Abstract
Infection with Listeria monocytogenes can cause meningitis and septicemia in newborn, elderly, or immunocompromised individuals. Pregnant women are particularly susceptible to Listeria, leading to a potentially fatal infection. Cytosolic Listeria activates the proinflammatory caspase-1 and induces the processing and secretion of interleukins IL-1β and IL-18 as well as caspase-1-dependent pyroptosis. This study elucidates the role of various inflammasome components of host macrophages in the proinflammatory response to infection with Listeria. Here, we have used macrophages from AIM2-, NLRC4-, NLRP3-, and ASC-deficient mice to demonstrate that AIM2, NLRC4, and NLRP3 inflammasomes as well as the adaptor protein ASC all contribute to activation of caspase-1 in Listeria-infected macrophages. We show that Listeria DNA, which escapes into the cytosol of infected macrophages, triggers AIM2 oligomerization, caspase-1 activation, and pyroptosis. Interestingly, we found that flagellin-deficient Listeria, like Francisella tularensis, is recognized primarily by the AIM2 inflammasome, as no caspase-1 activation or cell death was observed in AIM2-deficient macrophages infected with this Listeria mutant. We further show that prior priming of NLRC4-deficient macrophages with LPS is sufficient for Listeria-induced caspase-1 activation in these macrophages, suggesting that TLR4 signaling is important for activation of the AIM2 and NLRP3 inflammasomes by Listeria in the absence of NLRC4. Taken together, our results indicate that Listeria infection is sensed by multiple inflammasomes that collectively orchestrate a robust caspase-1 activation and proinflammatory response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mammalian cells respond rapidly via an innate immune response to diverse danger signals elicited by pathogens, pathogen-derived molecules, and even to self-derived molecular danger signals, which arise from tissue damage. Recognition of pathogen-associated molecular patterns (PAMPs) is achieved by pattern-recognition receptors (PRRs) that constantly survey the extracellular space and the cytoplasm for the presence of danger [1]. The PRRs include toll-like receptors (TLRs) that sense PAMPs on the cell surface or in the endosomes and NLRs that recognize microbial molecules in the host cytosol [1–3]. After stimulus recognition, both TLRs and NLRs induce the activation of multiple signaling pathways, which lead to innate and adaptive immune responses [1, 3].
The NLR family is composed of 23 family members in humans, whereas the mouse genome contains at least 34 NLR-encoding genes [2, 3]. Several NLRs, including NLRP1 (also called NALP1), NLRP3 (also called cryopyrin or NALP3), and NLRC4 (also called IPAF), can assemble inflammasomes, which are molecular platforms responsible for activation of the proinflammatory cysteine protease caspase-1 [3]. We and others have recently identified AIM2 (absent in melanoma 2) as a novel inflammasome involved in the recognition of cytosolic DNA during viral and bacterial infection [4–9]. AIM2 is particularly crucial for innate immunity against infection with Francisella tularensis, as infection of AIM2-deficient mice with F. tularensis resulted in greater bacterial burdens and higher mortality than infection of wild-type mice [8]. The increased lethality in F. tularensis-infected AIM2-deficient mice was associated with decreased production of caspase-1-generated cytokines in these mice [8]. AIM2 is an interferon-inducible HIN-200 family member that contains an amino-terminal pyrin domain and a carboxy-terminal HIN-200 oligonucleotide/oligosaccharide-binding domain. Recognition of cytoplasmic DNA by the HIN-200 domain triggers AIM2 oligomerization, recruitment of ASC, and the activation of caspase-1 [4, 8]. The interaction of AIM2 with ASC also leads to the activation of the ASC pyroptosome, which induces pyroptotic cell death [4, 8, 10].
Listeria monocytogenes (LM) is a Gram-positive bacterium and a facultative intracellular pathogen that causes life-threatening disease, particularly in immunocompromised individuals, pregnant women, and neonates [11]. Listeria infection of macrophages proceeds via phagocytosis followed by escape from the phagosome into the cytosol through the action of the pore-forming toxin listeriolysin O (LLO) [12, 13]. Intracellular secretion of a pore-forming toxin appears to be unique to Listeria infection.
