Abstract
Beneficial bacteria that live on salamander skins have the ability to inhibit pathogenic fungi. Our study aimed to identify the specific chemical agent(s) of this process and asked if any of the antifungal compounds known to operate in analogous plant–bacteria–fungi systems were present. Crude extracts of bacteria isolated from salamander skin were exposed to HPLC, UV-Vis, GC-MS, and HR-MS analyses. These investigations show that 2,4-diacetylphloroglucinol is produced by the bacteria isolate Lysobacter gummosus (AB161361), which was found on the red-backed salamander, Plethodon cinereus. Furthermore, exposure of the amphibian fungal pathogen, Batrachochytrium dendrobatidis (isolate JEL 215), to different concentrations of 2,4-diacetylphloroglucinol resulted in an IC50 value of 8.73 μM, comparable to crude extract concentrations. This study is the first to show that an epibiotic bacterium on an amphibian species produces a chemical that inhibits pathogenic fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mutualisms between bacteria and metazoa are ubiquitous and ecologically important. Some well-studied mutualisms involve antifungal bacteria and their hosts, including bacteria that reside on plant roots and on the cuticle of leaf cutter ants (Currie et al. 2006; Raaijmakers et al. 2002). Amphibians, which inhabit moist surroundings hospitable to fungi, not only produce defensive secretions that inhibit fungi and other skin microbes (Simmaco et al. 1998; Rollins-Smith et al. 2005), but also harbor a distinct resident cutaneous bacterial community (Harris et al. 2006; Lauer et al. 2007) that resists removal by thorough rinsing. A specific chemical agent of fungal inhibition, however, has remained unidentified. Therefore, this study aimed to isolate an antifungal substance from the cutaneous bacterial community of an amphibian species and to test for its inhibitory activity against a pathogenic fungal species.
Our previous work demonstrated that bacteria isolated from skin of different amphibian species and grown in a low nutrient medium on a Petri dish inhibit fungi that are pathogenic to amphibian adults and embryos (Harris et al. 2006; Lauer et al. 2007). While these results suggest that antifungal bacteria produce an antibiotic, inhibition by resource competition can also occur. Crude extract removed from bacteria grown in broth culture, however, inhibited a fungal pathogen (Walters 2007), providing evidence that skin bacteria had produced one or more antifungal chemicals. Of the various antifungal species identified by sequencing of the 16S rRNA gene (∼1,400 bp), an isolate with 99% genetic match to Lysobacter gummosus (AB161361, GenBank) was chosen for this study because of its consistently strong antifungal properties. This species was isolated from the red-backed salamander, Plethodon cinereus, and from the four-toed salamander, Hemidactylium scutatum.
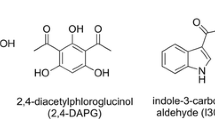
To begin our study, we focused on antibiotics known to be produced by Pseudomonas fluorescens, a relative of Lysobacter. The bacterium P. fluorescens is common in soil, on plant roots, and on amphibian skin, and is inoculated onto pea, wheat, and sugar beet roots to protect them from diseases (Timms-Wilson et al. 2005). This widespread bacterium P. fluorescens produces many different antibiotics. In this study, three of these were investigated and compared to the L. gummosus antibiotics: pyoluteorin, pyrrolnitrin, and 2,4-diacetylphloroglucinol (=2,4-DAPG; Whistler et al. 1998; Ligon et al. 2000; Sako et al. 2001; Brodhagen et al. 2004; Fig. 1).
Materials and Methods
Bacteria Isolates
Pure cultures of L. gummosus (AB161361) were obtained from the salamander Plethodon cinereus after a thorough rinsing of the animals to remove transient bacteria. Adult salamanders were obtained from James Madison University Arboretum, Harrisonburg, VA, USA. All L. gummosus isolates were stored in 20% glycerol solution and frozen at −20°C. When needed, L. gummosus was cultured onto R2A solid medium and allowed to grow at room temperature for 72 h. Isolates were cultured this way three times before extraction. Bacterial species identifications were made by sequencing a portion of the 16S rRNA gene and comparing the sequence with the GenBank database (www.ncbi.nlm.nih.gov/blast).
