C (p.S861T) (Table 3).
The c.2582G>C (p.S861T) alteration, located in exon 7 of IQSEC2, results in both a deleterious missense alteration and is predicted to abolish the native splice donor site. The alteration was not observed among 6,503 healthy individuals in the NHLBI Exome Sequencing Project (ESP) database. The S861 amino acid is highly conserved among available vertebrate species (Fig. 3). This alteration is predicted to be deleterious by PolyPhen and SIFT in silico analyses (Grohmann et al. 2004). Further, the alteration is located in the sec7 domain of IQSEC2, responsible for catalyzing nucleotide exchange onto ARF (ADP-ribosylation factor) substrates and it has been demonstrated that missense changes in this domain result in significantly diminished GTP binding to ARF6 (Shoubridge et al. 2010). Further, the c.2582G>C (p.S861T) alteration is at the last nucleotide of exon 7 and is predicted to abolish the native splice donor site by in silico prediction (Net2Gene and BDGP). Alterations that disrupt the canonical splice donor site are typically deleterious in nature (Richards et al. 2008).
Bioinformatics and manual filtering for patient #2 revealed 65 genes (73 unique alterations) post-family history inheritance model filtering Manual review to rule out sequencing artifacts and genes lacking significant clinical overlap with the patient’s evaluated phenotype resulted in 7 genes (9 unique alterations). Among these, one notable gene (one unique alteration) with potential clinical relevance was detected: IQSEC2 c.2052_2053delCG (p.C684X) (Table 4).
The deleterious c.2052_2053delCG (p.C684X) alteration, located in exon 5 of the IQSEC2 gene, results from a deletion of two nucleotides at positions 2052 and 2053, causing a translational frameshift with a predicted alternate stop codon at codon 684 (Fig. 4).
Co-segregation analysis in both patient families revealed that the mothers did not carry the same alteration, establishing a de novo mutation origin (Table 1).
Among the first 200 probands sequenced at Ambry Genetics, 33 (17 %) were referred for undiagnosed disorders including seizures/epilepsy. Of these individuals, 22 probands were identified to carry pathogenic mutations in genes reported to be associated with seizures/epilepsy phenotypes. Among this cohort, pathogenic IQSEC2 mutations account for approximately 9 % (2/22) of these positive findings.
Discussion
Historically, IQSEC2 has been associated with X-linked non-syndromic intellectual disability and mental retardation. This condition is characterized by early onset limited intellectual functioning and limited adaptive behavior (Shoubridge et al. 2010). Nevertheless, some affected individuals with IQSEC2 mutations present with additional clinical findings including seizures, autistic traits, psychiatric problems, and very limited motor and language development (Tran Mau-Them et al. 2013; Rauch et al. 2012, and Shoubridge et al. 2010).
Herein, diagnostic exome sequencing identified two novel deleterious IQSEC2 alterations in two males with developmental delays, seizures, hypotonia, vision impairments, plagiocephaly, autistic-like features, behavioral abnormalities, absent language skills, and abnormal MRI findings. In addition to overlapping features already reported in association with IQSEC2 mutations, these probands have additional phenotypic traits including mild beaking of the nasal tip, mild face hypoplasia, prognathia with slight maxillary hypoplasia, round facies, widely spaced nipples, hyperextensibility of the joints, absent deep tendon reflexes, bilateral coxa valga, and “doughy” skin (Fig. 5). Both probands, however, did not appear to manifest behavioral disturbances which have been identified in previous case reports. Given their young age, there is the potential for additional psychiatric findings to manifest in the future. These two cases, in addition to those reported by Tran Mau Them et al., suggest that other phenotypic features including seizures, strabismus or visual impairment, hypotonia, and overall brain volume loss are present in patients with IQSEC2 mutations.
The IQSEC2 gene alterations identified in both of our probands provide an underlying explanation for the patients’ symptoms; especially since no other likely candidate gene alterations were identified. Diagnostic exome sequencing established a molecular diagnosis for these two patients, in whom traditional testing methods were uninformative. Additionally, truncating mutations have been identified in both of our probands as well as in two male probands with chromosomal duplications involving IQSEC2. Compared to patients with missense alterations in the gene, these mutations appear to present with a more severe clinical phenotype.