Caspase-1 activation is required for the clearance of Listeria in murine infection [14]. Listeria is known to be recognized by TLR2, NOD1, and NOD2 [15–20], which can lead to NF-κB-dependent proinflammatory gene expression. Recent reports have suggested that Listeria activates caspase-1 through multiple NLRs [21]. There are conflicting reports regarding the contributing roles of NLRP3, AIM2, and NLRC4 inflammasomes in Listeria-infected macrophages [21–26]. In this study, we used AIM2-deficient mouse macrophages, together with mouse macrophages deficient in NLRP3, NLRC4, and ASC and demonstrate that all three inflammasomes (NLRC4, AIM2, and NLRP3 inflammasomes) and the ASC adaptor protein are involved in caspase-1 activation in Listeria-infected macrophages. We further show that Listeria flagellin and TLR4 signaling play critical roles in caspase-1 activation in Listeria-infected AIM2- and NLRC4-deficient macrophages, respectively.
Materials and Methods
Reagents
Polyclonal anti-AIM2 antibody was custom raised in rabbits at Invitrogen against a mixture of recombinant mouse and human AIM2 proteins. The anti-IL-1β monoclonal antibody was obtained from the NCI preclinical repository, biological resource branch. Other antibodies used against mouse NLRP3 (polyclonal anti-NLRP3 PYD; our lab), mouse ASC (polyclonal anti-mouse ASC; from Dr. Junji Sagara), and mouse caspase-1 p20 (monoclonal anti-mouse caspase-1 p20; from Dr. Junying Yuan) were described previously [10, 27, 28]. Anti-β-actin monoclonal antibody was obtained from Santa Cruz Biotechnology. Hoechst 33342 was obtained from Invitrogen. Nigericin was obtained from Sigma-Aldrich. Ultrapure LPS was obtained from Invivogen. CytoTox96 LDH-release kit was from Promega.
Mouse Macrophage Cell Culture
J2 retrovirus (carrying v-myc and v-raf oncogenes)-immortalized macrophage cell lines were used in all experiments. Immortalized bone marrow-derived macrophage cell lines deficient in NLRC4 (NLRC4-KO), NLRP3 (NLRP3-KO) or ASC (ASC-KO) were obtained from Dr. Eicke Latz. Immortalized wild-type (WT) and AIM2-deficient (AIM2-KO) macrophage cell lines were generated by infecting primary bone marrow cells with J2 recombinant retrovirus (a kind gift from Dr. Howard Young) as described previously [29]. Briefly, mouse bone marrow cells were isolated from femurs of WT and AIM2-KO mice siblings and cultured in DMEM for 3–5 days in DMEM (Gibco) supplemented with 12% (vol/vol) conditioned medium from L929 mouse fibroblasts, 10% (vol/vol) fetal bovine serum (FBS), and 1% (vol/vol) penicillin-streptomycin (15140; Gibco) for the induction of macrophage differentiation. Subsequently, cells were incubated with ψCREJ2 cell supernatant (a source of the J2 transforming retrovirus; [30]) with 8 μg/ml Polybrene (Sigma) and 12% (vol/vol) L929 conditioned medium for 24 h at 37°C. The supernatant was then replaced with fresh DMEM containing 12% (vol/vol) L929 conditioned medium and 10% FBS. Cells were slowly weaned off L929 conditioned medium until they were growing in DMEM, 10% FBS without L929 conditioned medium. NLRP3-KO-AIM2-GFP-N1 cell line was generated as described previously [8]. All immortalized macrophage cell lines were grown in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. All cells were grown at 37°C with 5% CO2.
Bacterial Culture and Infection of Macrophages with Listeria
Wild-type (10403S) and mutant Listeria strains containing deletions in the flagellin gene (flaA, DP-L4650) or the hly gene (Δhly, LLO-deficient, DP-L4027) were obtained from Dr. Daniel Portnoy. Bacteria were grown to logarithmic phase in brain heart infusion at 37°C with continuous shaking at 250 rpm. On the day of infection, a 1/50 dilution of the overnight culture was prepared and allowed to grow at 37°C with shaking to about A 600 = 0.5, which corresponds to ∼109 CFU/ml. Bacterial cultures were diluted to the desired concentration in OPTI-MEM and used to infect macrophages at different multiplicities of infection (MOI). Unless indicated otherwise, all Listeria infections were done with LPS-nonprimed macrophages. After 1 h of infection at 37°C, macrophages were washed twice with PBS, and fresh OPTI-MEM containing 50 µg/ml gentamycin was added to limit the growth of extracellular bacteria. After culture for an additional 4 h, culture supernatants and cell pellets were collected and assayed as indicated.