Preparation and Extraction of Crude Organic Samples
To extract bacterial products, broth cultures were grown. The L. gummosus isolate was inoculated into separate 250 ml flasks containing 1/10 strength Nutrient Broth medium (NB, Becton, Dickinson and Company, Sparks, MD, USA). Each liquid culture, as well as a 1/10 strength NB control, was kept at room temperature for 72 h in a Lab-Line incubator shaker. Bacterial cells were in stationary phase when they were removed from the nutrient broth to leave a “crude extract” as follows. The cultures were poured into 50 ml Falcon tubes and centrifuged at 5,000×g for 5–10 min. The supernatant was filtered through a 25 mm sterile syringe filter with 0.22 μm pores into new Falcon tubes to remove any remaining bacteria. The supernatant was acidified to pH < 2 by using 1 M HCl. Each Falcon tube was frozen at −20°C until needed for chemical analysis.
To extract organic compounds, approximately 90 ml of crude extract supernatant from the L. gummosus broth culture were poured into a separatory funnel. The aqueous layer was extracted × 4 with 35 ml of ethyl acetate (EtOAc). The combined organic layers were dried with Na2SO4 and filtered. The EtOAc solvent was evaporated in vacuo, and a crude recovery calculated. The crude samples (3–9 mg) were then kept frozen in a −20°C freezer until further analysis. A total of five samples were extracted.
HPLC Analysis of Extract
Each crude sample was dissolved in high-performance liquid chromatography (HPLC)-grade methanol (2 ml) before analysis. Standard solutions of 2,4-DAPG, pyrrolnitrin, and pyoluteorin were also prepared in HPLC-grade methanol (1 mg/ml). 2,4-DAPG and pyrrolnitrin were used as purchased (Sigma-Aldrich, Saint Louis, MO, USA and Maybridge Trevillett, Tintagel, UK); pyoluteorin was synthesized in three steps, slightly modifying a method described by Rao and Reddy (1990).
RP-HPLC analysis (Agilent Technologies, 1200 series, Wilmington, DE, USA) was used to determine the retention times of the antifungal standards as well as to analyze the components of bacterial samples. The HPLC diode array detector was programmed to observe and record absorbance at 270 and 310 nm. These two wavelengths best correlate to the λ max of the standards: 2,4-DAPG at 270 nm, pyrrolnitrin at 252 nm (broad), and pyoluteorin at 310 nm (Sako et al. 2001; Brodhagen et al. 2004). The samples were injected (20 μl) into the HPLC equipped with a C18 reverse phase column (5 μm; 4.6 × 150 mm; Agilent Technologies) and eluted at 1 ml/min. The initial eluent, 10% acetonitrile/water (v/v, both acidified with 0.1% acetic acid), ran for 2 min. This was followed by a linear gradient to 100% acetonitrile (acidified with 0.1% acetic acid), over an 18-min-period. This final solvent was eluted for another 3 min. Samples showing absorbance at 270 or 310 nm were collected.
GC-MS Analysis of Extract
The standards and HPLC eluates were further analyzed by using coupled gas chromatography–mass spectrometry (Thermo/Finnigan Polaris Q, Somerset, NJ, USA; column: Restek, Rtx-5 crossbond 5% diphenyl-95%dimethyl polysiloxane, length: 30 m, i.d.: 0.25 mm; particle size 0.25 μm); 1.0 μl-samples in HPLC grade methanol were injected (injector temperature: 200°C; EI full scan, mass range 50–500 amu, ion source 200°C). Column temperature began at 180°C for 2 min and increased to 280°C within 10 min. Retention times and mass spectra of the substances were compared to chemical standards of pyoluteorin, pyrrolnitrin, and 2,4-DAPG.
