Abstract
The subtropical and tropical regions of the Pacific Ocean are less productive than other oceanic regions. Although particle association should be an important strategy for heterotrophic prokaryotes to survive in such environments, we have little information on particle-associated (PA) prokaryotes in these regions. The specific aim of this study was to determine bacterial and archaeal community structures in the PA assemblage in comparison to the free-living (FL) assemblage in the North Pacific Subtropical Gyre, the South Pacific Subtropical Gyre, and an eastern equatorial region of the Pacific Ocean. Community profiles and phylogenetic identities were obtained by denaturing gradient gel electrophoresis, 454-pyrosequencing, and cloning followed by Sanger sequencing of 16Sr RNA gene amplicons. The distribution patterns of some abundant groups in three regions and two lifestyles (PA and FL) are shown in this study. Also, the PA community structures of bacteria differed from the FL ones and exhibited higher diversity than the FL ones, while the archaeal community structures did not show significant differences between PA and FL assemblages. We found that specific phylotypes of Gammaproteobacteria and Flavobacteria were abundant in PA bacterial assemblages, suggesting that they prefer to attach and consume particulate organic matter. In summary, the surface seawater PA assemblages represent very different bacterial and archaeal community structures between three different oceanic regions, each of which had distinct PA and FL community structures. These results imply that environmental factors determine microbial community structures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Subtropical and tropical ocean surface water is nutrient-depleted because of strong thermal stratification and less productive ecosystems. The southeastern Pacific is extremely oligotrophic and has the clearest seawater in the world, where nutrient concentrations are below detectable levels by standard analytical methods (Claustre and Maritorena 2003; Morel et al. 2007). Recent studies using highly sensitive analytical methods have indicated geographical variability in inorganic macronutrients at the nanomolar level in the oligotrophic ocean (Hashihama et al. 2009). In the North Pacific Subtropical Gyre (NPSG), phosphate concentrations are very low, whereas they are higher in the South Pacific Subtropical Gyre (SPSG) (Moutin et al. 2008, Hashihama et al. 2009). In contrast, inorganic nitrogen and phosphorous concentrations in the tropical ocean of the equatorial Pacific are relatively high as a result of an upwelling system (Murray et al. 1995). However, primary productivity is not high enough to use up abundant macronutrients because of a short supply of iron in the dust from the Eurasian continent, a so-called high-nutrient low-chlorophyll (HNLC) area (Miller and Wheeler 2012).
Marine bacteria mediate nutrient regeneration and thus play a critical role in biogeochemical cycles (Karl 2002). In nutrient-limited surface water, degradation of organic matter and nutrient regeneration by marine microbes is important and more directly linked to phytoplankton primary production than in nutrient-replete parts of the ocean. For example, the f ratio in a subtropical gyre generally ranged from 0.05 to 0.1, where primary production runs mostly on recycled nitrogen with some addition from nitrogen fixation (Miller and Wheeler 2012).
Attachment to particles is the first step in organic matter degradation by marine bacteria. Particle-associated (PA) bacteria exhibit higher enzymatic activity than free-living (FL) bacteria and play an important role in nutrient regeneration (Simon et al. 2002). Some studies have suggested that bacterial community structures are dissimilar between PA and FL assemblages (Acinas et al. 1997; DeLong et al. 1993; Moesender et al. 2001). Bacteroidetes, Planctomycetes, and Gammaproteobacteria species have been abundantly observed in PA fractions from some oceanic regions (DeLong et al. 1993; Crespo et al. 2013; Ortega-Retuerta et al. 2013). Although microbe–particle association is essential in organic matter degradation and nutrient regeneration, little is known about the diversity and function of PA bacteria in tropical and subtropical oceanic environments.
Furthermore, marine archaea are a major component of the marine ecosystem (Massana et al. 1997; Fuhrman and Ouverney 1998). Although relative archaeal abundance in total prokaryotes reportedly increases with depth, a recent study has suggested that heterotrophic archaea, including the uncultured marine Euryarchaeota group II (MGII), play significant roles in protein and lipid degradation with the aid of rhodopsin-based phototrophy in surface seawater (Iverson et al. 2012). Association to particles can be an important mechanism in organic matter degradation by these heterotrophic archaea. However, little is known about the archaea associated with particles, especially in oligotrophic environments. Previous studies have shown no differences in archaeal community structures between PA and FL assemblages (Acinas et al. 1997; Galand et al. 2008); however, Orsi et al. (2015) recently reported that MGII populations in PA fractions are phylogenetically distinct from those in FL fractions.