Immunoblot Analysis
Proteins in cell culture supernatants were precipitated by adding an equal volume of methanol and 0.25 volumes of chloroform as described [4, 31]. Protein samples were separated by 12.5% SDS-PAGE and were transferred onto PVDF membranes. Western blots were probed with rat anti-mouse caspase-1 p20 antibody. In addition, cell lysates were mixed with SDS sample buffer and separated by 12.5% SDS-PAGE and analyzed by immunoblotting with the appropriate antibodies as indicated.
LDH Release Assay
After infection with Listeria, cell culture supernatants were harvested and assayed for lactate dehydrogenase (LDH) activity using the CytoTox96 LDH-release kit (Promega) according to the manufacturer’s protocol.
Confocal Microscopy
NLRP3-KO-AIM2-GFP-N1 macrophages seeded on 35-mm glass-bottomed culture dishes were infected for 5 h with unstained or Hoechst-stained Listeria (MOI = 50). Cells infected with unstained Listeria were then stained with Hoechst 33342; cells infected with Hoechst-stained Listeria were not stained after infection. Live cell images were collected on a Nikon confocal microscope in the Kimmel Cancer Center Bioimaging facility as described previously [8].
Results
Listeria Flagellin Is Essential for Caspase-1 Activation in AIM2-deficient Macrophages
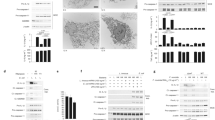
To study the involvement of the AIM2 inflammasome in innate signaling by Listeria, macrophages from WT and AIM2-KO mice were infected with wild-type (WT), flagellin-deficient (flaA), or LLO-deficient (Δhly) Listeria for 5 h. Inflammasome activation was assessed by Western blot for processed caspase-1 (p20) in culture supernatants. Cell death was evaluated by LDH release. As shown in Fig. 1a, b, WT Listeria at an MOI of 50 induced more caspase-1 activation and cell death in WT than in AIM2-KO macrophages, but Δhly Listeria, which cannot escape the phagosome, did not induce any caspase-1 activation or cell death. Interestingly, we observed that flagellin-deficient flaA Listeria triggered obvious caspase-1 activation and cell death in WT but not in AIM2-KO macrophages, indicating that Listeria flagellin is required for activating caspase-1 through other inflammasomes in the absence of AIM2. This also implies that the flagellin-deficient flaA Listeria mutant, like F. tularensis, is only recognized by the AIM2 inflammasome. Consistent with this, flaA Listeria at MOIs of 20 and 50 induced caspase-1 processing and cell death in a dose-dependent manner in WT macrophages but not in AIM2-KO cells (Fig. 1c, d). LPS plus Nigericin (a pore-forming toxin), which activate caspase-1 through the NLRP3 inflammasome, induced caspase-1 processing and cell death in both WT and AIM2-KO macrophages, demonstrating that AIM2-KO macrophages are not defective in caspase-1 activation by AIM2-independent stimuli. In addition, upregulation of NLRP3 and pro-IL-1β proteins was observed in macrophages infected with all three Listeria strains compared to the uninfected macrophage controls. However, AIM2 protein expression was unchanged following Listeria infection. Since NLRP3 and pro-IL1β proteins are upregulated through activation of the transcriptional NF-κB pathway [28], these observations indicate that the three Listeria strains do not have a defect in their ability to activate the NF-κB signaling pathway.
