Abstract
Objective
Interleukin (IL)-17 plays an important role in the pathogenesis of asthma. We investigated the association between single-nucleotide polymorphism (SNP) of IL-17 (rs2275913, IL-17 G-152A) and asthma-related traits. Its effect on IL-17 production was also attractive.
Methods
One hundred and sixty eight childhood asthmatic patients, 144 bronchiolitis patients, and 205 healthy controls were recruited in this study. SNP rs2275913 was genotyped by polymerase chain reaction–restriction fragment length polymorphism. Peripheral blood mononuclear cells (PBMCs) from parts of healthy controls with different genotype were isolated and cultured with phytohaemagglutinin (PHA) for detection of IL-17 in the supernatants.
Results
SNP rs2275913 was associated with asthma (P = 0.03) in genotype frequency test. Children with homozygous A were 2.29 times more likely to have asthma than others (95% confidence interval 1.39–3.78, P = 0.001). The strength of associations was moderately higher by allergy comorbidity. Furthermore, SNP rs2275913 A allele was associated with abnormal lung function and serum total IgE in asthmatics, although the production of IL-17 by PHA-induced PBMC seemed to be not different among individuals with different genotypes. The distribution of SNP rs2275913 in bronchiolitis was marginally statistically different with controls and demonstrated a tendency close to that in asthma. Higher Streptococcus pneumoniae and Moraxella catarrhalis detection rates were shown in bronchiolitis patients with homozygous A allele than those with other genotypes (20.8% vs. 3.7%, P < 0.01 and 20.8% vs. 6.2%, P = 0.03).
Conclusion
The preliminary results demonstrate that IL-17 SNP rs2275913 was associated with several asthma-related traits and confers genetic susceptibility to childhood asthma. It may be used to develop markers to assess the risk of asthma, especially in the bronchiolitis population. It may be a potential bridge to connect the bacterial colonization and the onset of asthma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asthma is an immune-mediated disease, known to be associated with excessive T-helper type 2 (Th2) immune responses, characterized by airway hyper-reactivity and eosinophilic inflammation in airway [1, 2]. Recently, it has been observed that neutrophils were particularly prominent in acute, severe exacerbations of asthma, also prominent in childhood asthma [3, 4]. IL-17, which was implicated in neutrophilic inflammation, was involved in the pathogenesis of asthma. A study on animal model indicated that IL-17 was essential during antigen sensitization to establish allergic asthma [5]. The level of IL-17 was elevated in the bronchoalveolar lavage fluid and airway tissues of asthma both in animal model and patients [6–10].
IL-17 is the signature cytokine of Th17, the newly identified subset of CD4+ T-helper cells, which has an indispensable role in the clearance of specific types of pathogens. The pathogenic role of IL-17-producing Th17 cells in autoimmune diseases has been well proven [11]. From a genetic perspective, IL-17 as a candidate gene has also been indicated in the correlation of gene single-nucleotide polymorphisms (SNP) rs2275913 with ulcerative colitis among the Japanese [12] and rheumatoid arthritis among Caucasians [13].
It is well known that asthma is attributed to the interaction of multiple genetic and environmental factors. There have been more than 100 candidate genes, and many were factors in Th2 immune network [14]. Few reports on the correlation of IL-17 polymorphism and asthma were reported so far. IL-17 gene is located on 6p12.1, the genomic region which has been reported to be linked to asthma and asthma-related phenotypes in multiple genome scans [15–17]. In the current study, we investigated the association of selected SNP of IL-17 (rs2275913, G-152A) with childhood asthma and several asthma-related traits. Its effect on the production of IL-17 was also attractive. Given that there could be more potential asthmatic patients in infants hospitalized with bronchiolitis [18], we also tested the distribution of the SNP in bronchiolitis patients and its relationship with the clinical features.
Methods
Sample Composition and Clinical Evaluation
A case–control association study was performed. Five hundred and seventeen subjects were recruited from Children’s Hospital of Chongqing Medical University, including children with asthma (asthma group, n = 168) and bronchiolitis (bronchiolitis group, n = 144) as well as healthy subjects without allergic diseases (control group, n = 205). The Ethics Committee of Chongqing Medical University approved the protocol, and written informed consent was obtained from all participants or their guardians.
