Abstract
A novel 6-ethoxy chromone-3-carbaldehyde benzoyl hydrazone (L) and its Ln(III) complexes, [Ln = Sm (1), Eu (2), Gd (3), Tb (4)], have been synthesized and characterized. The fluorescence properties of the Eu(III) and Sm(III) complexes in solid state and Eu(III) complex in different solutions (DMF, DMSO, methanol and acetonitrile) were investigated. At the same time, the DNA-binding properties of the two complexes are investigated using UV-Vis absorption spectroscopy, fluorescence spectroscopy, viscosity measurement. All the experimental evidences indicate that the two complexes can bind to CT-DNA via an intercalation mechanism. Furthermore, antioxidant activity tests in vitro showed that the complexes have significant antioxidative activity against hydroxyl free radicals from the Fenton reaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There has been considerable interest in the binding studies of small molecules to DNA. A more complete understanding of how to target DNA site would be valuable in the rational design of sequence-specific DNA binding molecules of application in chemotherapy and highly sensitive DNA probe [1–3]. Given the structural flexibility and variable dimensionality, transition-metal complexes play a key role in the development of new bound complex at a DNA site in recent years [4–10]. As both spectroscopic tags and functional models for the active centers, these complexes have been broadly used as foot-printing agents of both proteins and DNA. It was reported that the complexes’ intercalating ability may be varied from the coordination geometry, which are attributed to the metal ion type and its valence, and the ligand donor atom type [11–16]. Biological activity and also numbers of other applications resulted in strongly increasing interest for rare earth in the last decade [17–20]. One of the most studied applications is the usage of the rare earth complexes to address DNA/RNA by non-covalent binding and/or cleavage [21, 22].
Currently, due to their high antitumor activity and low toxicity, natural products are being paid attention. Chromones are ubiquitous in nature especially in plants [23].These natural products are potentially antibacterial, anticancer, antioxidant, anti-inflammatory, and antiallergenic agents since they stimulate or inhibit a wide variety of enzyme systems as pharmacological agents [24, 25]. Our previous work showed that the rare earth complexes of chromone hydrazone ligands have certain antioxidant and cytotoxic activity, and can bind to CT-DNA by intercalation [26–29], and the substitute position in the ligand can affect the binding ability of the complexes with DNA. Based on these, it occurred us further design different substitute ligand to study how different substitute affects the binding ability of complexes with DNA.
Here, we synthesized a novel chromone derivatives hydrazone ligand (6-ethoxy chromone-3-carbaldehyde benzoyl hydrazone) and its rare earth complexes. We also explored the interactions of Eu(III) and Sm(III) complexes with DNA using spectroscopic and hydrodynamic methods. The results should be extremely useful for understanding the mode of the complex with DNA as well as laying a foundation for the rational design of novel, powerful agents for probing and targeting nucleic acids. This study will be very helpful to examine the DNA conformation and structure.
Experimental
Chemicals
Acetic anhydride, hydroquinone were produced in China. All other chemicals used were of analytical grade and purchased from commercial vendors. The Ln(III) nitrates were derived from their oxide (99.9%) acquired from Nong Hua (PRC).
DNA sample
Calf thymus DNA (CT-DNA) was obtained from Sigma, UV-Vis spectrometer was employed to check DNA purity 1.8–1.9:1 and concentration (ε = 6,600 M−1 cm−1 at 260 nm) [30, 31]. All the experiments involving interaction of the metal complexes with DNA were carried out in doubly distilled water buffer containing 5 mM Tris and 50 mM NaCl, and adjusted to pH 7.2 with hydrochloric acid.
Physical measurements
The metal ions were determined by EDTA titration using xylenol orange as an indicator. Carbon, nitrogen and hydrogen analyses were determined using a Vario EL analyzer. Molar conductivity measurements were carried out with a DDS-11C type conductivity bridge using 1.0 × 1 0−3 mol /L solutions in methanol at 25 ℃. IR spectra were recorded on a Therrno Mattson FT-IR instrument using KBr discs in the 4,000–400 cm−1 region. The UV-visible spectra were recorded on a Perkin-Elmer Lambda-35 UV-Vis spectrophotometer. 1H NMR spectrum was measured on a Varian Mercury 200-MHz spectrometer in solution with TMS as internal standard. Electrospray ionization (ESI) mass spectrometry was recorded on APEX II FT-ICR MS using methanol as mobile phase. Fluorescence spectra were obtained on a Shimadzu RF-5301 spectrophotometer at room temperature. The melting points of the compounds were determined on a XT4-100x microscopic melting point apparatus (Beijing, China). The UV-Vis absorption of antioxidative activity experiment was performed in phosphate buffer with a 721-E spectrophotometer (Shanghai, China)
Synthesis of the ligand (L)
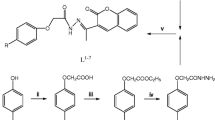
The organic compounds 1–3 (Fig. 1) were prepared according to the literature [32].