Implications for Clinical Practice and Diagnosis
Until now, molecular diagnosis depended tremendously upon a recognizable pattern of phenotypic features presenting in a clinical setting, and the ability of the skilled physician to correlate a suspicion with appropriate laboratory testing options (Glusman 2013). As is in both probands presented here, extensive physical, biochemical, and genetic testing over a lengthy period of time often failed to lead patients and clinicians alike to an appropriate diagnosis. Prior to pursuing diagnostic exome sequencing, patient #1 had >43 and patient # 2 had >24 different biochemical and molecular tests performed over approximately 3–4 years respectively in search of an underlying diagnosis. These tests were negative and/or uninformative in both cases, costing each patient an estimated total of \$10,000–25,000 in individual gene sequencing costs alone (not including biochemical analysis and other diagnostic assessments performed). Each individual’s unsuccessful diagnostic odyssey ultimately led to the desperate decision to pursue a more novel diagnostic approach through exome sequencing technology. Ultimately, once diagnostic exome sequencing was pursued, a pathogenic mutation was identified in both cases in the same gene that would have otherwise escaped a traditional clinical work-up.
This new generation of diagnostic sequencing allows for tremendous acceleration in the clinical diagnostic process, and additionally expands the analyses of genes that would not generally be considered obvious test candidates. Although IQSEC2 gene sequencing is not a test commonly ordered on a clinical basis, a few laboratories do offer IQSEC2 analysis as part of a non-syndromic X-linked intellectual disability panel test. As the phenotypic spectrum associated with this gene continues to grow, it is suggested that IQSEC2 be added to both X-linked intellectual disability and epilepsy panels in order to broaden the spectrum of clinical availability. Critics of diagnostic exome sequencing have voiced concerns about the lengthy turn-around-times (approximately 10–15 weeks for First-Tier) in comparison to the average X-linked intellectual disability gene panel (8–12 weeks). First-tier exome sequencing involves analysis of ~20 % of the human genome (~4,400 of ~20,000 genes). However, when considering these timeframes in the context of an overall diagnostic process spanning the course of several years, the positive implications for clinical management are significant. This is especially true when most first-tier biochemical and molecular analyses such as karyotype, chromosomal microarray, metabolic work-up, etc. are uninformative.
The IQSEC2-associated phenotype is variable and includes expansive features including severe seizures and dysmorphism, suggestive of a genetic syndrome. Historically, IQSEC2 has been reported in association with a non-syndromic intellectual disability phenotype, undoubtedly creating confusion when dysmorphism, seizures, and a wide range of phenotypic variability are present as well. Therefore, a clinician faced with the daunting task of diagnosing a patient presenting such a complex phenotype would reasonably forego an X-linked intellectual disability panel in search for an epilepsy disorder, complex single-gene, or metabolic condition. This phenomenon was true in both cases presented here. In addition to the emotional, psychosocial and physical investment often placed in the diagnostic process, the overall costs of gene-after-gene and biochemical analysis is extremely high, and it has sparked discussions regarding diagnostic efficiency in the clinical realm. Hypothetically, if diagnostic exome sequencing had been pursued as an early genetic test in both of our cases, the diagnostic process would have been completed in a matter of a few months versus several years.
As we discovered, since pathogenic IQSEC2 mutations account for 9 % of all positive diagnostic exome results with seizure/epilepsy genes at our laboratory, it is obvious that this gene plays a more significant clinical role than previously anticipated. As we know from the scientific literature, IQSEC2 has not been commonly identified as a seizure disorder gene. Nevertheless, as we show here, seizures are a major phenotypic feature seen in both our probands as well as in probands described in previous case reports (Tran Mau-Them et al. 2013). Therefore, it is encouraged that care providers in both pediatric genetics and neurology clinics consider diagnostic exome sequencing in cases with significant clinical overlap. Genetic counselors, especially, should be aware of the growing number of IQSEC2 cases being reported in association with severe seizures and should consider diagnostic exome sequencing in these cases, especially in the absence of a positive family history. As diagnostic exome sequencing becomes more widely utilized, it is our hypothesis that a significantly greater number of IQSEC2 cases will be identified.