Listeria flagellin is important for activation of the AIM2 inflammasome. a–b WT and AIM2-KO macrophages were left uninfected (Control) or infected for 5 h with WT (wild type), flaA (flagellin-deficient) or Δhly (LLO-deficient) Listeria monocytogenes (LM) at MOI 50. c–d WT and AIM2-KO macrophages were left uninfected or infected for 5 h with different MOIs of flaA Listeria, or primed with 200 ng/ml of LPS for 4 h, then pulsed with 5 μM Nigericin for 45 min as indicated. Culture supernatants (Sup) and cell lysates (Lysates) were harvested and analyzed by immunoblotting with the indicated antibodies (a and c). Culture supernatants were also analyzed by LDH release assay (b and d). Data are representative of at least three experiments
NLRC4 Is Required for WT Listeria-Induced Caspase-1 Activation in Nonprimed Macrophages
The above results suggest that activation of caspase-1 and cell death induced by WT Listeria is partially dependent on AIM2 since infection with WT Listeria only produced less caspase-1 processing and less LDH release in AIM2-KO macrophages compared to WT macrophages. Our results are in line with previous studies, which implicated multiple inflammasomes, including NLRC4 and NLRP3 in Listeria-induced caspase-1 activation in macrophages [21]. However, since the previous studies were performed with LPS-primed macrophages [21], whereas ours were performed without LPS priming of macrophages, we decided to reinvestigate the role of NLRC4 and NLRP3 in caspase-1 activation by Listeria in the absence of LPS priming. To this end, we infected WT, AIM2-KO, NLRC4-KO, and NLRP3-KO macrophages simultaneously under the same conditions with WT or flaA Listeria. Consistent with the previous findings [21], we found that WT Listeria induced less caspase-1 processing and cell death in NLRP3-KO macrophages compared to WT macrophages (Fig. 2a, b), suggesting that NLPR3 is partially involved in caspase-1 activation after Listeria infection and that absence of LPS priming reduces but does not abrogate caspase-1 activation or cell death in these macrophages. Similarly, the flaA Listeria mutant induced a smaller amount of caspase-1 processing and cell death in NLRP3-KO macrophages compared to WT macrophages, possibly through activation of the AIM2 inflammasome, since the flaA Listeria cannot activate NLRC4 because it lacks flagellin, which is thought to be sensed by NLRC4 [32, 33]. Notably, although only the flaA Listeria mutant, but not WT Listeria, was defective in caspase-1 activation in AIM2-KO macrophages, both strains were defective in inducing caspase-1 processing in LPS-nonprimed NLRC4-KO macrophages (Fig. 2a). These NLRC4-KO macrophages were also resistant to WT and flaA Listeria-induced cell death as most NLRC4-KO macrophages were viable after a 5-h infection with the two Listeria strains (Fig. 2b). Taken together, our results indicate that in the absence of prior LPS priming, the NLRC4 inflammasome is critical for caspase-1 activation and caspase-1 dependent cell death in WT Listeria-infected macrophages, whereas NLRP3 and AIM2 inflammasomes contribute downstream of NLRC4 to robust caspase-1 activation but are not essential. However, in flaA Listeria-infected macrophages, the AIM2, but not NLRP3, inflammasome is critical for caspase-1 activation and cell death.
NLRC4 and NLRP3 are required for activation of caspase-1 in Listeria-infected LPS-nonprimed macrophages. a WT, AIM2-KO, NLRC4-KO, and NLRP3-KO macrophages were left untreated or infected for 5 h with LM-WT or flaA Listeria at MOI 50. Culture supernatants and cell lysates were collected after infection and analyzed by immunoblotting with the indicated antibodies. b Release of LDH into culture supernatants of the macrophages in a was assayed with the CytoTox 96 LDH-release kit. Data are representative of at least three experiments
NLRC4 Is Not Critical for WT Listeria-Induced Caspase-1 Activation in LPS-Primed Macrophages
LPS priming has been shown to play critical roles in NLRP3 inflammasome activation [34]. Since WT Listeria-induced inflammasome activation was almost completely impaired in LPS-nonprimed NLRC4-KO macrophages (Fig. 2), we decided to test whether LPS priming alters the response of NLRC4-KO macrophages to WT Listeria infection. To this end, WT, AIM2-KO, NLRC4-KO and NLRP3-KO macrophages were primed with 200 ng/ml of LPS for 4 h prior to infection with WT or flaA Listeria for 5 h. Interestingly, LPS priming greatly enhanced caspase-1 processing and cell death in NLRC4-KO macrophages (Fig. 3), suggesting that prior LPS priming of macrophages can bypass the requirement for NLRC4. Notably, prior LPS priming of AIM2-KO macrophages did not alter their response to flaA Listeria infection, and they showed no caspase-1 activation or cell death after infection with this mutant (Fig. 3a, b). These results indicate that AIM2 is essential for caspase-1 activation and cell death by the flaA Listeria mutant in both LPS-primed and nonprimed macrophages. On the other hand, NLRC4 is only critical for activation of caspase-1 and cell death by WT and flaA Listeria in LPS-nonprimed macrophages (Fig. 2).