Asthma was diagnosed by the presence of characteristic symptoms—recurrent wheezing more than three times and the reversibility of airway obstruction or airway hyperresponsiveness to methacholine indicated by lung function test, based on previously published protocols [19]. The data on lung function [including forced vital capacity (FVC), forced expiratory volume in the first second (FEV1), and peak expiratory flow (PEF) values], skin prick test, and serum total IgE were obtained when asthma was diagnosed or before inhaled corticosteroids was taken regularly. Skin prick testing was conducted with a battery of 13 common aeroallergens, including Dermatophagoides, mix pollen, Aspergillus fumigatus, cat, dog, etc. (ALK-Abelló, Denmark). Histamine (10 mg/ml) was used as positive control and saline as negative control. Skin prick tests were considered positive if at least one wheal reaction of more than 3 mm diameter after subtraction of negative control was observed in skin prick tests, as previously described [20, 21]. Allergic status was based on a positive skin test to at least one allergen of the common aeroallergens. Serum total IgE levels were determined by human IgE enzyme-linked immunosorbent assay (ELISA) kit (BioCheck, USA). A serum IgE level was considered elevated if it exceeded the highest reference value of 150 IU/ml. Healthy subjects without symptoms and history of allergic diseases were enrolled in the study as controls.
The diagnosis of bronchiolitis was made in the presence of a history of upper respiratory tract infection followed by acute onset of respiratory distress with cough, breathlessness and wheeze, and clinical signs of chest overinflation, tachypnoea, rhonchi, or crepitations occurring during a winter epidemic of bronchiolitis, as previously described [22]. Only children with their first episode of wheezing were included. Children with underlying bronchopulmonary dysplasia and congenital heart disease were excluded. With the nasopharyngeal aspirates, RSV was identified by immunofluorescence and bacterial culture was performed. Demographic information and clinical data were recorded on admission.
DNA Extraction and Genotyping
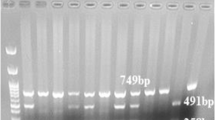
The genomic DNA was prepared from peripheral blood leukocytes (for asthma and control) or sputum cell (for bronchiolitis) by genomic DNA isolation kits (Qiagen, Germany) according to the manufacturer's instructions. A fragment of IL-17 gene including SNP rs2275913 was amplified in MyCycler Thermal Cycler System (BIO-RAD, USA) using the following primers: forward, CAGAAGACCTACATGTTACT; reverse, GTAGCGCTATCGTCTCTCT (designed by software Primer premier 5) in a 20 μl total volume reaction containing 50 ng genomic DNA, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl, 1× PCR buffer, 0.1 mM of each primer (Invitrogen, USA), 0.25 mM dNTPs, and 1 U Taq polymerase (Tiangen, China). Samples were denatured for 5 min at 95°C and then cycled 32 times through the following steps: 30 s at 94°C, 40 s at 58.5°C, and 30 s at 72°C. Following a final 5-min extension at 72°C, the PCR product was cooled to 22°C. The PCR product (20 μl) was then digested at 37°C for a minimum of 4 h using XmnI restriction enzyme (New England Biolabs, USA). Control samples with a known cleavage were included to ensure digestion had occurred. Digests were subsequently run on 2% agarose gel using ethidium bromide to visualize the fragments. The A allele was cleaved by XmnI digestion, while the G allele was not digested, as shown in Fig. 1. The homozygous G allele appeared as a single 344-bp band, the homozygous A allele as bands of 213 and 131 bp, and heterozygotes exhibited all three bands (344, 213, and 131 bp). All genotypes of SNP rs2275913 were confirmed by direct sequencing of the amplified IL-17 gene fragment (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems, USA).