The organic compound 4: A mixture of 3 (7.60 g, 50 mmol), 60 mmol of the proper alkyl halide, (10.46 g, 75.8 mmol) of anhyd K2CO3 and acetone (220 ml) was heated at reflux for 2–3 d, with stirring. The mixture was filtered off, washed with acetone and the brown filtrate was concentrated to a solid. The solid was chromatographed on a silica gel column, eluting with acetate—petroleum ether (1: 20) to give, after removal of solvents from the eluate collected, the nearly pure yellow crystals of compound 4 Yield: 70%. Mp: 50 ℃ ( [33] mp 52 ℃). Elemental analysis: C10H12O43 Calcd: C, 66.65; H,6.71. Found: C, 66.60; H,6.80. 1H NMR (CDCl3-d 1, 200 MHz): 8.63(1H, s, 2-H), 8.86 (1H, s, CH = N), 7.94–7.41 (8H, m, PhH, 5, 7, 8-H), 4.15 (2H, q, CH2), 1.37 (3H, t, CH3). IR (KBr pellet, cm−1): ν C=O (ketone): 1,643.
The organic compound 5 (Fig. 1): Compound 4 (3.60 g, 20 mmol) was dissolved in DMF (40 ml), the mixture was added POCl3 (20 ml), at 0 ℃. Then the solution was stirred at room temperature overnight. The solution was allowed to stir at 100 ℃ for 1 h. The mixture was added to ice water yielding a precipitate, which was filtered off, washed with water and dried. Then the solid was chromatographer on a silica gel column, eluting with ethyl acetate-petroleum ether (1:30) to give, after removal of solvents from the eluate collected, the nearly pure yellow compound 5 .Yield: 60%. Mp: 134–135 °C. Elemental analysis: C12H10O4 Calcd: C, 66.05; H, 4.62. Found: C, 66.60; H, 4.70. 1H NMR (CDCl3-d 1, 200 MHz):δ10.41(1H, s, CHO), 8.53 (1H, s, 2-H), 7.64–7.33 (3H, m, 5, 7, 8-H), 4.16 (2H, q, CH2), 1.47 (3H, t, CH3). IR (KBr pellet, cm−1): ν C=O (aldehyde) : 1,700, ν C=O (ketone):1,640.
The ligand L (Fig. 1) was synthesized as follows. An ethanol solution containing benzoyl hydrazone (1.36 g, 10 mmol) was added dropwise to another ethanol solution containing 5 (2.31 g, 10 mmol). The mixture was stirred for 1 h at room temperature yielding a precipitate, which was filtered off, washed with ethanol and dried in a vacuum. The solid was recrystallised from ethanol to yield a yellow solid and then dried. Yield: 80%. Mp: 159 °C. Elemental analysis: C19H16N2O4 Calcd: C, 67.85; H, 4.79, N, 8.33. Found: C, 67.80; H, 4.85; N, 8.27. ESI-MS [CH3OH, m/z]: [M + H]+: 337.1194, calcd: 337.1188. 1H NMR (DMSO-d 6 , 200 MHz):δ11.93 (1H, s, NH), 8.63(1H, s, 2-H), 8.81 (1H, s, CH = N), 7.94–7.41 (8H, m, PhH, 5, 7, 8-H), 4.16 (2H, q, CH2), 1.37 (3H, t, CH3). IR (KBr pellet, cm−1) : ν C=O(carbonyl): 1,650, ν C=O(hydrazonic): 1,626, ν C=N: 1,602. UV-vis (methanol):λmax (nm) = 213, 237, 287, 321.
Synthesis of the Ln(III) complexes
The ligand (L) (0.1 mmol) was dissolved in ethanol (15 ml) at 70 °C. To this solution was added dropwise Ln(NO3)3·6H2O (Ln = Sm, Eu, Gd, Tb) (0.05 mmol). After stirring for 1 h at 70 °C, the precipitate was separated from the solution by suction filtration, purified by washing several times with ethanol and dried for 24 h in a vacuum. The yields were in the range of 50–60 %.