LPS priming restores Listeria-induced inflammasome activation in NLRC4-KO macrophages. a WT, AIM2-KO, NLRC4-KO, and NLRP3-KO macrophages were primed with 200 ng/ml of LPS for 4 h before infection for 5 h with WT or flaA deletion mutant Listeria at MOI 50. Culture supernatants and cell lysates were collected after infection and analyzed by immunoblotting with the indicated antibodies. b Release of LDH into culture supernatants of the macrophages in a was assayed with the CytoTox 96 LDH-release kit. Data are representative of at least three experiments
ASC Is Indispensable for Caspase-1 Activation in Listeria-Infected Macrophages
Genetic studies have demonstrated that ASC is a critical adaptor protein for NLRC4, NLRP3, and AIM2 inflammasomes, linking microbial and endogenous ‘danger’ signals to caspase-1 activation [8, 35]. To assess whether ASC is essential for the activation of caspase-1 and cell death in Listeria infection, WT or ASC-KO macrophages were infected with different MOIs (10, 20, and 50) of WT or flaA Listeria for 5 h. Culture supernatants and cell lysates were prepared and immunoblotted with anti-caspase-1 and other antibodies as indicated. Cell death was evaluated by LDH release. WT Listeria induced dose-dependent caspase-1 processing and cell death in WT macrophages (Fig. 4a, b). flaA Listeria also induced dose-dependent caspase-1 processing and cell death in WT macrophages but to a lesser extent (Fig. 4a, b). In contrast, both WT and flaA Listeria did not induce any caspase-1 processing or cell death in ASC-KO macrophages even at MOI 50 (Fig. 4c, d). We therefore concluded that although Listeria-induced caspase-1 activation and cell death is dependent on multiple inflammasomes, ASC appears to be critical for Listeria-induced activation of each of these inflammasomes.
ASC is indispensable for caspase-1 activation in Listeria-infected macrophages. WT (a) or ASC-KO (c) macrophages were left untreated or infected for 5 h with WT or flaA Listeria at the indicated MOI. Culture supernatants and cell lysates were collected after infection and analyzed by immunoblotting with the indicated antibodies. Data are representative of at least three experiments. b and d Release of LDH into culture supernatants of the macrophages in a and c, respectively
Listeria DNA Triggers AIM2 Aggregation
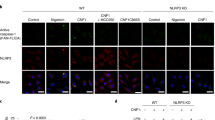
To provide additional support for the involvement of AIM2 in sensing cytoplasmic DNA generated by Listeria infection in macrophages, we engineered NLRP3-KO macrophages to express AIM2-GFP-N1 stably, infected the cells with WT Listeria, and stained for DNA using Hoechst 33342. NLRP3-KO-AIM2-GFP-N1 macrophages were used in our experiments to exclude the possibility of involvement of NLRP3 in the response to cytosolic DNA. AIM2 protein (green) was evenly distributed in the cytoplasm and nuclei of uninfected cells (Fig. 5a). Infection of the cells with Listeria resulted in AIM2 oligomerization (Fig. 5b), as shown by the clustering of AIM2-GFP fusion protein. These AIM2-GFP clusters co-localized with DNA-specific Hoechst 33342 staining, which is consistent with a role for bacterial DNA in AIM2 clustering. The infected NLRP3-KO-AIM2-GFP-N1 macrophages containing the AIM2-DNA clusters showed distinct features of caspase-1 dependent (pyroptotic) cell death.
Cytoplasmic DNA derived from Listeria triggers AIM2 oligomerization. NLRP3-KO-AIM2-GFP-N1 macrophages were left uninfected (a) or infected for 5 h with WT Listeria at MOI 50 (b) and then stained with the DNA-specific blue Hoechst dye. c NLRP3-KO-AIM2-GFP-N1 macrophages were infected for 5 h with Hoechst-labeled WT Listeria at MOI 50. Live cell images were then collected by confocal microscopy. The white arrow in the blue, green, and merged channels indicate the Hoechst-labeled Listeria cytoplasmic DNA. Data are representative of at least three experiments
To evaluate whether cytosolic DNA derived from Listeria was responsible for aggregation of AIM2-GFP, we pretreated Listeria with Hoechst stain prior to infection of NLRP3-KO-AIM2-GFP-N1 macrophages. Infection with the Hoechst-labeled Listeria resulted in aggregation of AIM2-GFP around the Hoechst-labeled DNA (Fig. 5c). Because only Listeria DNA was labeled with Hoechst dye, these results provide direct evidence that Listeria infection is sufficient to promote aggregation of AIM2 and that Listeria DNA colocalizes with AIM2 aggregates. Collectively, our results indicate that cytosolic DNA generated by infection of macrophages with Listeria induces AIM2 aggregation, which leads to the activation of caspase-1 and cell death.