Intensive time-series observations of microbial community structure in the subtropical Pacific Ocean have been conducted at the Station ALOHA (22°45′N, 158°00′W) (Brown et al. 2009; Giovannoni and Vergin 2012). They have revealed that Alphaproteobacteria (SAR11 and SAR116), Gammaproteobacteria (SAR86), and Cyanobacteria (Prochlorococcus) are abundant bacterial groups and MGII is an abundant archaeal group in surface seawater. These groups were ubiquitous and considered to be adapted to relatively low nutrient oceanic environments. Currently, our knowledge on microbial diversity and community structure in the tropical and subtropical Pacific largely relies on the data obtained from the Station ALOHA (Brown et al. 2009; Giovannoni and Vergin 2012). Although long-term time-series observations have revealed temporal patterns in microbial community structure, diversity, and roles in various biogeochemical processes (Giovannoni and Vergin 2012), we still do not know whether these patterns are applicable to other areas in the tropical and subtropical Pacific.
Recent advances in highly sensitive nutrient analysis have revealed variability in nitrogen and phosphorous concentrations at the nanomolar level across the tropical and subtropical Pacific Ocean. Therefore, we can expect regional differences in microbial diversity and community structures even among similar oligotrophic environments. Previous studies have revealed that there are some Prochlorococcus ecotypes and SAR11 exhibiting specific spatial distributions in the open ocean (Zwirglmaier et al. 2008; Brown et al. 2012). As for the SPSG, some studies have reported on bacterial abundance, production, and community structures using fingerprinting during the BIOSOPE (Biogeochemistry and Optics South Pacific Experiment) project cruise (Lami et al. 2007; Obernosterer et al. 2008; Van Wambeke et al. 2008) and the KNOX02RR expedition (Walsh et al. 2015). Rusch et al. (2007, 2010) found that the Rhodospirillaceae and Prochlorococcus HNLC groups are specific to the equatorial Pacific. However, information on the bacterial and archaeal assemblages in these regions remains scarce. Additionally, we have almost no information about PA prokaryotes in these regions. We know little about what kinds of taxa dominate PA assemblages and whether they form distinctive patterns among different oceanic regions.
The objective of this study was to determine and compare the bacterial and archaeal community structures in PA and FL assemblages in the tropical and subtropical Pacific Ocean. We hypothesized that bacteria and archaea community structures differ among PA assemblages, in which the three oceanic regions, i.e., the NPSG, the SPSG, and an eastern equatorial region, represent distinct PA community structures.
2 Materials and methods
2.1 Sample collection and treatment
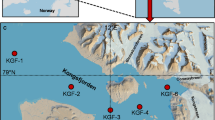
During the R/V “Hakuho-maru” cruises KH-11-10 (December 2011–January 2012) and KH-12-1 (January–March 2012), surface seawater samples were collected in acid-cleaned buckets at nine stations in the NPSG, SPSG, and the equatorial region (EQ) (Fig. 1).
Five liters of seawater were serially filtered through 3.0-µm pore-size Nuclepore polycarbonate membrane filters (Whatman, NJ, USA) and 0.22-µm pore-size Sterivex GS cartridge filter units (Millipore, MA, USA) with a peristaltic pump to obtain PA and FL bacteria, respectively. Immediately after filtration, the membrane and the Sterivex cartridge filter were stored at −80 °C until further analysis. Total DNA extraction was performed using ChargeSwitch® Forensic DNA Purification Kits (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions with slight modifications as described in a previous study (Cui et al. 2015).
2.2 Environmental parameters
Temperature and Salinity were measured with a mercury thermometer and a salinometer (Portasal 8410A; Guildline, ON, Canada), respectively. Samples for chlorophyll a (Chl a) concentration were filtered onto Whatman GF/F filter papers. Chl a concentration was measured by the fluorometric technique with a Turner Design 10-AU fluorometer (Welschmeyer 1994) after extraction with N,N-dimethylformamide (Suzuki and Ishimaru 1990). With the NPSG and SPSG samples, nitrate plus nitrite (N + N) and soluble reactive phosphorus (SRP) concentrations were measured by a highly sensitive colorimetric system with an AutoAnalyzer (AutoAnalyzerII; Technicon, USA) and long capillary guide cells (Liquid Waveguide Capillary Cell; World Precision Instruments, USA) (Hashihama et al. 2009). N + N and SRP concentrations were measured in the EQ samples based on standard methods using an AutoAnalyzer (TRAACS 2000; Bran + Luebbe, UK). The total number of prokaryotes was obtained by the 4′,6-diamidino-2-phenylindole (DAPI) counting technique (Porter and Feig 1980). Seawater samples (50-mL aliquots) were fixed with 2% paraformaldehyde (final concentration) for ~6–12 h at 4 °C in the dark and filtered onto 0.2-µm pore-size polycarbonate filters (Millipore). The filters were stored at −80 °C until processing. Each filter was stained with DAPI mix solution (Wilhartitz et al. 2007). The stained samples were immediately examined for counting with an epifluorescence microscope (Axioplan2 imaging; Zeiss, Germany).
