Abstract
Tannins are characterized by protein-binding affinity. They have astringent/bitter properties that act as deterrents, affecting diet selection. Two groups of salivary proteins, proline-rich proteins and histatins, are effective precipitators of tannin, decreasing levels of available tannins. The possibility of other salivary proteins having a co-adjuvant role on host defense mechanisms against tannins is unknown. In this work, we characterized and compared the protein profile of mice whole saliva from animals fed on three experimental diets: tannin-free diet, diet with the incorporation of 5% hydrolyzable tannins (tannic acid), or diet with 5% condensed tannins (quebracho). Protein analysis was performed by one-dimensional gel electrophoresis combined with Matrix-Assisted Laser Desorption Ionization-Time of Flight mass spectrometry to allow the dynamic study of interactions between diet and saliva. Since abundant salivary proteins obscure the purification and identification of medium and low expressed salivary proteins, we used centrifugation to obtain saliva samples free from proteins that precipitate after tannin binding. Data from Peptide Mass Fingerprinting allowed us to identify ten different proteins, some of them showing more than one isoform. Tannin-enriched diets were observed to change the salivary protein profile. One isoform of α-amylase was overexpressed with both types of tannins. Aldehyde reductase was only identified in saliva of the quebracho group. Additionally, a hypertrophy of parotid salivary gland acini was observed by histology, along with a decrease in body mass in the first 4 days of the experimental period.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tannins are plant secondary metabolites (PSMs) with an enormous structural diversity and are found worldwide in many families of higher plants. One of the characteristic properties of these natural high-molecular-mass polyphenols is their high capacity to bind proteins, forming complexes that tend to precipitate (Haslam 1998). In the mouth, the precipitates are perceived via mechanoreceptors as rough, puckering, or drying oral sensations characteristic of astringency (Breslin et al. 1993; Prinz and Lucas 2000). The astringent properties, together with the frequently associated bitter taste (Lesschaeve and Noble 2005), may result in the avoidance of some plants or plant parts by herbivores. Taste perception is generally considered to be an adaptive response in assessing nutritional value and/or coping with toxicity in potential foods (Le Magnen 1986). Animals with a relatively high occurrence of bitter and potentially toxic compounds in their diet (e.g., browsing herbivores) are believed to have developed a high bitter-taste threshold and tolerance (Glendinning 1994). The cause for this tolerance can be rooted in physiological postingestive adaptative mechanisms, namely, modifications of saliva composition or flow rate, which can contribute to mitigate the negative biological effects of PSMs.
Saliva contains a complex mixture of proteins with different biological roles in digestion, host defense, and lubrication (Humphrey and Williamson 2001). Its composition is diverse among animal species and changes with circadian rhythm (Hardt et al. 2005a) and diet (Katsukawa and Ninomiya 1999; Neyraud et al. 2006), among other factors. As the secretion of salivary fluid and proteins is controlled by autonomic nerves (Proctor and Carpenter 2007), saliva plasticity represents a rapid mechanism that allows animals to adapt to random dietary changes, thus conferring an important advantage. Rats and mice that consumed tannin-rich diets showed both parotid gland hypertrophy and increased synthesis and accumulation of salivary proline-rich proteins (PRPs; Mehansho et al. 1983, 1985, 1987; Jansman et al. 1994). These molecular and cellular effects produced by high-tannin diets, both on the salivary gland tissue and on the saliva of mice and rats, are indistinguishable from those produced by recurrent stimulation with the beta-adrenergic agonist isoproterenol (Ann et al. 1987, 1997). Recently, Gho et al. (2007) observed the induction of a group of salivary polypeptides (designated by isoproterenol-induced polypeptides) both by tannins and isoproterenol. Mice salivary PRP induction was suggested as acting as a countermeasure against tannins through the formation of tannin–protein complexes that remain insoluble in the conditions found in the digestive compartments (Hagerman et al. 1998; Lu and Bennick 1998). Apart from these protein species, little is known about possible changes in other salivary proteins that are induced by tannins, namely proteins that do not form insoluble complexes with these PSMs. Besides PRPs, isoproterenol also induces changes in the expression levels of salivary proteins, such as cystatins (Shaw and Yu 2000) and amylase (Gallacher and Petersen 1983), among others. Therefore, the possibility of changes in relative amounts of other proteins cannot be excluded. The characterization of such modifications may be useful for better understanding mammalian physiological countermeasures. Rodents have been traditionally used as an animal model for studies of the anatomy of the salivary glands and physiology, and several proteins have been reported as constituents of their saliva: PRPs (Lin and Ann 1991), namely the parotid salivary protein, (Madsen and Hjorth 1985) and amylase (Hagenbüchle et al. 1980). However, to our knowledge, a systematic characterization of rodent saliva proteome has not been reported. The most complete rodent salivary protein profile reported to date is a two-dimensional map of rat parotid saliva, in which the identification of detected proteins was inferred from determined molecular masses in the gel (Williams et al. 1999).
Peptide mass fingerprinting (PMF) that uses matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectra is a current strategy for the identification of proteins expressed in certain physiological conditions, allowing comparisons of different treatments. Here, our first objective was to identify salivary proteins from mice whole saliva by using MALDI-TOF mass spectrometry after separation by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently to evaluate if ingestion of tannins induced changes in the expression of these proteins. We tested the influence of hydrolyzable (tannic acid) and condensed (quebracho) tannins on saliva samples from which insoluble tannin–protein complexes had been removed before the analysis, to better assess proteins expressed in lower concentrations. Light microscopy was also used to study morphological changes in parotid glands.