UV-Vis absorption titration
Absorption titration experiment was performed by maintaining the metal complex concentration constant (5 μM) and gradually increasing the concentration of nucleic acid. The compounds were dissolved in a mixed solvent of 0.5% CH3OH and 99.5% Tris-HCl buffer (5 mM Tris-HCl, 50 mM NaCl, pH 7.2). The reference solution was the corresponding Tris-HCl buffer solution. While measuring the absorption spectra, equal amount of DNA was added to both compound solution and the reference solution to eliminate the absorbance of DNA itself. Each sample solution was scanned in the range 190–400 nm.
Fluorescence titration
To compare quantitatively the affinity of the compound bound to DNA, the intrinsic binding constants K b of the two compounds binding to DNA were obtained by the fluorescence titration method. The samples were excited at their corresponding isosbestic points. All the measurements were made at room temperature with 5 nm for both entrance and exit slit. Fixed amounts of the compound (5 μM) were titrated with increasing amounts of DNA, over a range of DNA concentrations from 2 μM to 26 μM. An excitation wavelength of 326 nm was used, and total fluorescence emission intensity was monitored at 438 nm for Eu(III) complex and 435 nm for Sm(III) complex.
Viscosity measurements
Viscosity experiments were carried out on a Ubbelohde viscometer, immersed in a thermostatic water bath maintained at 25.0 ± 0.1 ℃. DNA concentration was kept constant (5 μM) and gradually increased the concentration of tested compound. Flow time was measured with a stopwatch. Each sample was measured three times and an average flow time was calculated.
Hydroxyl radical scavenging activity
The hydroxyl radical in aqueous media was generated through the Fenton reaction [34]. Solution of the compound to be tested was prepared in DMF (1×10−3 M). 5 ml assay mixture contained following reagents: phosphate buffer (0.15 mM, pH=7.4), safranin (0.1 mM), EDTA-Fe(II) (1 mM), H2O2 (88 mM), the tested compound (1, 2, 3, 4 and 5μM). The tested samples were allowed to react for 1 h at a constant 37.0°C in a water bath, after which the absorbance was measured at 520 nm against a blank. All the tests were run in triplicate and expressed as the mean±standard deviation (SD).
Results and discussion
Characterization of complexes
The complexes were prepared by direct reaction of ligand with the appropriate mole ratios of Ln(III) nitrate in ethanol. The yields were good to moderate. Analytical data for the complexes, presented in Table 1, conform to Ln(L)2(NO3)3·2H2O (Ln = Sm, Eu, Gd, Tb). The four metal complexes are air stable for extended at least six months and soluble in methanol, DMSO and DMF; slightly soluble in ethanol and chloroform; insoluble in benzene, water and diethyl ether. The molar conductivity values of the complexes in methanol are in the range of 100–121 scm2mol−1, indicating that the complexes are 1:1 electrolytes [35]. The mass spectrum of Eu(III) complex shows peaks at m/z of 823.1279, (calcd: 823.1276), 412.0697 (calcd: 412.0677), which can be assigned to the ion pair {[EuL2·(NO3)2·NO3]-3NO3-2H}+, {[EuL2·(NO3)2·NO3] -3NO3-H}2+, respectively.
IR spectra
On the basis of the similar IR spectra of the complexes (Table 2), it may be assumed that all of them have similar coordination structures. The ν(C = O) (carbonyl) and ν (C = O) (hydrazonic) vibrations of the free ligand are at 1,650 and 1,626 cm−1, respectively; for the complexes these peaks shift to 1,633 and 1,611 cm−1, △ν (ligand-complex ) is equal to 17 and 15 cm−1. The band at 577 cm−1 is assigned to ν (M-O). These shifts and the new band demonstrate that the oxygen of carbonyl has formed a coordinative bond with the rare earth ions [36]. The band at 1,602 cm−1 for the free ligand is assigned to the ν (C = N) stretch, which shifts to 1,574 cm−1 for its complex. Weak bands at 427 cm−1 are assigned to ν (M-N). These shifts and the new band further confirm that the nitrogen of the imino-group bonds to the rare earth ions [37]. For all complexes, bands at 1,384 cm−1 in the spectra of complexes indicate that free nitrate groups (D3h) exist [38]. Additionally, the two intense absorption bands in the spectra associated with the asymmetric stretching appear in the range of 1,325–1,327 cm−1 (ν 4) and 1,487–1,489 cm−1 (ν 1), clearly establishing that there are some coordinated NO -3 groups (C2v) in the complexes[39]. The differences between the two bands lie in 160–162 cm−1, suggesting that the coordinated nitrate groups in the complexes are bidentate [40]. In addition, broad bands at 3,219 cm−1 indicate that water molecules are existent in the complexes [40], in agreement with the results of the elemental analysis.
According to the data of the elemental analyses, molar conductivity, IR spectra and mass spectra of Eu(III) complex, as well as the similar structure previously reported [26], the composition of the complexes can be inferred: [Ln(L)2(NO3)2]NO3·2H2O (Ln = Sm, Eu, Gd, Tb).