2.3 Polymerase chain reaction (PCR) amplification of 16S rRNA gene
To reveal the prokaryotic community structures in each sample, PCR amplicons of the 16S rRNA gene were subjected to three analytical methods, denaturing gradient gel electrophoresis (DGGE), 454-pyrosequencing, and cloning followed by Sanger sequencing (Table 1). PCR primer sets, amplification conditions, and DGGE conditions are summarized in Tables 2 and supplementary table A1. For PCR amplification of bacterial 16S rRNA gene in the DGGE analysis and the cloning, each 20 µL of the reaction mixture contained 0.25 µM of each primer with 10% (final concentration) 10× Ex Taq buffer, 8% (final concentration) dNTP mixture, 0.4% (final concentration) Taq DNA polymerase (Takara ExTaq Hot Start Version; Takara Biotechnology, Japan), and 1 µL template DNA. For the archaeal 16S rRNA gene amplification, each 20 µL of the reaction mixture contained 0.5 µM of each primer and 2 µL of template DNA, and the rest of the chemicals were the same as the reaction mixture for bacteria. A slightly different reaction mixture was used for 454-pyrosequencing. Each 50 µL of the reaction mixture contained 0.2 µM of each primer with 10% (final concentration) 10× Ex Taq buffer, 10% (final concentration) dNTP mixture, either 0.16 (for V1V2 primers) or 0.4 µg µL−1 (for V3V4 and PrK34 primers) (final concentration) bovine serum albumin (Takara), 0.4–0.8% (final concentration) Taq DNA polymerase (Takara), and 0.4–0.8 ng µL−1 (final concentration) template DNA.
2.4 DGGE analysis
The V3–V4 hypervariable region of the bacterial and archaeal 16S rRNA genes was amplified by touch-down PCR using the specific primer sets shown in Table 2. DGGE of the PCR products was performed with an IngenyphorU DGGE-System (Ingeny International, GP Goes, Netherlands). After electrophoresis, the gels were stained with SYBR Gold for 30 min and photographed with a Versa Doc (Bio-Rad, CA, USA). DGGE band position and intensity were determined using the QuantityOne image analysis software (Bio-Rad) (supplementary Figs. A1, A2). The band detection threshold was 1% in intensity to the most intensive band in a gel.
2.5 454-Pyrosequencing
We used three primer sets to increase the microbial community structure credibility: 27F_mod (Højberg et al. 2005) and 338R (Francés et al. 2004) targeting the bacterial V1–V2 hypervariable region (V1V2), 341F and 805R (Herlemann et al. 2011) targeting the bacterial V3–V4 region (V3V4), and PRK341F and PRK806R (Bowman et al. 2012) targeting the V3–V4 hypervariable region (Prk34) of both the bacterial and archaeal 16S rRNA genes (Table 2). The PCR amplicons were sequenced using a 454 Life Science GS Junior sequencer (Roche). The sequences obtained were quality-checked, trimmed, and binned using the Mothur software package (v.1.36.1) (Schloss et al. 2009). In brief, the samples were denoised less than 25% of average quality score and checked for chimeras using the command, chimera.uchime, implemented in Mothur (Edgar et al. 2011). The resulting high quality sequences were clustered into operational taxonomic units (OTUs) at a 97% identity level. Representative sequences from each OTU were assigned to taxonomic categories in Mothur based on the SILVA database (v.119) (Pruesse et al. 2007) and GreenGenes database (gg_13_5_99) (DeSantis et al. 2006). One of the three primer sets (PRK341F and PRK806R) was applicable to both bacteria and archaea and thus gave archaeal taxonomic assignments and relative abundance for each taxon. Additionally, phylogenic analysis was performed to determine detailed archaeal clades.
To estimate species richness and diversity in each sample, nonparametric abundance-based estimators of the Chao1 index and inverse Simpson index were calculated based on the standardized OTU definition of 97% sequence identity. The standardized OTUs were obtained from sequences randomly re-sampled to the same number of reads across the samples to equalize the sequencing effort.
2.6 Cloning and Sanger sequencing
To obtain detailed information on the phylogenetic diversity of FL and PA bacteria from the three regions, 16S rRNA gene fragment clone libraries were constructed and sequenced. The bacterial 16S rRNA gene was amplified with a universal bacterial primer set: 27F and 1492R (Lane 1991) (Table 2). The 16S rRNA gene amplicon clone libraries were constructed using a TOPO-TA cloning kit (Invitrogen). Positive clones were checked by colony PCR using vector primers (M13f and M13r). Bidirectional sequencing was performed with an automatic sequencer 3730xl (Applied Biosystems, CA, USA) using a universal primer set, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), after sequencing with a BigDye® Terminator Cycle Sequencing Kit (Applied Biosystems). The sequences obtained were clustered at a distance of 0.03 in CD-HIT-EST (Li and Godzik 2006).