HR-MS Analysis of Extract
The crude L. gummosus extract and 2,4-DAPG were analyzed by high resolution mass spectrometry (HR-MS). ESI mass spectra were obtained with an LCT-TOF mass spectrometer (Micromass, Harvard University Mass Spectrometry Facility).
Inhibitory Assay
To quantify the antifungal properties of 2,4-DAPG, a liquid growth assay was developed by using the fungal pathogen Batrachochytrium dendrobatidis. In a 96-well microtiter plate, 12 concentrations of 2,4-DAPG (ranging from 475 to 0.232 μM) as well as positive and negative controls were examined. Five replicates were performed per assay. Each well contained 125 μl of 1% tryptone medium plus 100 μl of fungal zoospores in 1% tryptone medium at a concentration of ≈1*106 zoospores/ml (determined by using a hemocytometer). The plates were incubated for 1 day at 23°C before 25 μl of a solution of 2,4-DAPG in dimethyl sulfoxide (DMSO) were added. A volume of 25 μl neat DMSO was added to the positive control wells; heat-killed zoospores were used in the negative control. The optical density at 490 nm (OD490) of each plate was measured with a BioRad microplate reader (model 680, Hercules, CA, USA), on days 0 and 8. Inhibition was determined by comparing the difference of the final and initial growth to the positive control by using a Dunnett’s test. The logarithmic trend line was determined by linear regression and used to calculate the IC50.
Results
RP-HPLC analysis of the organic extract of L. gummosus revealed a peak at 13.46 min, as observed at 270 nm (Fig. 2). This peak matched the HPLC retention time of the 2,4-DAPG standard (13.46 min). Furthermore, the UV spectra of extract and standard were similar, each showing a λ max of 270 nm (Fig. 3). On GC–MS, the 2,4-DAPG standard displayed a retention time of 6.76 min; the corresponding mass spectrum indicated for M+: 210.1 m/z and 211.1 m/z (M+H). The extract of L. gummosus showed a peak with a retention time of 6.78 min, and a mass spectrum with 210.1 m/z (M+) and 211.2 m/z (M+H).
HPLC chromatogram (270 nm) comparing Lysobacter gummosus crude extract (solid line, left scale) and 2,4-DAPG standard (dashed line, right scale). Both samples had similar retention times on the HPLC chromatograms (13.46 min). The L. gummosus extract had several additional peaks, including several very large peaks that were artifacts from the nutrient broth
HR-MS analysis of the 2,4-DAPG standard showed a peak at 211.0603 m/z, corresponding to the calculated (M+H) peak of 211.0607 m/z. The L. gummosus extract showed a peak at 211.0612 m/z.
Analogous investigations (HPLC, UV-Vis, GC-MS) of L. gummosus extracts for pyoluteorin and pyrrolnitrin did not provide evidence for the presence of these compounds in the crude sample.
The growth assay (Fig. 4), which used Batrachochytrium dendrobatidis, indicated a 2,4-DAPG IC50 of 8.73 μM and a minimum inhibitory concentration (MIC) of 136.13 μM. These concentrations were calculated by using a regression of log-transformed OD490 values on 2,4-DAPG concentration (F = 1118.22, df = 1,124, P < 0.001). All 2,4-DAPG concentration treatments differed significantly in OD490 values from the positive control with the exception of the 0.23 μM and the 0.46 μM concentrations (a posteriori Dunnett’s t tests, P < 0.05 for all comparisons).