Fluorescence spectra
Fluorescence arises from the presence of ligand-to-metal energy transfer [41]. The ability of transfer energy from ligand to metal is important in the design of lanthanide supramolecular photonic devices [42, 43]. The emission spectrum of the Eu(III) and Sm(III) complexes in solid state is shown in Fig. 2a, b. The Eu(III) and Sm(III) complexes show the strong red fluorescence in solid state, the spectra data are shown in Table 3. This indicates that the ligand L is a good organic chelator to absorb and transfer energy to lanthanide ions. However, the Tb(III) complexes only exhibit the free ligand band emission, which indicates that the energy would not be transferred efficiently between the ligand triplet level and the resonance levels of the Tb(III) ions.
The influences of several solvents on the fluorescence intensity of the Eu(III) complex was investigated (Fig. 3). From the emission spectrum data of complex in solid state (Table 3), it can be seen that the intensity of 5D0 → 7F2 transition is stronger than that of 5D0 → 7F1, and the most intensity ratio value η = 5D0 → 7F2 /5D0 → 7F1 is 2.84. However, from Table 4, the values for (5D0 → 7F2 / 5D0 → 7F1) are 2.45 (methanol), 2.96 (acetonitrile), 1.83 (DMF) and 0.64 (DMSO). It has been reported that the 5D0 → 7F2 transition is an electric dipole transition, which can be detected as a relatively strong peak only when Eu(III) does not lie in centrosymmetric ligand field, on the contrary, the 5D0 → 7F1 transition is a magnetic dipole transition, and its intensity becomes the most intensive only when Eu(III) ion is the centre of ligand field [44]. It also could be seen from Fig. 4 that in methanol solution the Eu(III) complex has the strongest luminescence, and then in acetonitrile, DMF and DMSO solution. We also see that the emission spectra of the Eu(III) complex exhibit different bands in different solvents. Interestingly, the complex exhibits five bands in solid, corresponding to 5D0 → 7F0, 5D0 → 7F1, 5D0 → 7F2, 5D0 → 7F3 and 5D0 → 7F4 transitions. Comparing to the fluorescence of solid complex, the 5D0 → 7F0, 5D0 → 7F3 and 5D0 → 7F4 were disappeared in the solvents. This is due to the coordinating effects of solvents, namely solvate effect [45]. The influence of solvent on the fluorescence intensities of the Sm(III) complex was even more apparent. The fluorescence intensity of the Sm(III) complex in DMF, DMSO was completely quenched. The data does not appear in this paper. These phenomena were also seen in other Eu(III) complexes previously reported [27–29, 46].
a Electronic spectra of Eu(III) complex (5 μM) in the presence of increasing amounts of CT-DNA, [DNA] = 0–24 μM. Arrow shows the absorbance changes upon increasing DNA concentration. b Electronic spectra of Sm(III) complex (5 μM) in the presence of increasing amounts of CT-DNA. [DNA] = 0–20 μM. Arrow shows the absorbance changes upon increasing DNA
Consequently, it can be concluded that the structure of the Eu(III) and Sm(III) complexes change in the above organic solvents, which leads to the abnormal fluorescence intensity change. These indicate that solvent molecules have strong coordination effect, and that the environment plays an important role in determining the fluorescence intensity of the complexes [47].
UV-Vis absorption titration
The binding of intercalative drugs to DNA has also been characterized classically through absorption titrations, following the hypochromism and red shift associated with binding of the colored complex to the helix [48]. These spectral characteristics are attributable to a mode of binding that involves an interaction between an aromatic chromophore and the base pairs of DNA. The magnitudes of the red shift and the degree of hypochromism are furthermore commonly found to correlate with the strength of the intercalative interaction [48]. Figure 4a, b show absorption titration for the Eu(III) and Sm(III) complexes as a function of DNA addition. In the UV region, the intense absorption bands with maxima of 202, 288 and 315 nm were attributed to intraligand Eu(III) complex. The spectra show that addition of DNA yields hypochromism and red shift in the three bands. The absorption bands of Eu(III) complex at 202, 288 and 315 nm exhibited hypochromism of about 16.22%, 62.59% and 48.73% , the red shift of about 1, 5 and 2 nm, respectively. For Sm(III) complex the absorption bands at 202 and 315 nm exhibited hypochromism of about 11.58 and 67.59%, red shift of about 1 and 2 nm, respectively.