2.7 Accession number
Bacterial 16S rRNA gene sequences have been deposited in the DDBJ (DNA Data Bank of Japan; http://www.ddbj.nig.ac.jp/) Sequence Read Archive under the project number DRA005203 (454-pyrosequencing) and accession numbers from LC192963 to LC193128 (cloning and Sanger sequencing).
2.8 Statistical analysis
To reveal differences in community structure among samples and the factor that explained the differences, nonmetric multidimensional scaling (NMDS) with environmental fitting was carried out using a Bray–Curtis distance matrix in the R software v.3.0.3. (R Development Core Team 2012). The relationship between environmental factors and microbial community structures determined from the DGGE banding patterns was tested. A permutation test using “MASS” (Venables and Ripley 2002) and “vegan” (Oksanen et al. 2013) in R was also performed to test whether the clustering of ordination plots was significant.
2.9 Phylogenic analysis and identification of major phylotypes
We conducted the phylogenetic analysis in MEGA v.6 (Tamura et al. 2013) to determine the phylogenetic positions of the sequences obtained from the cloning and Sanger sequencing. Furthermore, the sequences obtained from the 454-pyrosequencing were searched in the database consisting of 16S rRNA gene full-length sequences obtained from the cloning and Sanger sequencing using the local BLAST program (Altschul et al. 1990). When a 454-pyrosequencing OTU matched a Sanger sequencing OTU with >97% identity and represented a relative abundance percent of >1%, the OTU was defined as a major phylotype.
3 Results
3.1 Environmental variables
Sea surface temperature ranged from 21.9 °C in SPSG to 26.7 °C in NPSG (Table 3). Chl a concentration ranged from 0.045 to 0.123 µg L−1 in NPSG, from 0.022 to 0.064 µg L−1 in SPSG, and from 0.166 to 0.370 µg L−1 in EQ (Table 3). N + N and SRP concentrations were lower in NPSG and SPSG than those in EQ (Table 3). N + N concentrations were consistently low in NPSG and SPSG showing 3–7 nM or less. SRP concentrations ranged from 3 to 48 nM in NPSG, and from 115 to 264 nM in SPSG.
3.2 Bacterial and archaeal community structures revealed by DGGE
The DGGE images and identified bands are shown in supplementary Figs. A2 and A3. The NMDS ordination plot with the permutation test (Fig. 2) showed that that region, i.e., NPSG, EQ, and SPSG, made a significant contribution to the differences in bacterial and archaeal community structures in the tropical and subtropical Pacific Ocean (n = 1000, P < 0.05). However, the difference between tropical and subtropical regions did not make a significant contribution to their community sturucture variability (n = 1000, P > 0.05). The PA bacterial communities significantly differed from the FL ones (permutation test, n = 1000, P < 0.05), whereas the PA and FL archaeal communities did not (permutation test, n = 1000, P > 0.1) (Fig. 2).
Ordination plots by nonmetric multidimensional scaling (NMDS) of a bacterial and b archaeal community structures in the tropical and subtropical Pacific Ocean surface waters. DGGE band profiles were used to calculate the distance matrix. Sample name information is shown in Table 1
3.3 Bacterial and archaeal diversity and community structures revealed by 454-pyrosequencing
Taxonomic assignments and relative abundance of each bacterial and archaeal taxon were obtained through 454-pyrosequencing of the 16S rRNA gene (Fig. 3). Three different primer sets were used for the analysis. The number of sequences obtained by each primer set, total OTU numbers, species richness (Chao1), and diversity (inverse-Simpson) indices are shown in Table 4. There was no significant difference in taxon composition at the phylum level among the data from the three primer sets (permutation test; n = 1000, P = 0.58). Additionally, species richness and diversity in PA were always higher than in FL (Table 4).
Figure 3 shows the relative abundance of bacterial taxa at the phylum and class levels. There was no significant difference in taxonomic composition at the phylum level among the data from three primer sets (permutation test; n = 1000, P = 0.58). Alphaproteobacteria was the dominant taxon in the FL bacterial community (FL, 23–65%; PA, 11–44%) in all regions. Cyanobacteria were also abundant in the FL bacterial community in NPSG (28–56%), but were less abundant in SPSG (4–14%). Bacteroidetes (FL, 3–17%; PA, 4–51%) were more abundant in the PA than in the FL bacterial community. Verrucomicrobia (FL, 0.03–2%; PA, 0.05–12%) and Deltaproteobacteria (FL, 0.1–6%; PA, 2–17%) were more prevalent in the PA than in the FL bacterial community; however, neither taxa was found in the data obtained from the V1V2 primer set (Fig. 3).
Archaeal sequences accounted for only 0.02–1.4% of total reads. Phylogenetic analysis showed that most of archaeal OTUs were clustered into four groups: MGII subgroups A and B, and other clades (Fig. A2). The number of archaeal reads obtained in this study was very low, ranging from 1 to 41 per sample. Therefore, we could not test whether there were significant differences in relative abundance of these archaeal subgroups between PA and FL assemblages or between regions.