Discussion
Four methods indicate that 2,4-DAPG was produced by the amphibian skin bacterium L. gummosus. RP-HPLC analyses repetitively resulted in nearly identical retention times of a peak of the bacterial extract and the 2,4-DAPG standard (Fig. 2); UV-Vis comparison demonstrated a nearly identical spectrum and λ max (270 nm) of this peak and 2,4-DAPG (Fig. 3). Furthermore, GC-MS and HR-MS analyses support the presence of 2,4-DAPG in L. gummosus extracts. Although the exact concentration of 2,4-DAPG prevalent on an individual amphibian skin was not investigated in this study, the concentration calculated from the liquid cultures used in this study was about 2 μM. Estimation of 2,4-DAPG concentrations of amphibian skins will be the subject of further investigations.
Biologically, bacteria living on amphibian skin may act as part of the innate immune system of amphibia by preventing pathogens from colonizing. Amphibian species that attend their embryos, such as H. scutatum, may use skin bacteria and their antibiotics to keep their embryos free of fungus. Additional studies will be necessary to determine whether there are co-evolved mutualisms between amphibian hosts and skin bacteria. The investigation of other strong antifungal metabolites from a variety of epibiotic bacterial species obtained from frogs and salamanders are the focus of ongoing research. In addition, it would be of interest to determine the relative role of amphibian antimicrobial secretions and those of their bacterial symbionts in deterring pathogens. The study of skin bacteria and their antibiotics may also lead to an understanding of why some species succumb to an emerging infectious disease of the skin, chytridiomycosis, which is caused by Batrachochytrium dendrobatidis, and others do not.
References
Brodhagen, M., Henkels, M. D., and Loper, J. E. 2004. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 70:1758–1766.
Currie, C. R., Poulsen, M., Mendenhall, J., Boomsma, J. J., and Billen, J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83.
Harris, R. N., James, T. Y., Lauer, A., Simon, M. A., and Patel, A. 2006. The amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth 3:53–56.
Lauer, A., Simon, M. A., Banning, J. L., André, E., Duncan, K., and Harris, R. N. 2007. Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia. 2007:630–640.
Ligon, J. M., Hill, D. S., Hammer, P. E., Torkewitz, N. R., Hofmann, D., Kempf, H. J., and van PEE, K. H. 2000. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manage. Sci. 56:688–695.
Raaijmakers, J. M., Vlami, M., and de Souzal, J. T. 2002. Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek 81:537–547.
Rao, K. V., and Reddy, G. C. 1990. Synthesis and herbicidal activity of the halo analogues of pyoluteorin. J. Agric. Food Chem. 38:1260–1263.
Rollins-Smith, L. A., Reinert, L. K., O’Leary, C. J., Houston, L. E., and Woodhams, D. C. 2005. Antimicrobial peptide defense in amphibian skin. Integrative and Comparative Biology 45:137–142.
Sako, M., Kihara, T., Okada, K., Ohtani, Y., and Kawamoto, H. 2001. Reductive cleavage of heteroaryl C-halogen bonds by iodotrimethylsilane: facile and regioselective dechlorination of antibiotic pyrrolnitrin. J. Org. Chem. 66:3610–3612.
Simmaco, M., Mignogna, G., and Barra, D. 1998. Antimicrobial peptides from amphiban skin: what do they tell us? Biopolymers (Peptide Science) 47:435–450.
Timms-Wilson, T. M., Kilshaw, K., and Bailey, M. J. 2005. Risk assessment for engineered bacteria used in biocontrol of fungal disease in agricultural crops. Plant Soil 266:57–67.
Walters, R. L. 2007. Antibiotic production by cutaneous bacteria isolated from two species of salamanders. MSc thesis. James Madison University, Harrisonburg.
Whistler, C. A., Corbell, N. A., Sarniguet, A., Ream, W., and Loper, J. E. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigma σs and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635–6641.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brucker, R.M., Baylor, C.M., Walters, R.L. et al. The Identification of 2,4-diacetylphloroglucinol as an Antifungal Metabolite Produced by Cutaneous Bacteria of the Salamander Plethodon cinereus . J Chem Ecol 34, 39–43 (2008). https://doi.org/10.1007/s10886-007-9352-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-007-9352-8