Methods and Materials
Animals
Thirty-one inbred male Balb/c mice, 4-week-old, were obtained from the licensed bioterium of Instituto Gulbenkian de Ciência (Oeiras, Portugal). The animals were housed in mice cages, type IV (Techniplast; (10 to 11 mice per cage), according to European Union (EU) recommendations and revision of Appendix A of European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (ETS No. 123) and maintained on a 12:12 h, light/dark cycle at a constant temperature of 22°C with ad libitum access to water and to a standard diet with 21.86% crude protein (dry basis) in the form of pellets (RM3A-P; Dietex International, UK). Animals were individually marked and submitted to a 7-day acclimation period to minimize stress associated with transportation. This period was followed by a 7-day pretrial period to allow adaptation to the ground diet used during the feeding trials. The standard pellet diet was ground daily with a blender to obtain a meal with visibly homogeneous fine-sized particles. Before the feeding-trial period, animals were individually weighed and allocated to three experimental groups, each group having no significant differences in body mass (24.5 ± 3 g).
All procedures involving the animals were approved by the scientific committee, supervised by a Federation of European Laboratory Animal Science Associations (FELASA)-trained scientist and conforming to the regulations of the Portuguese law (Portaria 1005/92), following European Union Laboratory Animal Experimentation Regulations.
Feeding Trials
Immediately after the pretrial period, we started the 10-day experimental period, the first day of which we consider day 1. The control group (n = 10) received a tannin-free diet, the same standard ground diet as in the pretrial period. The tannic acid group (TA; n = 10) and the quebracho group (Q; n = 11) received the standard ground diet plus tannic acid (Merck Ref 1.00773.100; hydrolyzable tannin) or quebracho extract (Tupafin-Ato, SilvaChimica SRL; 72% ± 1.5 of condensed tannins), respectively, added to obtain a mixture with 5 g tannin/100 g wet weight of the standard diet. Food and water were provided ad libitum, and the diets were prepared daily with a blender, as described for the pretrial period. Body mass changes during the first 8 days were determined daily.
Saliva and Salivary Gland Collection and Sample Preparation
On day 11, individual mice whole saliva was collected. The mice remained in the cages, with food and water available, until the time of collection. Saliva production was induced with an intraperitoneal injection of pilocarpine (Sigma; 5 mg kg−1 body weight), dissolved in 1 ml 9% (m/v) sodium chloride, and prepared immediately before use, as described by Muenzer et al. (1979). Saliva was individually collected by aspiration from all the mice, directly from their mouths with a micropipette. Saliva samples were frozen immediately in liquid nitrogen and stored at −80°C until required. Before protein quantification, samples were centrifuged at 16,000×g for 5 min at 4°C to remove particulate matter and salivary proteins that could be precipitated. Only the soluble fraction was used for further analyses. All animals were injected intraperitoneally with anesthetic (xylazine hydrochloride combined with ketamine hydrochloride) and euthanized with an overdose. Both parotid salivary glands were dissected, washed briefly with phosphate buffer 0.1 M, pH 7.4, and fixed in 10% neutral buffered formalin, to carry out further routine histological procedures.
Protein Quantification and Gel Electrophoresis
Total protein concentration was determined by the bicinchoninic acid (BCA) protein assay method (Pierce) by using a microplate reader (SpectroMAX 340, Molecular Devices, Union City, CA, USA). One-dimension SDS-PAGE was run with 20 μg of saliva total protein after reduction with dithiothreitol (USB) and alkylation with iodoacetamide (Sigma). Denatured protein samples were loaded on bis-tris polyacrilamide 4–12% gradient precast gels (100 × 100 × 1 mm; Nupage Invitrogen) with MES SDS (Nupage Invitrogen) used as a running buffer. Molecular mass markers (BioRad, Ref 161-0317) were run simultaneously with the samples in each gel to calibrate molecular masses of the protein sample bands. Protein bands were stained with colloidal Coomassie blue (Neuhoff et al. 1988) and with Comassie blue R-250 following Beeley et al (1991). Digital images of the gels were acquired with a densitometer with internal calibration (Molecular Dynamics, Amersham Biosciences Europe GmbH, Freiburg, Germany), and gels were subjected to linescan analysis by using Imagequant Software 5.0 (Amersham Biosciences Europe GmbH, Freiburg, Germany). Sensitivity 9.0 and kernel 4.0 were the software parameters used to assign the detected significant bands in the protein profiles obtained. Only bands present in at least 50% of individuals were considered.
Protein Identification
Protein bands were excised from all gels and the polypeptides subjected to digestion with a sequence grade modified trypsin (Promega) according to Pandey et al. (2000). Sample peptides were assayed for peptide mass fingerprinting in a Voyager-DE STR (Applied Biosystems, Foster City, CA, USA) MALDI-TOF mass spectrometer. Peptide co-crystallization was achieved by applying 0.5 μl of the peptide digest on the MALDI plate and adding an equal volume of recrystallized matrix α-cyano-4-hydroxycinnamic acid 10 mg/ml prepared in acetonitrile 70% (v/v) with 0,1% trifluoroacetic acid (v/v). The mixture was allowed to air dry (dried droplet method). Average spectra were obtained in the mass range between 800 and 4,000 Da, using three spectra acquired with 500 laser shots in the positive-ion reflectron mode. Spectra were processed and analyzed by the Data Explorer (version 4.0, Applied Biosystems, Foster City, CA, USA). Monoisotopic peptide masses were used to search for protein identification with Mascot software (Matrix Science, UK; Perkins et al. 1999). Searches were performed in the National Center for Biotechnology Information nonredundant protein-sequence database. A minimum mass accuracy of 100 ppm, one missed cleavage in peptide masses, and carbamidomethylation of Cys and oxidation of Met as fixed and variable amino acid modifications, respectively, were considered. Criteria used to accept the identification were significant homology scores achieved in Mascot, significant sequence-coverage values, and similarity between the protein molecular mass calculated from the gel and for the identified protein (Roxo-Rosa et al. 2006).