According to the previous literature [26–29], we can also find that the UV-visible spectra of other Ln(III) complexes reported by Wang [26–29] do not differ significantly with the substitutions on the chromone hydrazone ligand. Base on the spectral characteristics, we presume that there are some interactions between the complexes and DNA.
Fluorescence titration
Figures 5 and 6 display a well-behaved titration of the Eu(III) and Sm(III) complexes with CT-DNA. Increases in emission are apparent with DNA addition. The results of the emission titrations suggest that the two compounds get into a hydrophobic environment inside the DNA and avoid the quenching effect of solvent water molecules. Moreover, the similarity in spectroscopic perturbations indicates that two complexes bind to DNA in a similar fashion. These phenomena were also seen in other Ln(III) complexes reported [26–29].
To compare quantitatively the affinity of the compound bound to DNA, the intrinsic binding constants K b of the two compounds based upon these fluorescence titrations may be made with according Eq. 1 [32]:
Where C t is the total compound concentration; F is the observed fluorescence emission intensity at a given DNA concentration; F 0 is the intensity in the absence of DNA, and F max is the fluorescence of the totally bound compound. Binding data were cast into the form of a Scatchard plot [49] of r/C f versus r, where r is the binding ratio, C b /[DNA]t and C f is the free compound concentration. The binding constants are (3.17 ± 0.06) × 106 M−1, (1.84 ± 0.04) × 106 M−1 for Eu(III) and Sm(III) complex, respectively. This value is lower than the one reported for Sm(III) complex [26] (Kb = (3.70 ± 0.47) × 106 M−1)[26].
After corrections for the differing affinities of Sm(III) complex with different ligand in [26] and this paper, it appears from these data that the substitutions of ligand can affect the binding affinity that complex binds to DNA. This variation likely reflects the differing ability of these ligands to stack and overlap well with the base pairs. In this paper, the -OH of phenyl was substituted by CH3CH2O-. Compared with the hydrogen, the CH3CH2- is a large group in space, which is not favorable to corresponding ligand intercalation with DNA. Hence, it is reasonable to understand why the complexes show a low magnitude of the red shift in UV-Vis absorption as well as lower intrinsic binding constants compared with our previously reported complexes [26].
Viscosity measurements
Photophysical studies have proven quite useful in determining binding modes of metal complexes to DNA [50], but not sufficient clues to support a binding mode. The modes of binding of the complexes were further investigated by hydrodynamic measurements such as viscosity. A classical intercalation model demands that the DNA helix lengthens as base pairs are separated to accommodate the bound ligand, leading to the increase of DNA viscosity. In contrast, a partial, non-classical intercalation of ligand could bend (or kink) the DNA helix reduce its effective length and, concomitantly, its viscosity [51, 52]. As a means for further clarifying the binding of these compounds with DNA, viscosity studies were carried out. Data are presented as (η/η 0 )1/3 versus R, where R = [Compound]/[DNA]; ηandη 0 are the relative viscosities of DNA in the presence and absence of compound, respectively. The relative viscosity values were calculated from the flow time of DNA-containing solution t and the flow time of buffer alone t 0 , using the following Eq. 2 [53]:
The results of the viscosity measurements are shown in Fig. 7. On increasing the amount of Sm(III) and Eu(III) complexes, the relative viscosity of DNA increase steadily, this is similar to our previously reported complexes [26–29]. The results suggest that complexes may bind to DNA by intercalation.
Hydroxyl radical scavenging activity
Generation of reactive oxygen species (ROS) is a normal process in the life of aerobic organisms. Oxidative damage to DNA has been suggested to contribute to aging and various diseases including cancer and chronic inflammation. Antioxidants are substances that delay or prevent the oxidation of cellular oxidizable substrates. They exert their effects by scavenging ROS, activating a battery of detoxifying proteins, or preventing the generation of ROS [54]. So ,it is vital to develop efficient scavengers. Since among all reactive oxygen species, the hydroxyl radical is by far the most potent and therefore the most dangerous oxygen metabolite, elimination of this radical is one of the major aims of antioxidant administration [55].
The hydroxyl radicals in aqueous media were generated through the Fenton system. The hydroxyl radical bleached the safranin, so decreased the absorbance of the reaction mixture indicating a decrease in hydroxyl radical scavenging ability. The scavenging ratio for hydroxyl radical was calculated from the following Eq. 3:
Where A sample = the absorbance in the presence of the tested compound; A 0 = the absorbance in the absence of the tested compound; A control = the absorbance in the absence of the tested compound, EDTA-Fe(II) and H2O2.