Genotyping by agarose gel electrophoresis of PCR–RFLP. A fragment of IL-17 gene including SNP rs2275913 was amplified and products were digested with XmnI restriction enzyme, subsequently run on 2% agarose gel using ethidium bromide to visualize the fragments. The A allele was cleaved by XmnI digestion while the G allele was not digested. The homozygous G allele appeared as a single 344-bp band, the homozygous A allele as bands of 213 and 131 bp, and heterozygotes exhibited all three bands (344, 213, and 131 bp)
Cell Culture and Cytokine Determination
Six subjects with AA genotype, 12 subjects with AG genotype, and nine subjects with GG genotype were recruited from healthy controls. Peripheral blood mononuclear cells (PBMCs) isolated through Ficoll-Hypaque (Sigma, USA) density centrifugation were washed and resuspended (2.0 × 106 cells/ml) in complete RPMI-1640 medium (GIBCO, USA) containing 0.01 mol/l HEPES (Invitrogen, USA), 100 U/ml penicillin, and 100 U/ml streptomycin supplemented with 10% fetal bovine serum and maintained at 37°C in 5% CO2. Cells were stimulated with 10 μg/ml phytohaemagglutinin (PHA; Sigma, USA) for 72 h and then the supernatants were collected for detection of IL-17 by a commercial ELISA kit (Biosource, USA). The sensitivity for the assay was 2 pg/mL.
Statistical Analysis
The quantitative dates were expressed as mean ± standard deviation (SD). Student’s t test was used to compare the difference. Cytokine levels were expressed as median and minimum to maximum, and non-parametric test was used. The chi-square test was performed to compare proportions of subjects with different clinical features among subjects with different genotypes.
The genotype and allele frequencies were obtained by direct counting. Hardy–Weinberg equilibrium was tested between cases and controls separately. Comparisons of the distributions of the allele and genotype frequencies were performed using the chi-square test. The relative risk associated with rare alleles was estimated as an odds ratio (OR) with a 95% confidence interval (CI). A P value of 0.05 or less was considered statistically significant. All statistical analyses were performed using SPSS version 13.0 and SHEsis software [23].
Results
The Characteristics of the Subjects and the Frequencies of the Genotypes
The mean age was significantly lower in the bronchiolitis group than in the control group. The gender between these two groups was also different (P = 0.001). No difference between the asthma and control group (P > 0.05) was observed. The genotypes were successfully obtained from all of the subjects. Allele and genotype frequencies of bronchiolitis and healthy control subjects were in Hardy–Weinberg equilibrium, respectively, except asthma subjects, as presented in Table I. The A allele and AA genotype frequencies of SNP rs2275913 in the asthma group were significantly higher than that in the control group (P = 0.03, P = 0.003). They were marginally and statistically significantly higher in the bronchiolitis group than in the control group (P = 0.04, P = 0.08).
The Association of SNP rs2275913 in IL-17 Promoter Region with Childhood Asthma
The A allele of SNP rs2275913 was significantly related to the increasing risk of developing childhood asthma (OR = 1.38, 95% CI = 1.03–1.84, P = 0.03). In addition, children with homozygous A allele were 2.29 times more likely to have asthma than others (95% CI 1.39–3.78, P = 0.001). The strength of associations was moderately higher by allergy morbidity (Table II). Non-allergic asthma was not presented because allergy information of all subjects was not completely recorded.
The Association of SNP rs2275913 in IL-17 Promoter Region with Serum Total IgE and Lung Functions of Asthmatic Subjects
No differences in serum total IgE levels and FVC, FEV1, and PEF values were observed among subjects with different genotype (Table III). But when concerning the proportions of subjects with abnormal values of serum total IgE and lung function [IgE > 150 IU/ml; FVC/FEV1/PEF (% predicted value) < 80%] in asthmatic patients, we found that the proportions of subjects with abnormal total serum IgE or FVC and FEV1 values were higher in subjects with the AA genotype than in those with other genotypes (Table IV).
The Influence of SNP rs2275913 in IL-17 Promoter Region on IL-17 Production in PBMCs
PBMCs from subjects with different genotypes were isolated and cultured for 72 h in the presence of PHA. Figure 2 presents the IL-17 levels in the supernatants. There were no significant differences in IL-17 levels among individuals with different genotypes [AA = 47.3 (0–245.6) pg/ml; AG = 8.7 (0–202.0) pg/ml; GG = 83.6 (8.0–141.3) pg/ml] or between AA genotype and AG plus GG genotypes.