Histology
To confirm that the doses of tannins used were enough to induce significant changes in glandular morphology, salivary glands were observed through light microscopy with a Nikon Eclipse 600 microscope (Kanagawa, Japan). After embedding the fixed parotid glands in paraffin wax, using routine procedures, a series of sections of 5 mm thick were cut with a microtome, and the slides were stained with hematoxylin and eosin. Three animals from each group were used, and for each animal, ten pictures from random parotid areas were collected with a Nikon DN100 camera (Kanagawa, Japan) at ×200 magnification. For each animal, the area and perimeter of 150 parotid acini were randomly chosen and measured by using SigmaScan Pro5.0 software (SPSS, Chicago, IL, USA).
Statistical Analysis
Body mass data were tested for normality and homocedasticity by Kolmogorov–Smirnov and Levene tests, respectively. Body mass was analyzed according to a general linear model procedure with two fixed factors and one nested factor:
where Y ijkl are the observed values for body mass, μ is the observed mean value, T i is the fixed effect of treatment, W k is the fixed effect of weighing day, T(A) i (j) is the nested effect, and ε ijkl is the random error or residual effect.
Means significantly different were submitted to post hoc comparisons of means (Tukey–Hsu test) and regarded as significantly different when P < 0.05.
All statistical analysis procedures were performed by NCSS 2001 software package (Kaysville, UT, USA).
Results
Body Mass
Animals were weighed daily to search for changes in body masses that reflect physiological adaptation of the animals to tannin diets, including the induction of PRPs (Mehansho et al. 1983; Skopec et al. 2004). We observed that tannic acid and quebracho groups lost weight during the first 3–4 days, respectively, after which they recovered, whereas the control group increased in body mass in the first 2 days and remained nearly stable after that. Statistically significant differences were observed on days 3 and 4, with the animals from the tannin groups presenting lower mean body masses than animals from control group (Table 1). Despite the increases in body masses after day 4, for animals from either tannic acid or quebracho groups, that increase was less pronounced in the tannic acid group, with animals remaining with lower body masses until the end of the trial.
Histology
A dramatic increase in the acinar area and the perimeter of the parotid salivary gland was observed in the animals that received tannin-enriched diets over a period of 10 days (Fig. 1). Levels of 5 g of hydrolyzable or condensed tannin per 100 g wet weight in the diet seem to produce a hypertrophy of the secretor tissue (Table 2).
Saliva Protein Concentration
Total protein saliva concentration, after the 10 days of the feeding trial, was significantly higher in the control group than in the quebracho and in tannic acid groups (Table 3).
Saliva Protein Profile
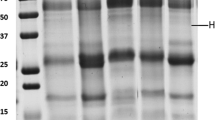
We characterized the saliva profile of the control group by using the Imagequant 5.0 software to assign significative bands in the profiles obtained. These results are in agreement with visual observation of the gel pattern. Despite slight interindividual variations in whole saliva composition, 21 protein bands (from a to u) were present consistently in all animal saliva protein profiles (n = 10). Fig. 2a shows a typical 1-DE pattern of the control group saliva and also an overlay of the ten linescans resulting from the analysis of the control group saliva protein profiles. The molecular masses of the proteins were calculated by gel analysis software, after superimposing the linescan of the molecular mass markers on each sample linescan.
With regard to the control group, interindividual protein profile variability was assessed for the TA (n = 10) and Q (n = 11) groups. Only slight variation was found among the animals on the tannin-fed diets. By comparing the saliva protein profile of the control group with the TA group, we identified one additional band (band β, Fig. 2b) around molecular mass 51 kDa that was not visible in the control saliva protein profile. The saliva protein profile of the Q group, when compared with the control saliva protein profile, presented two additional bands (Fig. 2c). There was an intense protein band presented in the saliva protein profile of the TA group around 51 kDa (band β), and an additional band (band γ) of reduced intensity and molecular mass around 20 kDa
For each dietary group, a typical one-dimensional (1-D) gel pattern and a linescan analysis of salivary proteins are represented on the left hand side and on the right side, respectively. Molecular markers are represented on the right side of the 1-D gel and on the top on the linescan. The letters a-u, β and γ represent the protein bands and the individual peaks assigned by the software. The graphics represent an overlay of a ten gel linescan analysis of the control group; b a linescan analysis of the control group and of the TA group; c a linescan analysis of the control group and of the Q group
.
Protein Identification
Identification of all protein bands excised from the control group can be seen in Table 4. The only band whose identification was not possible was band p, probably due to the presence of a low amount of protein. From the 21 protein bands excised, we identified ten different polypeptides. The other ten bands were assigned to four of the already identified polypeptides: four additional bands to salivary amylase 1, one to carbonic anhydrase VI, four to parotid secretory protein (PSP), and one to androgen-binding protein. This was attributed to the existence of post-translational modifications and/or protein fragments. Band u was assigned to a mixture of β and γ subunits of the androgen-binding protein.
The two protein bands that were detected additionally in the saliva profile of the animals submitted to a tannin-enriched diet (β and γ) were also used for protein identification following the PMF approach. As seen in Table 4, these two bands were identified as amylase 1 and aldehyde reductase, respectively. Aldehyde reductase was observed only in the Q group.