Figure 8a depicts the inhibitory effect of the complexes on hydroxyl radial. The inhibitory effect of the complexes is marked and suppression ratio increases with increasing concentration in the range of tested concentration. Their orders of scavenging activities on hydroxyl radicals were: 4 > 3> 1 > 2. Moreover, mannitol is a well-known natural antioxidant, so we also studied the scavenging activity of mannitol against hydroxyl radical using the same model. Compared with Fig. 8a, b when arrived at similar suppression ratio, concentration of the four complexes is far less than that of mannitol. The results showed that the complexes have significant antioxidative activity against hydroxyl free radicals from the Fenton reaction.
a Scavenging effect of metal complexes and ligand on hydroxyl radicals. Values are expressed as mean standard deviation (n = 3). Eu(III)complex (1) ◆; (A) Sm(III) complex (2) ▲; Gd(III) complex (3) ■; Tb(III) complex(4) ●. b Scavenging effect of mannitol (▲) on hydroxyl radicals. Experiments were performed in triplicate. Values are expressed as mean standard deviation (n = 3)
Conclusion
In summary, a novel ligand, 6-ethoxy chromone-3-carbaldehyde benzoyl hydrazone, and its Ln(III) complexes have been prepared and characterized. Comparing the luminescence spectra of the Eu(III) and Tb(III) complexes, we found that the ligand L in Eu(III) complex is more effective in energy-transfer than in Tb(III) complex that does not emit its characteristic luminescence. Furthermore, the DNA-binding properties of the Eu(III) and Sm(III) complexes were investigated by spectra titration and viscosity measurements. The two complexes all appear to intercalate into DNA. The conclusion is based upon the effects of hypochromism, the increases in emission intensities and the increased degree of viscosity. This work also find that substituting group in chromone Schiff bases structure can affect the DNA binding affinity. Furthermore, the rare earth complexes have active scavenging effect on hydroxyl radical. Based on those results, one may design and readily prepare a different repertoire of shapes for interaction with DNA. The ligand may be varied in a synthetically convenient fashion to produce a family of substitutionally inert DNA binding molecules with a range of ligand functionalities. These and similar complexes may therefore be useful in many potential practical applications, like fluorescence mark, fluorescence analysis , nucleic acid molecular probes and so on .
Abbreviations
- CT-DNA:
-
calf thymus DNA
- DMF:
-
N, N-dimethylformamide
- DMSO:
-
Dimethylsulphoxide
- L:
-
6-ethoxy chromone-3-carbaldehyde-(4′-hydroxy) benzoyl hydrazone
- Tris:
-
Tris (hydroxymethyl)-aminomethane
- NMR:
-
Nuclear magnetic resonance
- UV-Vis:
-
Ultraviolet and visible
References
Erkkila KE, Odom DT, Barton JK (1999) Recognition and reaction of metallointercalators with DNA. Chem Rev 99:2777–2795
Trawick BN, Daniher AT, Bashkin JK (1998) Inorganic mimics of ribonucleases and ribozymes: from random cleavage to sequence-specific chemistry to catalytic antisense drugs. Chem Rev 98:939–960
Sigman DS, Mazumder A, Perrin DM (1993) Chemical nucleases. Chem Rev 93:2295–2316
Dandliker PJ, Holmlin RE, Barton JK (1997) Oxidative thymine dimer repair in the DNA helix. Science 275:1465–1468
Hall DB, Holmlin RE, Barton JK (1996) Oxidative DNA damage through long-range electron transfer. Nature 382:731–735
Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK (1990) A molecular light switch for DNA: Ru(bpy)2(dppz)2+. J Am Chem Soc 112:4960–4962
Lincoln P, Norden B (1996) Binuclear ruthenium(II) phenanthroline compounds with extreme binding affinity for DNA. Chem Commun: 2145–2146
Hartshorn RM, Barton JK (1992) Novel dipyridophenazine complexes of ruthenium (II): exploring luminescent reporters of DNA. J Am Chem Soc 114:5919–5925
Sitlani A, Long EC, Pyle AM, Barton JK (1992) DNA photocleavage by phenanthrenequinone diimine complexes of rhodium(III): shape-selective recognition and reaction. J Am Chem Soc 114:2303–2312