The influence of SNP rs2275913 on IL-17 production in PBMC. Six subjects with AA genotype, 12 subjects with AG genotype, and nine subjects with GG genotype were recruited from healthy controls. PBMCs were isolated and cultured in the presence of 10 μg/ml PHA for 72 h. Production of IL-17 in the supernatants was detected by ELISA
The Association of SNP rs2275913 in IL-17 Promoter Region with Bronchiolitis and the Clinical Features
When patients with bronchiolitis were typed for SNP rs2275913, a marginally statistical difference of frequencies with controls was observed (Tables I and II). As shown in Table V, the mean age of the subjects, course of wheezing, and clinical severity were not significantly different in each group. The length of hospital stay was significantly higher in subjects with homozygous A allele than in subjects with other genotypes (7.9 vs 6.3 days; P = 0.02).
When considering virology and bacteriological finding, similar proportional results were obtained from RSV testing among the different genotype groups. A significantly higher Streptococcus pneumoniae and Moraxella catarrhalis detection rate was seen in bronchiolitis subjects with homozygous A allele compared with subjects with other genotypes (20.8% vs 3.7%, P < 0.01 and 20.8% vs 6.2%, P = 0.03).
Discussion
Our results suggested that promoter region polymorphism rs2275913 of IL-17 was associated with asthma phenotype. Children with homozygous A were 2.29 times more likely to have asthma than those with other genotypes (95% CI 1.39–3.78, P = 0.001). The strength of association was moderately higher by allergy comorbidity (OR 2.91, 95% CI 1.52–5.58, P = 0.001). The correlation was further supported by the results that the proportion of subjects with abnormal lung function or serum total IgE was much higher in asthmatic patients with homozygous A than those with other genotypes. Moreover, bronchiolitis subjects with homozygous A were more likely to have S. pneumoniae or M. catarrhalis colonization of the hypopharynx, which were linked to the development of childhood asthma [24].
IL-17 is a proinflammatory cytokine mainly produced by a newly defined Th cell lineage, termed “Th17”. Through its receptor IL-17RA and IL-17RC, IL-17 induces the expression of a series of inflammatory cytokines. It is indispensable in clearing particularly exocellular pathogens and aberrant responses of IL-17 leading to tissue inflammation, promoting autoimmune pathology such as rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, etc. [11]. The levels of IL-17 mRNA and/or proteins were reported to be elevated in the lungs, sputum, and bronchoalveolar lavage fluids from asthmatic patients, and correlated with the severity of neutrophilic inflammation [6–10] and hyperresponsiveness to methacholine in the airway [25], suggesting that the factors that influence IL-17 expression level are candidate factors for asthma susceptibility.
The SNP rs2275913, located at position 152 bp upstream of the starting site of IL-17 mRNA, has been reported as connected with ulcerative colitis among the Japanese [12] and rheumatoid arthritis among Caucasians [13]. Besides, IL-17 SNP rs3804513 in IL-17 intron 2 was associated with the radiographic progression of rheumatoid arthritis among the Japanese [26]. Our results indicated that SNP rs2275913 of IL-17 was associated with asthma and, furthermore, was associated with asthma-related traits. The homozygous A was a risk genotype and connected with abnormal lung function. It was consistent with the effect of IL-17 to potentiate bronchial fibroblast, epithelial cell, and smooth muscle cell activation, and enhances IL-6, IL-8, and other cytokine/chemokine production, then accumulating neutrophils and of which proteolytic enzymes may burden the airway [27]. Our data also demonstrated the association of homozygous A with allergy and elevated serum total IgE in asthmatic children. Allergy is the most important risk factor and indicator of asthma development, and a higher serum total IgE is a robust risk factor for asthma regardless of the specific IgE to allergen [28]. Hashimoto et al. reported that PBMC from atopic asthmatics produced more IL-17 when cultured with the allergen [29]. The implication of IL-17 with allergy is also supported by an animal model indicating that IL-17 was required during antigen sensitization to develop allergic asthma [5].
It is interesting to understand the effect of SNP rs2275913, which confers genetic susceptibility to childhood asthma, on the expression of IL-17. Our research demonstrated that SNP rs2275913 was not related to PHA-induced IL-17 production in PBMCs. To our best knowledge, there were no previous data about the effects of IL-17 variant on IL-17 production levels of PBMCs or other cell types. Notably, many kinds and subtypes of cells existed in PBMCs and the networks of regulating immune response and cytokine production were complicated; in addition, many polymorphisms may influence IL-17 expression. Thus, our protocol was not precise enough to illuminate the function of SNP rs2275913. Illustrating the exact mechanisms of the SNP in regulating IL-17 expression demands further investigations.