Discussion
Tannins act as feeding deterrents, due both to their aversive chemosensory properties (such as bitter taste and astringency) and the negative postingestive effects they exert (Glendinning 1994). A reduction in body mass in the first days of tannin administration has been reported previously (Mehansho et al. 1983, 1985; Jansman et al. 1994; Shimada et al. 2004, 2006). In mice and rats, this effect is reversed after 2–3 days, when a hypertrophy of parotid glands occurs, coincident with a dramatic increase in salivary PRPs production (Mehansho et al. 1983). In the present work, tannin-fed mice decreased in body mass in the first days (Table 1). This effect ceased after 3–4 days. Despite the body mass recovery of tannin-fed mice, animals from the tannic acid group remained with lower body masses until the end of the trial, which suggests a greater capacity to adapt to quebracho tannin. The parotid hypertrophy usually associated with tannin consumption was also observed in this experiment. This effect in parotid glands is associated with the expression of a number of isoproterenol-induced salivary proline-rich polypeptides (Lopez-Solis and Kemmerling 2005; Gho et al. 2007), which seems to be related to β1 adrenergic activity (Gonzalez et al 2000). We thus found those changes relevant, demonstrating that the tannin levels used were enough to induce an effect at glandular level.
This study helps explain changes in whole saliva protein composition that occur when mice consume tannin-enriched diets. Most studies suggest a defense response associated with the induction of salivary proteins (mainly PRPs) that precipitate PSMs, preventing them from exerting negative effects (Bennick 2002). We predicted that tannin consumption would also influence other salivary proteins. To ensure that the minor expressed-salivary proteins would not be masked by overexpression of the tannin-binding proteins, we removed the insoluble fraction that is expected to contain the tannin–protein complexes. We assumed that this was achieved since total protein concentration from the tannin-fed groups was lower compared with the control group. The lack of observation of pink bands in the Coomassie stained gels (according to Beeley et al. 1991) supports the idea that we efficiently removed the majority of PRPs.
Although a great number of salivary protein studies have been performed on mice, a global perspective that relates whole saliva protein profile to protein identification is presently unknown. Williams et al. (1999) provide a 2-DE protein profile, referring to the presence of PRPs, amylase, acidic epididymal glycoprotein, deoxyribonuclease, parotid secretory protein, and common salivary protein, although a protein identification was not performed but rather inferred from the position of protein spots. Our results showed the presence of ten different salivary proteins, some showing more than one isoform. This situation is documented in human saliva (Hirtz et al. 2005b), where a simultaneous low diversity in terms of variety of accessions and a high complexity in terms of number of protein bands identified in the same accession are reported. This degree of redundancy displayed by extensive salivary protein polymorphisms seems to be important in saliva plasticity. Moreover, the presence of several families of structurally and functionally closely related molecules indicates that these proteins have been subjected to evolutionary pressures that may reflect selection for improved function (Oppenheim at al. 2007).
The identified proteins have been described previously in saliva: acidic chitinase, mucin apoprotein, amylase 1, carbonic anhydrase VI, apolipoprotein A-I, parotid secretory protein, immunoglobulin heavy chain, submandibular gland 15 kDa protein, androgen-binding protein, and glandular kallikrein (Table 4). The first seven have multiple functions in saliva and are involved in the formation of enamel pellicle, have hydrolytic activity, and act as a first line of defense against microorganisms (Yao et al. 2003; Huang 2004; Vitorino et al. 2004; Hardt et al. 2005b). The submandibular gland 15 kDa protein is known as a gross cystic disease fluid protein that was initially described in humans as a protein secreted by the T47D human breast cancer cell line. This protein is produced in many exocrine glands such as sweat, salivary and lachrymal; however, its function remains unknown (Myal et al. 1998). Androgen-binding protein secreted by mice submandibular salivary glands has been hypothesized as having a major role in mate selection. This member of the secretoglobins family is secreted into saliva in two different dimer forms: an α subunit disulfide bridged to either a β or a γ subunit (Karn and Laukaitis 2003). However, cellular site(s) of synthesis, mode of function, and evolution patterns of this biologically important protein are otherwise unknown (Dlouhy et al. 1987). Glandular kallikrein, mainly secreted by submandibular glands, is a peptidase that belongs to the serine proteases family, which brings about maturation of growth factors and polypeptide hormones, by conversion of inactive precursors to biologically active peptides (Kim et al. 1991). Parasympatic stimulation that causes an increase in saliva flow will promote low outputs of this protein without degranulation of acinar or granular tubules cells (Shori and Asking 2001).
The control group electrophoresis gel pattern presents five different bands identified as salivary amylase 1, between masses 48 and 66 kDa—one intense band at a lower molecular mass (band g), and four weak or medium intense bands at higher molecular mass values. Different bands may correspond to different isoforms. Glycosylation and deamidation of amylase 1 have been described (Bank et al. 1991; Hirtz et al. 2005a). Glycosylation could be an explanation for the higher molecular mass of the amylase bands observed in the gel, compared to the amylase native form. Knowing that glycosylated asparagine residues almost always occur in the sequence Asn-X Ser/Thr, two potential N-glycosylation sites for mice α-amylase are 412–414 and 461–463 (peptide residue numbered taking into account that the 15 amino acid peptide signal is lost during the secretion process). These two sites were also referred for human α-amylase (Bank et al. 1991; Hirtz et al. 2005a). The peptide that contains the first site is detected for all our amylase bands, so a glycosylation should not occur in that position. On the other hand, N-glycosylation can occur in the second potential site, since the peptide that contains it systematically fails to be detected in mass spectra. Although some (Bank et al., 1991) have considered that N-glycosylation is more prone to occur in the first potential site, Hirtz et al. (2005a) found results similar to ours for human α-amylase. The lack of the specific detection of glycoproteins does not allow the confirmation of this hypothesis since the absence of peptide 458–466 can also be explained based on experimental limitations. Bank et al. (1991) proposed that amylase-deaminated isoforms correspond to the lower molecular-mass bands observed in the saliva SDS-PAGE profile. By using the software Findmod (http://www.expasy.ch/tools/findmod/), it is possible to assign two peaks in the salivary amylase mass spectra of band g as deaminated peptides, both in the control group and in the Q and AT groups. These post-translational modifications can occur at one of two residues of peptide 46–76 and at one of three residues of peptide 414–436. These modified peptides were also observed for band β, present only in the animal groups that were submitted to the tannin-enriched diet. The deamination of these residues does not seem to be characteristic of any of the amylase isoforms observed.