Discussion
Inflammasomes have emerged as the critical signaling molecules of the innate immune system. In this study, we have used AIM2-KO, NLRP3-KO, NLRC4-KO, and ASC-KO macrophages to investigate the roles of the AIM2, NLRP3, and NLRC4 inflammasomes in caspase-1 activation in Listeria-infected macrophages. Although a previous RNA interference knockdown study suggested that AIM2 is dispensable for caspase-1 activation in Listeria-infected human PBMCs [26], our study demonstrated clearly that AIM2 is associated with cytosolic Listeria DNA, as stable AIM2-GFP-N1-expressing NLRP3-KO macrophages infected with the Hoechst-labeled Listeria showed obvious co-localization of AIM2-GFP-N1 with Hoechst-labeled Listeria DNA. These observations suggest that cytoplasmic DNA derived from Listeria binds to AIM2 and induces its aggregation, which leads to caspase-1 activation and cell death as we showed recently in DNA-transfected or F. tularensis-infected macrophages [4, 8]. Our results are consistent with two recent reports, which indicated that Listeria can be partially sensed by the AIM2 inflammasome [9, 25]. In addition, we have used a flagellin-deficient (flaA) Listeria mutant to study the role of Listeria flagellin in caspase-1 activation. We found that flaA Listeria induces very little caspase-1 activation and cell death in AIM2-KO macrophages compared with WT cells, which suggests that Listeria flagellin is essential for AIM2-independent activation of caspase-1 and that the flaA Listeria, like F. tularensis, is only recognized by the AIM2 inflammasome [8].
Cytosolic flagellin has been shown to require NLRC4 for activation of caspase-1 in Salmonella-infected macrophages [32, 33] and in the regulation of Legionella infection [32]. However, the role of NLRC4 in Listeria infection is less clear. Some groups [25, 26] found no evidence that NLRC4 is critical for caspase-1 activation in Listeria-infected macrophages, while others observed a partial role for NLRC4 in caspase-1 activation in Listeria-infected macrophages [21]. In contrast with the results from the above mentioned studies, we found that caspase-1 processing is almost absent in LPS-nonprimed NLRC4-KO macrophages infected with WT and flaA Listeria, which demonstrates a critical role for NLRC4 in caspase-1 activation in Listeria-infected macrophages in the absence of LPS priming (Fig. 6). This could explain the seemingly contradictory results between our findings and the previous studies in which macrophages were primed with LPS prior to infection with Listeria. The WT Listeria strain (10403s) used in our experiments expresses flagellin at 37°C [36], which excludes the possibility that defective expression of flagellin under the conditions of the experiment is responsible for failure to activate caspase-1 in NLRC4-KO macrophages.
Our results show that LPS priming potentiates inflammasome activation by Listeria in NLRC4-KO and NLRP3-KO macrophages (Fig. 3), indicating that prior LPS priming of macrophages can bypass the requirement for NLRC4 and/or NLRP3. Recent studies documented that LPS priming triggered de novo protein synthesis via TLR4, which is a critical prerequisite for inflammasome activation by various danger signals [28, 37]. LPS priming may upregulate inflammasome or inflammasome-related gene expression to bypass upstream NLRC4 signaling. Our results also suggest that LPS priming may obscure the roles of various inflammasomes in regulating caspase-1 activation in response to Listeria infection. For example, while knockout of NLRP3 is not sufficient to abrogate caspase-1 activation by either wild-type or flagellin-deficient Listeria (Fig. 2), it does reduce caspase-1 activation significantly, suggesting that NLRP3 inflammasomes contribute to caspase-1 activation in response to Listeria infection, which is consistent with previous reports [21, 23, 25, 26].