In a recent study, J. Y. Wang and co-workers reported a correlation of IL-17 promoter region polymorphism rs8193036, but not rs2275913, with pediatric asthma in Taiwan Han Chinese population [30]. The distribution of SNP rs2275913 in Southwestern Han Chinese population seemed to be different from that in Taiwan Han Chinese population. The samples in our study are few, but probably, there are inheritance variations between the north and south Han Chinese population [31]. In any case, the correlation of SNP rs2275913 with several asthma-related phenotypes in our study also supports the association with asthma.
Infants hospitalized with bronchiolitis are at increased risk of both recurrent wheezing and childhood asthma [18]. It appears rational to suppose that there are more potential asthmatic subjects or more subjects with asthmatic genetic backgrounds in the bronchiolitis group than in healthy controls. Thus, we tested the distribution of SNP rs2275913 in bronchiolitis. They were marginally statistically different than those in the control group and demonstrated a tendency close to those in the asthma group, suggesting that IL-17 SNP rs2275913 was associated with wheeze and might be used to develop markers to assess the risk of recurrent wheeze and asthma, especially in the bronchiolitis population.
Considerable evidences have indicated that RSV infection contributes to the mechanisms of bronchiolitis progressing into asthma [18, 32]. Our research indicated that IL-17 SNP rs2275913 was not connected with RSV but S. pneumoniae and M. catarrhalis detection in the hypopharynx. Bronchiolitis subjects with homozygous A allele were more likely to have S. pneumoniae and M. catarrhalis colonization of the hypopharynx. Currently, Bisgaard and colleagues found that neonates colonized in the hypopharyngeal region with S. pneumoniae, Haemophilus influenzae, M. catarrhalis, or a combination of these organisms were at an increased risk of recurrent wheeze and asthma early in life [24]. The mechanism was unclear. It seemed paradoxical that S. pneumoniae colonization was related to the development of asthma, but IL-17, which has been proven to play a crucial role in clearing the extracellular pathogens [33] and protecting against the S. pneumoniae indispensably [34], was elevated in the airway of asthmatic patients [6–10]. Nevertheless, connecting the SNP rs2275913 with S. pneumoniae and M. catarrhalis detection further indicated the correlation with asthmatic predisposition to some extent.
In conclusion, our preliminary results demonstrated that IL-17 SNP rs2275913 was associated with several asthma-related traits and confer genetic susceptibility to childhood asthma in a southwestern Chinese population. The SNP identified in this study may be used to develop markers to assess the risk of asthma, especially in the bronchiolitis population. It demands expanding the samples to confirm and further investigations to illustrate the effect of SNP rs2275913.
References
Busse WW, Lemanske Jr RF. Asthma. N Engl J Med. 2001;344(5):350–62.
Umetsu DT, McIntire JJ, Akbari O, et al. Asthma: an epidemic of dysregulated immunity. Nat Immunol. 2002;3:715–20.
Douwes J, Gibson P, Pekkanen J, et al. Non-eosinophilic asthma: importance and possible mechanisms. Thorax. 2002;57:643–8.
Luo Z, Xiao L, Liu E. The significance on the cell dynamic changes in the induced sputum in the different periods in children with asthma. J Clin Pediatr. 2005;23(09):615–7. in Chinese.
Schnyder-Candrian S, Togbe D, Couillin I, et al. Interleukin-17 is a negative regulator of established allergic asthma. J Exp Med. 2006;203(12):2715–25.
Oboki K, Ohno T, Saito H, et al. Th17 and allergy. Allergol Int. 2008;57:121–34.
Bullens DM, Truyen E, Coteur L, et al. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Respir Res. 2006;7:135.
Molet S, Hamid Q, Davoine F, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108:430–8.
Chakir J, Shannon J, Molet S, et al. Airway remodeling-associated mediators in moderate to severe asthma: effect of steroids on TGF-β, IL-11, IL-17, and type I and type III collagen expression. J Allergy Clin Immunol. 2003;111:1293–8.
Sun YC, Zhou QT, Yao WZ. Sputum interleukin-17 is increased and associated with airway neutrophilia in patients with severe asthma. Chin Med J. 2005;118:953–6.
Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517.
Arisawa T, Tahara T, Shibata T, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28(1):44–9.