Although salivary proteins can bind tannins as a mode of defensive action, other physiological mechanisms, including enzymatic or immune responses, can not be excluded. The only effect common to both types of tannins in the salivary profile of proteins was an increase in amylase. The overexpression of amylase 1 induced in both groups (band β, Fig. 2) overlaps the medium molecular mass and less intense band of this protein in the control group (band f, Fig. 2). This suggests that only the medium molecular mass isoform of amylase 1 is overexpressed in the tannin-rich diet fed groups or that a new amylase isoform is induced in these groups. Furthermore, protein band g (Fig. 2), the most intense band in the control group, identified as amylase 1, did not show any significant expression level change with tannin treatment. Several differences can be observed between the tryptic peptide mass spectra of band β and of band g. We could not assign these peaks to described modifications of salivary amylase 1, but it suggests that a different isoenzyme of salivary amylase 1 is being expressed in the Q and AT groups.
The increase in salivary α-amylase levels may be a consequence of tannin stimulation of sympathetic pathways. Isoproterenol and tannins lead to similar changes in salivary glands, and it has been suggested that these changes are due to stimulation of the β1 adrenergic receptors (Waters et al. 1998). Beta adrenergic agonists are capable of stimulating salivary α-amylase release (Gallacher and Petersen 1983). There is a recurrent pattern across studies that shows that salivary α-amylase levels increase in response to physical and psychological stress (Chatterton et al. 1996; for a comprehensive review see Granger et al. 2007). As a result, salivary α-amylase has been used as a surrogate marker of the autonomic/sympathetic nervous system component of stress in humans. Other oral stimulators that influence texture and flavor perception have also been observed to increase α-amylase expression. Amylase increased with taste stimulation in rabbits (Gjorstrup, 1980) and humans (Neyraud et al. 2006), and a strong correlation between α-amylase and texture perception was observed by Engelen et al. (2007).
We suggest that, despite the primary biological function of salivary α-amylase being the digestion of polysaccharides, the increase of its expression is not a direct consequence of dietary carbohydrates but a result of adrenergic stimulation. Indirectly, this increase can also represent a co-adjuvant for the inhibition of tannin biological activity. Tannins are potent inhibitors of salivary amylase 1 (Kandra et al. 2004; McDougall et al. 2005), although affinity of α-amylase for tannins is not as high as PRPs affinity (De Freitas and Mateus 2001). Tannins have been described as responsible for an enlarged pancreas (Ahmed et al. 1991; Mahmood and Smithard. 1993), stimulation and secretion of an increased amount of amylase, which, to some extent, may counteract the inhibition of this enzyme during gut digestion. Although α-amylase in saliva and the pancreas are produced by independent sources, we believe that a similar function could be involved, and amylase content may increase when animals are fed with tannins, thus counteracting the inhibition of this enzyme in the mouth.
Salivary amylase is stored in large dense-core secretory granules that undergo stimulated secretion in response to extracellular stimulation. (Gorr et al. 2005). The α-amylase increase observed in the present study could result from the exocytosis of acinar secretory mature granules, as usually occurs in the first hours after isoproterenol administration (Henriksson 1982). Moreover, chronic stimulation with isoproterenol is reported to decrease (Madsen and Hjorth 1985; Ann et al. 1987) or maintain (Bedi 1993) amylase levels. To the best of our knowledge, this is the first reference of overexpression of salivary amylase 1 after the ingestion of polyphenols and, surprisingly, not all the amylase isoforms changed with tannins but only one. Band ß could be an induction of a newly produced isoform, or could represent an overexpression of one of the previously visible isoforms.
Another detected difference was the expression of protein band γ (n = 8) in the quebracho tannin-fed diet group. This protein was identified as aldehyde reductase. Aldehyde reductase belongs to the aldo-keto reductases (AKR) superfamily. They are carbonyl-reducing enzymes, along with the short-chain dehydrogenases/reductases, and are responsible for the reduction of aldehydes, ketones, and quinones to their corresponding hydroxyl derivatives (Hoffmann and Maser 2007). Although salivary levels of enzymes that belong to the human aldo-keto reductase superfamily were found to be high in humans subjected to a continuous intake of coffee, which has a high content of polyphenols (Sladeck 2003), we interpret this result with caution. First, this enzyme, although ubiquitous (present in plants, fungi, insects, fish, bacteria), has been found mainly in the intracellular media and in tissues. In mice, it is found in several tissues, such as the liver (Ahmed et al. 1978), lung (Nakayama et al. 1986), and ovary (Iwata et al. 1990). A search in the University of California at Los Angeles Human Salivary Proteome Project database (http://www.hspp.ucla.edu) found only two salivary proteins from the AKR family 1 in saliva (Aldose reductase-like and Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase). Secondly, quebracho used in this study is a plant extract that contains only 72% tannins. The presence of carbonyl-group-bearing substances, such as small phenolics, other PSMs, or even quinones, resulting from oxidation during the extraction process, is not to be overlooked. The presence of aldehyde reductase could be a consequence of chemical species other than tannin.