Ji LN, Zou XH, Liu JG (2001) Shape- and enantioselective interaction of Ru (II)/Co(III) polypyridyl complexes with DNA. Coord Chem Rev 216–217:513–536
Cory M, McKee DD, Kagan J, Henry DW, Miller JA (1985) Design, synthesis, and DNA binding properties of bifunctional intercalators. Comparison of polymethylene and diphenyl ether chains connecting phenanthridine. JACS 107:2528–2536
Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK (1989) Mixed-ligand complexes of ruthenium(II): factors governing binding to DNA. J Am Chem Soc 111:3051–3058
Mahadvan S, Palaniandavar M (1997) Spectroscopic and voltammetric studies of copper(II) complexes of bis(pyrid-2-yl)-di/trithia ligands bound to calf thymus DNA. Inorg Chim Acta 254:291–302
Xu H, Zheng KC, Deng H, Lin LJ, Zhang QL, Ji LN (2003) Effects of the ancillary ligands of polypyridyl ruthenium(II) complexes on the DNA-binding behaviors. New J Chem 27:1255–1263
Asadi M, Safaei E, Ranjbar B, Hasani L (2004) Thermodynamic and spectroscopic study on the binding of cationic Zn(II) and Co(II) tetrapyridinoporphyrazines to calf thymus DNA: the role of the central metal in binding parameters. New J Chem 28:1227–1234
Chaires JB (1998) Energetics of drug-DNA interactions. Biopolymers 44:201–215
Yang ZY, Yang RD, Li FS, Yu KB (2000) Crystal structure and antitumor activity of some rare earth metal complexes with Schiff base. Polyhedron 19:2599–2604
Cha A, Snyder GE, Selvin PR, Bezanilla F (1999) Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402:809–813
Wang ZM, Lin HK, Zhu SR, Liu TF, Zhou ZF, Chen YT (2000) Synthesis, characterization and cytotoxicity of lanthanum(III) complexes with novel 1, 10-phenanthroline-2, 9-bis-alpha- amino acid conjugates. Anticancer Drug Des 15:405–411
Xu HX, Zhang HY, Qu XG (2006) Interactions of the human telomeric DNA with terbium-amino acid complexes. J Inorg Biochem 100:1646–1652
Wang K, Li R, Cheng Y, Zhu B (1999) Lanthanides—the future drugs? Coord Chem Rev 190–192:297–308
Yang ZY, Wang B-D, Li YH (2006) Study on DNA-binding properties and cytotoxicity in L1210 of La(III) complex with PMBP-isonicotinoyl hydrazone. J Organomet Chem 691:4159–4166
Barton D, Ollis WD (1979) In comprehensive organic chemistry. Pergamon: Oxford 4:7
Wiley PF (1952) Chromones in the Mannich reaction. J Am Chem Soc 74:4326–4328
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro.ovrddot.organisms. J Mol Biol 3:208–218
Wang BD, Yang ZY, Crewdson P, Wang DQ (2007) Synthesis, crystal structure and DNA-binding studies of the Ln(III) complex with 6-hydroxy-chromone-3- carbaldehyde benzoyl hydrazone. J Inorg Biochem 101:1492–1504
Wang BD, Yang ZY, Li TR (2006) Synthesis, characterization, and DNA-binding properties of the Ln(III) complexes with 6-hydroxychromone-3-carbaldehyde 2-hydroxybenzoylhydra zone. Bioorg Med Chem 14:6012–6021
Wang BD, Yang ZY, Qin DD, Chen ZN (2008) Synthesis, characterization, cytotoxic activity and DNA-binding properties of the Ln(III) complexes with ethylenediiminobi(6-hydroxy chromone-3-carbaldehyde) Schiff-base. J Photochem Photobiol, A 194:49–58
Wang BD, Yang ZY (2008) Synthesis, characterization, DNA-binding properties of the Ln(III) complexes with 6-hydroxy chromone-3-carbaldehyde-(4′-hydroxy) benzoyl hydrazone. J Fluoresc 18:547–553
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro.ovrddot.organisms. J Mol Biol 3:208–218
Kumar CV, Asuncion EH (1993) DNA binding studies and site selective fluorescence sensitization of an anthryl probe. J Am Chem Soc 115:8547–8553
Yu JX, Liu FM, Lu WJ, Li YP, Zao XM, Liu YT, Liu C (2000) Synthesis of 3-(1′, 3′, 4′-oxadiazoline-2′-yl) chromones. Youji Huaxue 20:72–80 (in Chinese)
Boyer JL, Krum JE, Myers MC, Fazal AN, Wigal CT (2000) Synthetic utility and mechanistic implications of the fries rearrangement of hydroquinone diesters in boron trifluoride complexes. J Org Chem 65:4712–4714
Winterbourn CC (1981) Hydroxyl radical production in body fluids. Roles of metal ions, ascorbate and superoxide. Biochem J 198:125–131