Nordang GB, Viken MK, Hollis-Moffatt JE, et al. Association analysis of the interleukin 17A gene in Caucasian rheumatoid arthritis patients from Norway and New Zealand. Rheumatology. 2009;48(4):367–70.
Koppelman GH, te Meerman GJ, Postma DS. Genetic testing for asthma. Eur Respir J. 2008;32(3):775–82.
Wjst M, Fischer G, Immervoll T, et al. A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics. 1999;58:1–8.
Haagerup A, Bjerke T, Schiotz PO, et al. Asthma and atopy—a total genome scan for susceptibility genes. Allergy. 2002;57:680–6.
Wang JY, Lin CGJ, Bey MSJ, et al. Discovery of genetic difference between asthmatic children with high IgE level and normal IgE level by whole genome linkage disequilibrium mapping using 763 autosomal STR markers. J Hum Genet. 2005;50:249–58.
Singh AM, Moore PE, Gern JE, et al. Bronchiolitis to asthma: a review and call for studies of gene–virus interactions in asthma causation. Am J Respir Crit Care Med. 2007;175(2):108–19.
Polonikov AV, Ivanov VP, Solodilova MA, et al. Promoter polymorphism G-50T of a human CYP2J2 epoxygenase gene is associated with common susceptibility to asthma. Chest. 2007;132(1):120–6.
Sub-Committee on Skin Tests of the European Academy of Allergology and Clinical Immunology. Skin tests used in type I allergy testing Position paper. Allergy. 1989;44 Suppl 10:1–59.
Isidoro-García M, Dávila I, Laffond E, et al. Interleukin-4 (IL4) and interleukin-4 receptor (IL4RA) polymorphisms in asthma: a case control study. Clin Mol Allergy. 2005;3:15.
El-Radhi AS, Barry W, Patel S. Association of fever and severe clinical course in bronchiolitis. Arch Dis Child. 1999;81(3):231–4.
Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–8.
Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–95.
Barczyk A, Pierzchala W, Sozañska E. Interleukin-17 in sputum correlates with airway hyperresponsiveness to methacholine. Respir Med. 2003;97(6):726–33.
Furuya T, Hakoda M, Ichikawa N, Higami K, Nanke Y, et al. Associations between HLA-DRB1, RANK, RANKL, OPG, and IL-17 genotypes and disease severity phenotypes in Japanese patients with early rheumatoid arthritis. Clin Rheumatol. 2007;26(12):2137–41.
Lindén A. Rationale for targeting interleukin-17 in the lungs. Curr Opin Investig Drugs. 2003;4(11):1304–12.
Sherrill DL, Lebowitz MD, Halonen M, Barbee RA, Burrows B. Longitudinal evaluation of the association between pulmonary function and total serum IgE. Am J Respir Crit Care Med. 1995;152:98–102.
Hashimoto T, Akiyama K, Kobayashi N, et al. Comparison of IL-17 production by helper T cells among atopic and nonatopic asthmatics and control subjects. Int Arch Allergy Immunol. 2005;137 suppl 1:51–4.
Wang JY, Shyur SD, Wang WH, et al. The polymorphisms of interleukin 17A (IL17A) gene and its association with pediatric asthma in Taiwanese population. Allergy. 2009;64(7):1056–60.
Shuhua Xu, Xianyong Yin, Shilin Li, et al. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 2009;85(6):762–74.
Kalina WV, Gershwin LJ. Progress in defining the role of RSV in allergy and asthma: from clinical observations to animal models. Clin Dev Immunol. 2004;11(2):113–9.
Bettelli, Korn T, Oukka M, et al. Induction and effector functions of T(H)17 cells. Nature. 2008;453(7198):1051–7.
Lu YJ, Gross J, Bogaert D, et al. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 2008;4(9):e1000159.
Acknowledgments
This work was funded by the New Century Excellent Talents program from the Education Ministry of China (NCET-06-0775). We thank Qi Cheng Zheng for helping us in the collection of blood samples.
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, J., Deng, Y., Zhao, J. et al. The Polymorphism of IL-17 G-152A was Associated with Childhood Asthma and Bacterial Colonization of the Hypopharynx in Bronchiolitis. J Clin Immunol 30, 539–545 (2010). https://doi.org/10.1007/s10875-010-9391-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-010-9391-8