We conclude that amylase up-regulation is an unspecific adaptation of saliva to dietary tannins that could be a consequence of the stimulation of sympathetic pathways and of amylase inhibition. Indirectly, and through these mechanisms, tannin adverse effects could be reduced, probably due to preferential involvement of one amylase isoform more prone for this activity. Despite the similarities of results obtained for tannic acid and quebracho, we think that further studies with other forms of tannins would help to clarify whether a specific protein defense response exists for the type of tannin.
References
Ahmed, N. K., Felsted, R. L., Bachur, and N. R. 1978. Heterogeneity of anthracycline antibiotic carbonyl reductases in mammalian livers. Biochem. Pharmacol. 27:2713–2719.
Ahmed, A. E., Smithard, R., and Ellis, M. 1991. Activities of enzymes of the pancreas, and the lumen and mucosa of the small intestine in growing broiler cockerels fed on tannin-containing diets. Br. J. Nutr. 65:189–197.
Ann, D. K., Clements, S., Johnstone, E. M., and Carlson, D. M. 1987. Induction of tissue-specific proline-rich protein multigene families in rat and mouse parotid glands by isoproterenol. Unusual strain differences of proline-rich protein mRNAs. J. Biol. Chem. 262:899–904.
Ann, D. K., Lin, H. H., and Kousvelari, E. 1997. Regulation of salivary gland specific gene expression. Crit. Rev. Oral Biol. Med. 8:244–252.
Bank, R. A., Hettema, H. E., Arwert, F., Amerongen, A. V., and Pronk, J. C. 1991. Electrophoretic characterization of post-translational modifications of human parotid salivary alpha-amylase. Electrophoresis 12:1032–1041.
Bedi, G. S. 1993. The effect of adrenergic agonists and antagonists on the expression of proteins in rat submandibular and parotid glands. Crit. Rev. Oral Biol. Med. 4:565–571.
Beeley, J. A., Sweeney, D., Lindsay, J. C., Buchanan, M. L., Sarna, L., and Khoo, K. S. 1991. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis of human parotid salivary proteins. Electrophoresis 12:1032–1041.
Bennick, A. 2002. Interaction of plant polyphenols with salivary proteins. Crit. Rev. Oral Biol. Med. 13:184–196.
Breslin, P. S. A., Gilmore, M. M., Beauchamp, G. K., and Green, B. G. 1993. Psychophysical evidence that oral astringency is a tactil sensation. Chem. Senses. 18:405–415.
Chatterton Jr, R. T., Vogelsong, K. M., Lu, Y. C., Ellman, A. B., and Hudgensg, A. 1996. Salivary alpha-amylase as a measure of endogenous adrenergic activity. Clin. Physiol. 16:433–448.
De Freitas, V., and Mateus, N. 2001. Structural features of procyanidin interactions with salivary proteins. J. Agric. Food Chem. 49:940–945.
Denny, P. C., Mirels, L., and Denny, P. A. 1996. Mouse submandibular gland salivary apomucin contains repeated N-glycosylation sites. Glycobiology 6:43–50.
Dlouhy, S. R., Taylor, B. A., and Karn, R. C. 1987. The genes for mouse salivary androgen-binding protein (ABP) subunits alpha and gamma are located on chromosome 7. Genetics 115:535–543.
Engelen, L., Van Den Keybus, P. A., De Wijk, R. A., Veerman, E. C., Amerongen, A. V., Bosman, F., Prinz, J. F., and Van Der Bilt, A. 2007. The effect of saliva composition on texture perception of semi-solids. Arch. Oral Biol. 52:518–525.
Gallacher, D. V., and Petersen, O. H. 1983. Stimulus-secretion coupling in mammalian salivary glands. Int. Rev. Physiol. 28:1–52.
Gho, F., Pena-neira, A., and Lopez-solis, R. O. 2007. Induction of salivary polypeptides associated with parotid hypertrophy by gallotannins administered topically into the mouse mouth. J. Cell Biochem. 100:487–498.
Gjorstrup, P. 1980. Taste and chewing as stimuli for the secretion of amylase from the parotid gland of the rabbit. Acta Physiol. Scand. 110:295–301.
Glendinning, J. I. 1994. Is the bitter rejection response always adaptive? Physiol. Behav. 56:1217–1227.
Gonzalez, M. J., Peña, Y., Lillo, S., Alliende, C., and Lopez-solis, R. O. 2000. Cell-enlargement-related polypeptides are induced via beta(1)-adrenoceptors in mouse parotids. Exp. Mol. Pathol. 69:91–101.
Gorr, S.-U., Venkatesh, S. G., and Darling, D. S. 2005. Parotid secretory granules: crossroads of secretory pathways and protein storage. J. Dent. Res. 84:500–509.
Granger, D. A., Kivlighan, K. T., El-Sheikh, M., Godis, E. B., and Stroud, L. R. 2007. Salivary alpha-amylase in biobehavioral research: recent developments and applications. Ann. N Y Acad. Sci. 1098:122–144.
Hagerman, A. E., ice, M. E., and itchard, N. T. 1998. Mechanisms of protein precipitation for two tannins, pentagalloyl glucose and epicatechin (4-8) catechin (procyanidin). J. Agric. Food Chem. 46:2590–2595.
Hagenbüchle, O., Bovey, R., and Young, R. A. 1980. Tissue-specific expression of mouse α-amylase genes: Nucleotide sequence isoenzyme mRNAs from pancreas and salivary gland. Cell 21:179–187.