3.4 Phylogenetic analysis and identification of major phylotypes
A total of 165 clones were obtained from six clone libraries representing three stations (N2, S2, and E2) and PA and FL assemblages (supplementary Fig. A3, supplemntary Table A2). The resulting sequences clustered into 76 OTUs using a 3% cutoff value. The 17 most abundant OTUs (>1% relative abundance per sample) in these clone libraries were also abundant in the pyrosequencing data using three different primer sets (>1% relative abundance observed in at least one sample) (Table A3). The phylogenetic position of these OTUs was also determined based on maximum-likelihood trees (supplementary Figs. A4–A13). These phylotypes were clustered into 13 cultured and uncultured known phylogenetic groups such as SAR11, Prochlorococcus HL/HNLC clade, SAR86, Flavobacteria-NS2, -NS4, -NS9 and -Fluviicola clades, Alteromonas sp., SAR92, SAR116, Roseobacter clade, KI89A and HIMB59 related unclutured alphaproteobacteria clades. Additionally, their preferential region and PA and FL assemblages were defined based on their relative abundance in the 454-pyrosequencing data (Tables 5, 6, supplementary Table A3). Prochlorococcus (OTU02), the SAR11-1 clade (OTU01, OTU06), and the SAR116 clade (OTU21) were abundant in all three regions and both assemblages (PA and FL). The Roseobacter clade (OTU27) was abundant in EQ and both assemblages. The SAR86-1 group (OTU03) was abundant in the SPSG and EQ in FL assemblages. In contrast, the SAR86 unclassified group (OTU39) and SAR116 (OTU60) were abundant only in FL from the SPSG, whereas SAR92 (OTU16), KI89A (OTU36), Flavobacteria NS2 (OTU05), and NS9 (OTU19) were abundant in PA from SPSG. Alteromonas sp. (OTU08) was only abundant in PA from the NPSG. The genus Fluviicola (OTU17) was abundant in PA from the EQ.
4 Discussion
PA bacterial community structures were distinct from the FL ones. Bacterial community structure also differed significantly by region in both PA and FL assemblages (Fig. 2). For bacteria, PA and FL lifestyles were also important in determining community structure. We also found that the PA bacterial community exhibited higher diversity than the FL bacterial community (Table 4). Distinctive community structure and high PA bacterial diversity compared with the FL have been reported in other environments, e.g., a lagoon (LaMontagne and Holden 2003), a lake (Rösel et al. 2012), the Beaufort Sea (Ortega-Retuerta et al. 2013), the Puerto Rico Trench (Eloe et al. 2011), and the Mediterranean Sea (Crespo et al. 2013). The distinct and higher bacterial diversity in PA assemblages compared with that in FL assemblages indicates the existence of particle specialists (Ortega-Retuerta et al. 2013) and the niche associated with micro-gradients in substrate availability on particles (Ganesh et al. 2013). Our results suggest that this is also true in oligotrophic tropical and subtropical oceanic environments.
Previous studies suggested that most PA bacteria have a higher diversity, a different community from FL bacteria, and a unique phylotype even if samples were collected from different location and analyzed by several different methods. In order to collect particle-associated bacteria, previous studies used several different pore sizes of filtration (1.2, 1.6, 2, 3 µm, etc.). Since the size of particles possibly affects associating with bacterial community structures, the results of this study should be compared with those using 3.0-µm pore-size Nuclepore polycarbonate membrane filters to collect PA assemblages. It was reasonable that the studies of Crespo et al. (2013) and Ortega-Retuerta et al. (2013) used the same pore-size filters as this study and obtained similar results, showing distinctive patterns between the PA and FL assemblages. Also, some studies have shown that spatial and temporal variabilities of community structure were even larger than that between PA and FL lifestyles, suggesting the significance of regional difference in prokaryote community structures (Acinas et al. 1997; LaMontagne and Holden 2003). In the Beaufort Sea, possible factors determining regional differences in bacterial community structure were reportedly the quantity and quality of inorganic nutrients and organic matter such as amino acids (Ortega-Retuerta et al. 2013).