Implications for Psychosocial Considerations
Diagnostic exome sequencing significantly shortens the diagnostic process which has a substantial impact on the psychosocial experiences of patients and their families (Roper 2012). Although no cure currently exists for IQSEC2-associated disease, the quest for emotional closure traditionally prevails as a major driving force behind the search for positive test results. In some circumstances, multi-gene panels may be a more appropriate testing strategy, especially when clinical suspicion is high for a specific single-gene disorder included in that package. However, in conditions where phenotypic overlap with a variety of single-gene disorders is significant, genetic counselors can play a significant role in presenting diagnostic exome sequencing as the more appropriate option due to its more significant gene-coverage. With the increased usage of Next Generation sequencing in mainstream clinical practice, traditional genetic approaches to clinical diagnosis will undoubtedly be revolutionized (Raffan and Semple 2011). Researchers believe that these advanced testing technologies will ultimately decrease the number of patient with reported undiagnosed genetic syndromes by identification of atypical presentations of known diseases (Raffan and Semple 2011).
References
American Psychiatric Association (2000) Diagnostic and Statistical Manual of Mental Disorders, 4th ed, Text Revision (DSM-IV-TR), American Psychiatric Association, Washington, DC.
Chelly, J., Khelfaoui, M., Francis, F., Cherif, B., & Bienvenu, T. (2006). Genetics and pathophysiology of mental retardation. European Journal of Human Genetics, 14(6), 701–713.
Gitiaux, C., Bergounioux, J., Magen, M., Quijano-Roy, S., Blanc, T., Bonnefont, J. P., et al. (2013). Diaphragmatic weakness with progressive sensory and motor polyneuropathy: case report of a neonatal IGHMBP2-related neuropathy. Journal of Child Neurology, 28(6), 784–787.
Glusman, G. (2013). Clinical applications of sequencing take center stage. Genome Biology, 14, 303.
Gnirke, A., Melnikov, A., Maguire, J., Rogov, P., LeProust, E. M., Brockman, W., et al. (2009). Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotechnology, 27, 182–189.
Grohmann, K., Rossoll, W., Kobsar, I., Holtmann, B., Jablonka, S., Wessig, C., et al. (2004). Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1). Human Molecular Genetics, 13, 2031–2042.
Li, N., & Carrel, L. (2008). Escape from X-chromosome inactivation is an intrinsic property of the Jarid1c locus. PNAS, 105(44), 17055–17060.
Morleo, M., Iaconis, D., Chitayat, D., Peluso, I., Marzella, R., Renieri, A., et al. (2008). Disruption of the IQSEC2 transcript in a female with X;autosome translocation t(X;20)(p11.2q11.2) and a phenotype resembling X-linked infantile spasms (ISSX) syndrome. Molecular Medicine Reports, 1, 33–39.
Nagase, T., Ishikawa, K. I., Miyajima, N., Tanaka, A., Kotani, H., Nomura, N., et al. (1998). Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Research, 5, 31–39.
Protection of Human Subjects, 45 CFR 46.102(d) (2009). Department of Health and Human Services retrieved from http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.html#subparta
Raffan, E., & Semple, R. K. (2011). Next generation sequencing—implications for clinical practice. British Medical Bulletin, 99, 53–71.
Rauch, A., Wieczorek, D., Graf, E., Wieland, T., Endele, S., Schwarzmayr, T., et al. (2012). Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet, 380(9854), 1674–1682.
Richards, S. L., Bale, S., Bellissimo, D. B., Das, S., Grody, W. W., Hegde, M. R., et al. (2008). ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genetics in Medicine, 10(4), 294–300.