Hardt, M., Witkowska, H. E., Webb, S., Thomas, L. R., Dixon, S. E., Hall, S. C., and Fisher, S. J. 2005a. Assessing the effects of diurnal variation in the composition of human parotid saliva: quantitative analysis of native peptides using iTRAQ reagents. Anal Chem. 77:4947–4954.
Hardt, M., Thomas, L. R., Dixon, S. E., Newport, G., Agabian, N., Prakobphol, A., Hall, S. C., Witkowska, H. E., and Fisher, S. J. 2005b. Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry 44:2885–2899.
Haslam, E. 1998. Practical Polyphenols: From Structure to Molecular Recognition and Physiological Action. Cambridge University Press, Cambridge.
Henriksson, R. 1982. Beta 1- and beta 2-adrenoceptor agonists have different effects on rat parotid acinar cells. Am. J. Physiol. 242:G481–G482.
Hirtz, C., Chevalier, F., Centeno, D., ofidal, V., Egea, J. C., ossignol, M., Sommerer, N., and Periere, D. D. 2005a. MS characterization of multiple forms of alpha-amylase in human saliva. Proteomics 5:4597–4607.
Hirtz, C., Chevalier, F., Centeno, D., Egea, J. C., ossignol, M., Sommerer, N., and Périère, D. D. 2005b. Complexity of the human whole saliva proteome. J. Physiol. Biochem. 61:469–80.
Hoffmann, F., and Maser, E. 2007. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab. Rev. 39:87–144.
Huang, C. M. 2004. Comparative proteomic analysis of human whole saliva. Arch. Oral Biol. 49:951–962.
Humphrey, S. P., and Williamson, R. T. 2001. A review of saliva normal composition, flow and function. J. Prosthet. Dent. 85:162–169.
Iwata, N., Inazu, N., and Satoh, T. 1990. Immunological and enzymological localization of carbonyl reductase in ovary and liver of various species. J. Biochem. (Tokyo) 107:209–212.
Jansman, A. J., Frohlich, A. A., and Marquardt, R. R. 1994. Production of proline-rich proteins by the parotid glands of rats is enhanced by feeding diets containing tannins from faba beans (Vicia faba L.). J. Nutr. 124:249–258.
Kandra, L., Gyémánt, G., Zajacz, A., and Batta, G. 2004. Inhibitory effects of tannin on human salivary alpha amylase. Biochem. Biophy. Res. Comm. 319:1265–1271.
Karn, R. C., and Laukaitis, C. M. 2003. Characterization of two forms of mouse salivary androgen-binding protein (ABP): Implications for evolutionary relationships and ligand-binding function. Biochemistry 42:7162–7170.
Katsukawa, H., and Ninomiya, Y. 1999. Capsaicin induces S-like substances in submandibular saliva of the rat. J. Dent. Res. 78:1609–1616.
Kim, W. S., Nakayama, K., Nakagawa, T., Kawamura, Y., Haraguchi, K., and Murakami, K. 1991. Mouse submandibular gland prorenin-converting enzyme is a member of glandular kallikrein family. J. Biol. Chem. 266:19283–19287.
Le Magnen, J. 1986. Hunger. Problems in the Behavioural Sciences. Cambridge University Press, Cambridge.
Lesschaeve, I., and Noble, A. C. 2005. Polyphenols: factors influencing their sensory properties and their effects on food and beverage preferences. Am. J. Clin. Nutr. 81:330S–335S.
Lin, H. H., and Ann, D. K. 1991. Molecular characterization of rat multigene family encoding proline-rich proteins. Genomics 10:102–113.
Lopez-solis, R. O., and Kemmerling, U. 2005. Codominant expression of genes coding for different sets of inducible salivary polypeptides associated with parotid hypertrophy in two inbred mouse strains. J. Cell Biochem. 95:99–107.
Lu, Y., and Bennick, A. 1998. Interaction of tannin with human salivary proline-rich proteins. Arch. Oral Biol. 43:717–728.
Madsen, H. O., and Hjorth, J. P. 1985. Molecular cloning of mouse PSP mRNA. Nuc. Acids Res. 13:1–13.
Mahmood, S., and Smithard, R. 1993. A comparison of effects of body weight and feed intake on digestion in broiler cockrels with effects of tannins. Br. J. Nutr. 70:701–709.
Mcdougall, G. J., Shpiro, F., Dobson, P., Smith, P., Blake, A., and Stewart, D. 2005. Different polyphenolic components of soft fruits inhibit alpha-amylase and alpha-glucosidase. J. Agric. Food Chem. 53:2760–2766.
Mehansho, H., Hagerman, A. E., Clements, S., Butler, L. G., ogler, J., and Carlson, D. M. 1983. Modulation of proline-rich protein biosynthesis in rat parotid glands by sorghum with high tannin levels. Proc. Natl. Acad. Sci. USA. 80:3948–3952.
Mehansho, H., Hagerman, A. E., Cements, S., Butler, L., ogler, J., and Carlson, D. M. 1985. Indution of proline rich glycoproteins synthesis in mouse salivary glands by isopretrenol and by tannins. J. Biol. Chem. 260:4418–4423.
Mehansho, H., ogler, J., and Carlson, D. M. 1987. Dietary tannins and salivary proline rich peroteins: interation, inductions and defense mechanisms. Annu. Rev. Nutr. 7:423–440.
Muenzer, J., Bildstein, C., Gleason, M., and Carlson, D. M. 1979. Purification of proline-rich proteins from parotid glands of isoproterenol-treated rats. J Biol. Chem. 254:5623–5628.