Surface seawater in both the NPSG and SPSG were extremely oligotrophic with very low Chl a concentrations. In the EQ, Chl a concentration was also low, but not as low as in the other two regions. In this study, the difference of nitrate plus nitrite (N + N) concentration between subtropical (NPSG, SPSG) and tropical (EQ) regions was obvious (about 1000 times difference). If prokaryotic community structures correlate to N + N concentrations, their difference should be significant between subtropical (NPSG, SPSG) and tropical (EQ) regions. However, this was not the case when we performed permutation analysis of the ordination plots. Significant differences were seen in the community structure when we clustered them into three groups based on regions (NPSG, SPSG, EQ). Although the quality of organic matter likely influences heterotrophic bacterial community structure (Simon et al. 2012), we do not know if it is also found in these regions. Diazotrophs are abundant in the NPSG resulting in SRP depletion under low nitrate concentrations (Hashihama et al. 2009), which may lead to differences in organic matter chemical composition between this region and the SPSG. Also, SRP in surface water was depleted in NPSG but repleted in SPSG, and thus the availability of SRP was quite different between these regions. Bacteria show diverse abilities in acquiring phosphorous. For example, having either a high-affinity phosphate-binding protein or phosphonate C-P lyase system conveys a survival advantage in oligotrophic environments (Dyhrman et al. 2006). A limited supply of SRP may cause the preferential growth of some species that can use more diverse, dilute phosphorous sources than others.
In contrast to bacteria, there was no significant difference between PA and FL archaeal community structures, although the regional difference was significant (Fig. 2). This result contradicts a previous study by Orsi et al. (2015) which reported differences between PA and FL archaea communities. It is currently unclear why archaea did not differ between the PA and FL assemblages in this study. They do not seem to specialize on either the PA or the FL lifestyle. We assume that energy acquisition by rhodopsin phototrophy in MGII, the dominant surface archaea group, might reduce their dependency to heterotrophy or particulate organic matter. Such a mixotrophic nature enables them to survive without the lifestyle specialization found in bacteria.
Because the bacterial and archaeal community structures clustered into three regional groups, one of three stations in each region was selected as a representative station to evaluate the relative abundance of detailed taxonomic groups by means of 454-pyrosequencing. Because the 454-pyrosequencing reads are limited to short fragments, we used three different primer sets to check and compensate taxonomic resolution. The primer set targeting the V3–V4 hypervariable region was useful in comparing the results with those from the DGGE analysis; however, its taxonomic resolution was insufficient to differentiate Prochlorococcus ecotypes. The V1–V2 region differentiated Prochlorococcus ecotypes (supplementary Table A3; supplementary Fig. A5) but could not detect Verrucomicrobia, which was an abundant taxon in PA from both the subtropical and tropical Pacific Ocean (Fig. 3). A previous study also reported the absence of the Verrucomicrobia when using the V1–V2 region in soil environments (Bergmann et al. 2011). We attempted to check primer specificity using Probe Match (Cole et al. 2003). The V3V4 and prk34 primer sets exhibited better matching to higher percentages of Verrucomicrobia sequences in the database of the Ribosomal Database Project (RDP) than the V1–V2 primer sets (supplementary Fig. A14). This was consistent with our 454-pyrosequencing results (Fig. 3). The prk34 primer set has an advantage in reading both bacterial and archaeal sequences at the same time. However, in this study. we found a smaller number of archaeal sequences (0.02–1.4% of total reads) than we expected. More sequencing reads than we have had in this study would be required to have reliable community structures of archaea. Also, it is possible that primer mismatch caused an underestimation of some archaeal species and led to biased results. According to the study of Bowman et al. (2012), in silico analysis of the PRK34 primer set indicated a low-binding efficiency to marine Crenarchaeota.
The distribution patterns of the most abundant groups in three regions and two lifestyles (PA and FL) are shown in Table 5. Three groups, Prochlorococcus, SAR11-1, and SAR116, were found in both PA and FL assemblages in all three regions, suggesting their ubiquity in surface ocean environments, as has been reported in previous studies (Partensky et al. 1999; Morris et al. 2002; Grote et al. 2011). Unlike heterotrophic bacteria, Prochlorococcus, an autotrophic organism, does not have to attach to particles to consume organic matter. However, previous studies have suggested that Prochlorococcus DNA can be detected from sediment traps (Fontanez et al. 2015), indicating that Prochlorococcus attaches to sinking particles. One group, SAR86, primarily found in the SPSG FL, may be specialized to extremely oligotrophic conditions. Six groups, Gammaproteobacteria (SAR92, KI89A, Alteromonas) and Flavobacteria (NS2, NS9, Fluviicola) were abundant in PA, suggesting their niche preference for particle association. Many previous studies have already been suggested that Gammaproteobacteria and Flavobacteria were specialized to particles (e.g., DeLong et al. 1993). However, an interesting finding of this study is that there are some specific subgroups in these taxa. For example, SAR92, KI89A, Alteromonas (Gammaproteobacteria), NS2, NS9, and Fluviicola (Flavobacteria) exhibited different patterns of relative abundance between regions. Most of these subgroups were uncultured; therefore, further studies are required to elucidate the physiological differences among these potential ecotypes.