Our finding that NLRC4 knockout completely blocks caspase-1 activation in LPS-nonprimed macrophages (Fig. 2), but that caspase-1 activation can be largely restored by priming with LPS (Fig. 3), suggests strongly that NLRC4 functions upstream of the inflammasome that is primed with LPS (possibly NLRP3, which is known to be upregulated in response to LPS [28]) (Fig. 6). AIM2 is clearly necessary for caspase-1 activation by the flagellin-deficient Listeria mutant (Fig. 1), but is not sufficient to activate caspase-1 in an NLRC4 knockout background (Fig. 2), suggesting that NLRC4 functions upstream of the AIM2 inflammasome as well in LPS-nonprimed macrophages. Taken together, our results suggest that NLRC4 is necessary for priming of the AIM2 and NLRP3 inflammasomes as opposed (or in addition) to functioning as an independent inflammasome that activates caspase-1 in response to Listeria infection (Fig. 6) in LPS-nonprimed macrophages. The mechanism by which NLRC4 primes the AIM2 and NLRP3 inflammasomes is currently unknown, but we can speculate that NLRC4 is required for phagosomal maturation in Listeria-infected macrophages as previously seen with Legionella-infected macrophages [38] (Fig. 6). In NLRC4-KO macrophages, the maturation of the Legionella-containing phagosome is stalled and Legionella escapes degradation [38]. Similarly, we believe that in LPS-nonprimed NLRC4-KO macrophages, Listeria might not be able to induce phagosomal maturation and thereby cannot induce phagosomal destabilization, which is required for activation of the NLRP3 inflammasome by a mechanism reminiscent of the activation of NLRP3 inflammasome by silica crystals [31]. Likewise, phagosomal maturation is required for release of DNA into the cytosol, which is necessary for activation of the AIM2 inflammasome. Our recent results showed that phagosomal maturation is critical for activation of the AIM2 inflammasome by F. tularensis infection in WT macrophages [8]. Therefore, defective phagosomal maturation in LPS-nonprimed NLRC4-KO macrophages might prevent the escape of Listeria DNA into the cytosol, which prevent activation of the AIM2 inflammasome as well. It will be interesting to see if F. tularensis is also defective in activation of the AIM2 inflammasome in LPS-nonprimed NLRC4-KO macrophages.
The mechanism by which LPS priming restores AIM2 and NLRP3 inflammasome activation in NLRC4-KO macrophages could also be dependent on modulation of phagosomal maturation by LPS. Indeed, there is considerable evidence indicating that LPS priming can enhance killing of numerous intracellular pathogens including Listeria, Leishmania, and Mycobacterium through induction of superoxide and nitric oxide production [39], and modulation of phagosome maturation [40–43].
Our results clearly indicate the involvement of at least three different inflammasomes in caspase-1 activation in response to Listeria infection. However, these inflammasomes must somehow translate danger signals (cytoplasmic DNA, flagellin) into activation of caspase-1, maturation of proinflammatory cytokines, and cell death. ASC is an adaptor protein that links upstream receptors involved in pathogen recognition to caspase-1. NLRP3, NLRC4, and AIM2 are all known to interact with ASC [35]. Our data (Fig. 4) indicate that ASC is indispensable for caspase-1 activation in WT and flaA Listeria-infected macrophages. Our results suggest that signaling through NLRP3, NLRC4, and AIM2 inflammasomes in Listeria-infected macrophages converges on caspase-1 activation through agonist-induced assembly of an oligomeric complex of each inflammasome with the common ASC adapter protein.
In summary, all three major inflammasomes (NLRP3, NLRC4, and AIM2) are involved in caspase-1 activation in Listeria-infected macrophages. We provide direct evidence to demonstrate that Listeria DNA specifically binds to AIM2 and induces AIM2 oligomerization, which leads to activation of caspase-1 and cell death. We show that Listeria flagellin is essential for caspase-1 activation in the absence of AIM2 and that NLRC4 likely functions upstream of both AIM2 and NLRP3 in Listeria-infected LPS-nonprimed macrophages as opposed (or in addition) to its function in direct activation of caspase-1. In our working model (Fig. 6), Listeria infection triggers signaling from NLRC4 that primes both AIM2 and NLRP3 inflammasomes, leading to robust caspase-1 activation, which induces inflammation and pyroptosis. Flagellin and possibly other PAMPs serve as the primary agonists for activation of the NLRC4 inflammasome, while cytosolic DNA released from ruptured phagosomes serves as the primary agonist triggering caspase-1 activation by the AIM2 inflammasome. Future studies will focus on further elucidating the details and mechanism of crosstalk between different inflammasomes in Listeria or other pathogen infections.