Myal, Y., Iwasiow, B., Cosby, H., Yarmill, A., Blanchard, A., Tsuyuki, D., Fresnoza, A., Duckworth, M. L., and Shiu, R. P. C. 1998. Analysis of tissue- and hormone-specific regulation of the human prolactin-induceble protein/gross cystic disease fluid proein-15 gene in transgenic mice. J. Mol. Endocrinol. 21:217–223.
Nakayama, T., Yashiro, K., Inoue, Y., Matsuura, K., Ichikawa, H., Hara, A., and Sawada, H. 1986. Characterization of pulmonary carbonyl reductase of mouse and guinea pig. Biochim. Biophys. Acta 882:220–227.
Neuhoff, V., Arold, N., Taube, D., and Ehrhardt, W. 1988. Improved staining of proteins in polyacrilamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brillant Blue G-250 and R-250. Electrophoresis 9:255–262.
Neyraud, E., Sayd, T., Morzel, M., and Dransfield, E. 2006. Proteomic analysis of human whole and parotid salivas following stimulation by different tastes. J. Proteome Res. 5:2474–2480.
Oppenheim, F. G., Salih, E., Siqueira, W. L., Zhang, W., and Helmerhorst, E. J. 2007. Salivary proteome and its genetic polymorphisms. Ann N Y Acad. Sci. 1098:22–50.
Pandey, A., Andersen, J. S., and Mann, M. 2000. Use of mass spectrometry to study signaling pathways. Sci STKE 37:PL1.
Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567.
Prinz, J. F., and Lucas, P. W. 2000. Saliva tannin interactions. J. Oral Rehabil. 27:991–994.
Proctor, G. B., and Carpenter, G. H. 2007. Regulation of salivary gland function by autonomic nerves. Auton. Neurosci. 133:3–18.
Roxo-Rosa, M., Da Costa, G., Luider, T. M., Scholte, B. J., Coelho, A. V., Amaral, D. M., and Penque, D. 2006. Proteomic analysis of nasal cells from cystic fibrosis (CF) patients and non-CF control individuals: search for novel biomarkers of CF lung disease. Proteomics 6:2314–25.
Shaw, P. A., and Yu, W. A. 2000. Autonomic regulation of cystatin S gene expression in rat submandibular glands. Auton. Neurosci. 83:49–57.
Shimada, T., Saitoh, T., and Matsui, T. 2004. Does acclimation reduce the negative effects of acorn tannins in the wood mouse Apodemus speciosus?. Acta Theriol. 49:203–214.
Shimada, T., Saitoh, T., Sasaki, E., Nishitani, Y., and Osawa, R. 2006. Role of tannin-binding salivary proteins and tannase-producing bacteria in the acclimation of the Japanese wood mouse to acorn tannins. J. Chem Ecol. 32:1165–1180.
Shori, D. K., and Asking, B. 2001. Dynamics of protein and fluid secretion from the major salivary glands of rat: relevance of research findings to clinically observed defective secretion in cystic fibrosis. Pflugers Arch. 443:Suppl 1535–543.
Skopec, M. M., Hagerman, A. E., and Karasov, W. H. 2004. Do salivary proline-rich proteins counteract dietary hyrdolyzable tannin in laboratory rats. J. Chem. Ecol. 30:1679–1692.
Sladeck, N. 2003. Human aldehyde dehydrogenase: Potential pathological, pharmacological and toxicological impact. J. Biochem. Mol. Toxicol. 17:7–23.
STJEIN, V., AMEROGEN, A.V., Veerman, E.C.I., Kasanmoentalib, S., and OVerduk, B., 1999. Chitinase in whole human saliva and in whole saliva of patients with periodontal inflammation. Eur. J. Oral Sci. 107:328–337.
Vitorino, R., Lobo, M. J., Ferrer-correira, A. J., Dubin, J. R., Tomer, K. B., Domingues, P. M., and Amado, F. M. 2004. Identification of human whole saliva protein components using proteomics. Proteomics 4:951–962.
Waters, C. A., Morand, J. N., Schatzman, R. C., and Carlson, D. M. 1998. Induction of p34cdc2 in mouse parotid glands upon activation of beta 1-adrenergic receptors. Cell Mol. Biol. (Noisy-le-grand) 44:333–342.
Williams, K. M., Ekstrom, J., and Marshal, T. 1999. High resolution electrophoretic analysis of rat parotid salivary proteins. Electrophoresis 20:1373–1381.
Yao, Y., Berg, E., Costello, C. E., Troxler, R. F., and Oppenheim, F. G. 2003. Identification of proteins components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J. Biol. Chem. 278:5300–5308.
Acknowledgments
We acknowledge the generous offer of Tupafin Ato from the Silva Chimica Company (Italy) to Dr André M. Almeida to review this manuscript and Prof. Alfredo Pereira for support with the statistical analysis. This work was supported by POCTI FCT-CVT/33039/99-00 scientific project. Gonçalo da Costa and Elsa Lamy acknowledge a PhD grant from 3° Quadro Comunitário de Apoio of FCT—Fundação para a Ciência e a Tecnologia of the Ministério da Ciência Tecnologia e Ensino Superior.
Author information
Authors and Affiliations
Corresponding author
Additional information
G. da Costa and E. Lamy have contributed equally to this work.
Rights and permissions
About this article
Cite this article
da Costa, G., Lamy, E., Capela e Silva, F. et al. Salivary Amylase Induction by Tannin-Enriched Diets as a Possible Countermeasure Against Tannins. J Chem Ecol 34, 376–387 (2008). https://doi.org/10.1007/s10886-007-9413-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-007-9413-z