In conclusion, the PA microbial assemblages in surface seawater represent largely different bacterial and archaeal community structures, in which three contrasting oceanic regions, i.e., the NPSG, the SPSG, and an eastern equatorial region, exhibit distinct PA and FL community structures. Environmental factors such as organic and inorganic nutrient concentrations may have caused these differences. In future studies, increasing the 16S rRNA gene amplicon sequence data points will help to determine environmental factors significantly related to the change of prokaryotic community structures. Also, a comparative metagenomic analysis between these three regions or between PA and FL fractions may enable us to relate prokaryotic functional potentials and environmental factors or lifestyles.
References
Acinas SG, Rodríguez-Valera F, Pedrós-Alió C (1997) Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rDNA. FEMS Microbiol Ecol 24:27–40
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N (2011) The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455
Bowman JS, Rasmussen S, Blom N, Deming JW, Rysgaard S, Sicheritz-Ponten T (2012) Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J 6:11–20
Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, Fuhrman JA, Donachie SP (2009) Microbial community structure in the North Pacific ocean. ISME J 3:1374–1386
Brown MV, Lauro FM, Demaere MZ, Muir L, Wilkins D, Thomas T, Riddle MJ, Fuhrman JA, Andrews-Pfannkoch C, Hoffman JM, McQuaid JB, Allen A, Rintoul SR, Cavicchioli R (2012) Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8:595
Casamayor EO, Massana R, Benlloch S, Øvreas L, Díez B, Goddard VJ, Gasol JM, Joint I, Rodríguez-Valera F, Pedrós-Alió C (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4:338–348
Claustre H, Maritorena S (2003) The many shades of ocean blue. Science 302:3089–3121
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Crespo BG, Pommier T, Fernández-Gómez B, Pedrós-Alió C (2013) Taxonomic composition of the particle-attached and free-living bacterial assemblages in the Northwest Mediterranean Sea analyzed by pyrosequencing of the 16S rRNA. Microbiologyopen 2(4):541–552
Cui Y, Suzuki S, Omori Y, Wong SK, Ijichi M, Kaneko R, Kameyama S, Tanimoto H, Hamasaki K (2015) Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical Pacific Ocean. Appl Environ Microbiol 81:4184–4194
DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38:924–934
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brondie EL, Keller K, Huber T, Dalevi F, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Envrion Microbiol 72:5069–5072
Dyhrman ST, Chappell PD, Haley ST, Moffet JW, Orchard ED, Waterbury JB, Webb EA (2006) Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439:68–71
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH (2011) Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ Microbiol 3(4):449–458
Fontanez KM, Eppley JM, Samo TJ, Karl DM, DeLong EF (2015) Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front Microbiol 6:469
Francés R, Benlloch S, Zapater P, González JM, Lozano B, Muñoz C, Pascual S, Casellas JA, Uceda F, Palazón JM, Carnicer F, Pérez-Mateo M, Such J (2004) A sequential study of serum bacterial DNA in patients with advanced cirrhosis and ascites. Hepatology 39(2):484–491
Fuhrman JA, Ouverney CC (1998) Marine microbial diversity studied via 16S rRNA sequences: cloning results from coastal waters and counting of native archaea with fluorescent single cell probes. Aquat Ecol 32:3–15
Galand PE, Lovejoy C, Pouliot J, Vincent WF (2008) Heterogeneous archaeal communities in the particle rich environment of an arctic shelf ecosystem. J Mar Syst 74:774–782
Ganesh S, Parris DJ, DeLong EF, Stewart FJ (2013) Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J 8:187–211
Giovannoni SJ, Vergin KL (2012) Seasonality in ocean microbial communities. Science 335:671–676
Grote J, Bayindirli C, Bergauer K, Carpintero de Moraes P, Chen H, D’Ambrosio L, Edwards B, Fernández-Gómez B, Hamisi M, Logares R, Nguyen D, Rii YM, Saeck E, Shutte C, Widner B, Church MJ, Steward GF, Karl DM, DeLong EF, Eppley JM, Schuster SC, Kyrpides NC, Rappé MS (2011) Draft genome sequence of strain HIMB100, a cultured representative of the SAR116 clade of marine Alphaproteobacteria. Stand Genomic Sci 5:269–278
Hashihama F, Furuya K, Kitajima S, Takeda S, Takemura T, Kanda J (2009) Macro-scale exhaustion of surface phosphate by dinitrogen fixation in the western North Pacific. Geophys Res Lett 36:L03610
Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5(10):1571–1579
Højberg O, Canibe N, Poulsen HD, Hedemann MS, Jensen BB (2005) Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets. Appl Environ Microbiol 71:2267–2277
Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV (2012) Untangling genomes from metagenomes: revealing an uncultured class of marine euryarchaeota. Science 335:587–590
Karl DM (2002) Nutrient dynamics in the deep blue sea. Trends Microbiol 10:410–418
Lami R, Cottrell MT, Ras J, Ulloa O, Obernosterer I, Claustre H, Lebaron P (2007) High abundances of aerobic anoxygenic photosynthetic bacteria in the South Pacific Ocean. Appl Environ Microbiol 73:4198–4205