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nature Biotechnology, 29(1), 24–26.
Roper, H. H. (2012). On the future of genetic risk assessment. Journal of Community Genetics, 3, 229–236.
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Research, 29(1), 308–311.
Shoubridge, C., Tarpey, P. S., Abidi, F., Ramsden, S. L., Rujirabanjerd, S., Murphy, J. A., et al. (2010). Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nature Genetics, 42(6), 486–488.
Stenson, P. D., Mort, M., Ball, E. V., Howells, K., Phillips, A. D., Thomas, N. S., et al. (2009). The Human Gene Mutation Database: 2008 update. Genome Medicine, 1(1), 13.
Strømme, P. (2000). Aetiology in severe and mild mental retardation: a population-based study of Norwegian children. Developmental Medicine and Child Neurology, 42(2), 76–86.
Suthers, G. K., Turner, G., & Mulley, J. C. (1988). A non-syndromal form of X-linked mental retardation (XLMR) is linked to DXS14. American Journal of Medical Genetics, 30, 485–491.
The International HapMap Consortium. (2003). The international hapmap project. Nature, 426, 789–796.
Tran Mau-Them, F., Willems, M., Albrecht, B., Sanchez, E., Puechberty, J., Endele, S., et al. (2013). Expanding the phenotype of IQSEC2 mutations: truncating mutations in severe intellectual disability. European Journal of Human Genetics.
Acknowledgments
We are grateful to the patients and their families for their participation.
Comments
This manuscript is submitted solely to this journal and was not published elsewhere. We have submitted a similar version of the manuscript’s abstract to the American Society of Human Genetics conference for poster presentation at the 2013 educational conference.
Consent Process and IRB Exemption Status for Case Reports
Based on the definition of research as outlined in The Federal Policy for the Protection of Human Subjects (45 CFR 46.102(d)), case reports of 3 or fewer patients do not meet the criteria for human-subject research and do not need IRB review and approval since these reports do not involve the formulation of a research hypothesis, implementation of research investigation, or systematic and prospective data collection. Therefore, it was determined that our 2 cases reported here met the criteria for exemption from IRB review and approval under these Federal regulations. Proper documented informed consent meeting Federal criteria was obtained verbally and in writing from all patients and their family members involved in this case report, in compliance with the requirements outlined in The Federal Policy for the Protection of Human Subjects (Protection of Human Subjects 2009). Additionally, proper written informed consent for the use of photographs was also obtained by the clinicians involved in this case report.
Disclosures of Conflicts-of-Interest
Authors S. K. Gandomi, K.D. Farwell Gonzalez, M. Parra, L. Shahmirzadi, W. Zeng, and S. Tang are full-time salaried employees of Ambry Genetics, which is a commercial-based sequencing laboratory. These authors have no financial or commercial conflicts-of-interest to disclose beyond their basic employment with the laboratory. No outside funding was obtained nor was any other internal or external resource received for the analysis or publication process of this report. All laboratory data for this report was collected retrospectively after diagnostic testing had been completed for clinical purposes, and therefore the creation of this report does not additionally contribute to any relational or financial conflicts-of-interest with the referring providers.
Authors J. Mancuso, P. Pichurin, R. Temme, S. Dugan are associated with Mayo Clinic and Children’s Hospital & Clinics of Minnesota and do not have any conflicts-of-interest to disclose and are separate, independent entities from Ambry Genetics. They do not have any financial relationship with Ambry Genetics beyond a standard clinical nature.
Ambry Genetics has full control of the laboratory data associated with the DES testing process, and agrees to allow the journal to review the data if requested.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gandomi, S.K., Farwell Gonzalez, K.D., Parra, M. et al. Diagnostic Exome Sequencing Identifies Two Novel IQSEC2 Mutations Associated with X-Linked Intellectual Disability with Seizures: Implications for Genetic Counseling and Clinical Diagnosis. J Genet Counsel 23, 289–298 (2014). https://doi.org/10.1007/s10897-013-9671-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10897-013-9671-6