References
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20.
Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–7.
Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–32.
Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13.
Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8.
Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72.
Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60.
Fernandes-Alnemri T, Yu J, Juliana C, Solorzano L, Kang K, Wu J, et al. The AIM2 inflammasome is critical for innate immunity against F. tularensis. Nature Immunol. 2010;11:385–93.
Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:367–9.
Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–604.
Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–23.
Bielecki J, Youngman P, Connelly P, Portnoy DA. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990;345:175–6.
Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–7.
Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, Nakanishi K, et al. Roles of caspase-1 in Listeria infection in mice. Int Immunol. 2004;16:335–43.
Torres D, Barrier M, Bihl F, Quesniaux VJ, Maillet I, Akira S, et al. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect Immun. 2004;72:2131–9.
Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–4.
Park JH, Kim YG, Shaw M, Kanneganti TD, Fujimoto Y, Fukase K, et al. Nod1/RICK and TLR signaling regulate chemokine and antimicrobial innate immune responses in mesothelial cells. J Immunol. 2007;179:514–21.
Kim YG, Park JH, Shaw MH, Franchi L, Inohara N, Nunez G. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–57.
Machata S, Tchatalbachev S, Mohamed W, Jansch L, Hain T, Chakraborty T. Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J Immunol. 2008;181:2028–35.
Mosa A, Trumstedt C, Eriksson E, Soehnlein O, Heuts F, Janik K, et al. Nonhematopoietic cells control the outcome of infection with Listeria monocytogenes in a nucleotide oligomerization domain 1-dependent manner. Infect Immun. 2009;77:2908–18.
Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–64.
Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–8.
Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32.
Franchi L, Kanneganti TD, Dubyak GR, Nunez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J Biol Chem. 2007;282:18810–8.
Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 Inflammasome. Eur J Immunol. 2010 (in press).
Meixenberger K, Pache F, Eitel J, Schmeck B, Hippenstiel S, Slevogt H, et al. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J Immunol. 2010;184:922–30.
Yu JW, Fernandes-Alnemri T, Datta P, Wu J, Juliana C, Solorzano L, et al. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell. 2007;28:214–27.
Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–91.
Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, et al. BCL-3 and NF-kappaB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem. 2004;279:49995–50003.
Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature. 1985;318:667–70.
Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–56.
Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–82.
Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–75.
Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–30.
Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–7.
Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, Freitag NE, et al. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 2004;6:235–42.
Franchi L, Eigenbrod T, Nunez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–6.
Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, Brady G, et al. Regulation of legionella phagosome maturation and infection through flagellin and host ipaf. J Biol Chem. 2006;281:35217–23.
MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–50.
Yates RM, Hermetter A, Taylor GA, Russell DG. Macrophage activation downregulates the degradative capacity of the phagosome. Traffic. 2007;8:241–50.
Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111(Pt 7):897–905.
Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–6.
Tsang AW, Oestergaard K, Myers JT, Swanson JA. Altered membrane trafficking in activated bone marrow-derived macrophages. J Leukoc Biol. 2000;68:487–94.
Acknowledgments
We thank Drs Junying Yuan (Harvard University) for anti-mouse caspase-1 antibody, Junji Sagara for antibody to mouse ASC, Eicke Latz for the immortalized NLRP3-KO, NLRC4-KO, and ASC-KO macrophages, Daniel Portnoy for Listeria strains, Howard Young for the J2 retrovirus-producing ψCREJ2 cell line and Maria Covarrubias for technical assistance with confocal microscopy and Charles Scott for critical reading of the manuscript. This work is supported by grants from the National Institute of Health (AG14357 and AR055398 to E.S.A.) and a grant from GlaxoSmithKline.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, J., Fernandes-Alnemri, T. & Alnemri, E.S. Involvement of the AIM2, NLRC4, and NLRP3 Inflammasomes in Caspase-1 Activation by Listeria monocytogenes . J Clin Immunol 30, 693–702 (2010). https://doi.org/10.1007/s10875-010-9425-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-010-9425-2