Geary WJ (1971) Use of conductivity measurements in organic solvents for the characterization of coordination compounds. Coord Chem Rev 7:81–122
Lewis FD, Barancyk SV (1989) Lewis acid catalysis of photochemical reactions. 8. Photodimerization and cross-cycloaddition of coumarin. J Am Chem Soc 111:8653–8661
Raman N, Kulandaisamy A, Jeyasubramanian K (2002) Synthesis, spectral, redox and biological studies of some Schiff base copper(II), nickel(II), cobalt(II), manganese(II), zinc(II) and oxovanadium(IV) complexes derived from 1-phenyl- 2, 3-dimethyl-4-(4-iminopentan-2-one)-pyrazol-5-one and 2-aminophenol/2- aminothiophenol. Indian J Chem, Sect A: Inorg, Bio-inorg, Phys, Theor Anal Chem 41A:942–949
Hirashima Y, Kanetsuki K, Yonezu I, Kamakura K, Shiokawa J (1983) Lanthanide nitrate complexes with some polyethylene glycols and glymes. Bull Chem Soc Jpn 42:738–743
Carnall WT, Siegel S, Ferrano JR, Tani B, Gebert E (1973) A new series of anhydrous double nitrate salts of the lanthanides structural and spectral characterization. Inorg Chem 12:560–564
Nakamoto K (1978) Infrared and Raman spectra of inorganic and coordination compounds, 3rd edn. Wiley, New York
Piguet C, Hopfgartner G, Williams AF, Buenzli J-CG (1995) Self-assembly of the first heterodinuclear d-f triple helix in solution. J Chem Soc, Chem Commun: 491-493
Aime S, Fasano M, Terreno E (1998) Lanthanide (III) chelates for NMR biomedical applications. Chemical Society Reviews 27:19–29
Dagdigiam JV, Mckee V, Reed CH (1982) Structural comparison of a redox pair of copper (I/II) complexes having benzimidazole thioether ligands. Inorg Chem 21:1332–1342
Qiang S (1993) Chemistry of rare earths. Henan Technology & Science, Zhengzhou, pp 304–314 in Chinese
Liu HQ, Cheung TC, Che CM (1996) Cyclometallated platinum (II) complexes as luminescent switches for calf-thymus DNA. Chem Commun 9:1039–1040
Wang BD, Yang ZY, Zhang DW, Wang Y (2006) Synthesis, structure, infrared and fluorescence spectra of new rare earth complexes with 6-hydroxy-chromone-3-carbaldehyde benzoyl hydrazone. Spectrochim Acta, Part A: Mol Biomol Spectrosc 63:213–219
Zhou D, Li Q, Huang C, Yao G, Umetani S, Matsui M, Ying L, Yu A, Zhao X (1997) Room-temperature fluorescence, phosphorescence and crystal structure of 4-acyl pyrazolone lanthanide complexes: Ln (L) 3.2H2O. Polyhedron 16:1381–1389
Bloomfield VA, Crothers DM, Tinoco IJr (1974) Physical chemistry of nucleic acids. Harper and Row, New York, p 432
Satyanarayana S, Dabrowiak JC, Chaires JB (1992) Neither D- nor L-tris (phenanthroline) ruthenium (II) binds to DNA by classical intercalation. Biochemistry 31:9319–9324
Kumar CV, Barton JK, Turro NJ (1985) Photophysics of ruthenium complexes bound to double helical DNA. J Am Chem Soc 107:5518–5523
Satyanarayana S, Dabrowiak JC, Chaires JB (1992) Neither D- nor L-tris (phenanthroline) ruthenium (II) binds to DNA by classical intercalation. Biochemistry 31:9319–9324
Satyanarayana S, Dabrowiak JC, Chaires JB (1993) Tris (phenanthroline) ruthenium(II) enantiomer interactions with DNA: Mode and specificity of binding. Biochemistry 32:2573–2584
Eriksson M, Leijon M, Hiort C, Norden B, Graeslund A (1994) Binding of D- and L-[Ru(phen)3]2+ to [d(CGCGATCGCG)]2 Studied by NMR. Biochemistry 33:5031–5040
Halliwell B, Gutteridge JMC, Cross CE (1992) Free radicals, antioxidants, and human disease: where are we now? J Lab Clin Med 119:598–620
Udilova N, Kozlov AV, Bieberschulte W, Frei K, Ehrenberger K, Nohl H (2003) The antioxidant activity of caroverine. Biochem Pharmacol 65:59–65
Acknowledgements
This work is supported by the National Natural Science Foundation of China (20475023) and Gansu Natural Science Foundation (0710RJZA012).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, J., Yang, ZY., Wang, BD. et al. Synthesis, Characterization and DNA-binding Properties of Ln(III) Complexes with 6-Ethoxy Chromone-3-Carbaldehyde Benzoyl Hydrazone. J Fluoresc 19, 847–856 (2009). https://doi.org/10.1007/s10895-009-0482-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-009-0482-y