LaMontagne MG, Holden PA (2003) Comparison of free-living and particle-associated bacterial communities in a coastal lagoon. Microb Ecol 46:228–237
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acids techniques in bacterial systematics. Wiley, Chichester, pp 115–147
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Massana R, Murray AE, Preston CM, DeLong EF (1997) Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl Environ Microbiol 63:50–56
Miller BC, Wheeler PA (2012) Biological oceanography. 2nd edn. Wiley-Blackwell, Chichester, West Sussex, p 464
Moesender M, Winter C, Herndl GJ (2001) Horizontal and vertical complexity of attached and free-living bacteria in the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46:95–107
Morel A, Huot Y, Gentili B, Werdell PJ, Hooker SB, Franz BA (2007) Examining the consistency of products derived from various ocean color sensors in open ocean (Case 1) waters in the perspective of a multi-sensor approach. Remote Sens Environ 111:69–88
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810
Moutin T, Karl DM, Duhamel S, Rimmelin P, Raimbault P, Mooy BASV, Claustre H (2008) Phosphate availability and the ultimate control of new nitrogen input by nitrogen fixation in the tropical Pacific Ocean. Biogeosciences 5:95–109
Murray JW, Johnson E, Garside C (1995) A US JGOFS process study in the Equatorial Pacific (EqPac): Introduction. Deep-sea Res Part II 42:275–293
Obernosterer I, Catala P, Lami R, Caparros J, Ras J, Bricaud A, Dupuy C, Van Wambeke F, Lebaron P (2008) Biochemical characteristics and bacterial community structure of the sea surface microlayer in the South Pacific Ocean. Biogeosciences 5:693–705
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R package version 2.0-7. http://www.cran.r-project.org
Orsi WD, Smith JM, Wilcox HM, Swalwell JE, Carini P, Worden AZ, Santro AE (2015) Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J 9:1747–1763
Ortega-Retuerta E, Joux F, Jeffrey WH, Ghiglione JF (2013) Spatial variability of particle-attached and free-living bacterial diversity in surface waters from the Mackenzie River to the Beaufort Sea (Canadian Arctic). Biogeosciences 10:2747–2759
Partensky P, Hess WR, Vaulot D (1999) Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63:106–127
Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943–948
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
R Development Core Team (2012) R: a language and Environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org. ISBN 3-900051-07-0
Rösel S, Allgaier M, Grossart HP (2012) Long-Term characterization of free-living and particle-associated bacterial communities in lake Tiefwaren reveals distinct seasonal patterns. Microb Ecol 64:571–583
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, HoVman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC (2007) The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical PaciWc. PLoS Biol 5:e77
Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC (2010) Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA 107:16184–16189
Schäfer H, Muyzer G (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. In: Paul J (ed) Methods in microbiology, marine microbiology. Academic, London, pp 425–468
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Simon M, Grossart HP, Schweitzer B, Ploug H (2002) Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28:175–211
Simon M, Billerbeck S, Kessler D, Selje N, Schlingloff A (2012) Bacterioplankton communities in the Southern Ocean: composition and growth response to various substrate regimes. Aquat Microb Ecol 68:13–28
Suzuki R, Ishimaru T (1990) An improved method for the determination of phytoplankton chlorophyll using N,N-dimethylformamide. J Oceanogr Soc Jpn 46:190–194
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Van Wambeke F, Bonnet S, Moutin T, Raimbault P, Alarcon G, Guieu C (2008) Factors limiting heterotrophic bacterial production in the southern Pacific Ocean. Biogeosciences 5:833–845
Venables WN, Ripley BD (2002) Modern applied statistics with S-Plus, 4th edn. Springer, New York
Walsh EA, Smith DC, Sogin ML, D’Hondt SL (2015) Bacterial and archaeal biogeography of the deep chlorophyll maximum in the South Pacific Gyre. Aquat Microb Ecol 75:1–13
Welschmeyer NA (1994) Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol Oceanogr 39:1985–1992
Wilhartitz I, Mach RL, Teira E, Reinthaler T, Herndl GJ, Farnleitner AH (2007) Prokaryotic community analysis with CARD-FISH in comparison with FISH in ultra-oligotrophic ground- and drinking water. J Appl Microbiol 103:871–881
Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ (2008) Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10:147–161
Acknowledgements
We thank the captain and crew of the R/V “Hakuho-maru” for assistance with sample collection. We also thank the Center for Omics and Bioinformatics, Graduate School of Frontier Sciences, The University of Tokyo, for the 454-pyrosequencing. This research was supported by JSPS KAKENHI Grant Numbers 24121004 and 24121003, as part of the NEOPS (New Ocean Paradigm on Its Biogeochemistry, Ecosystem and Sustainable Use) project in Grants-in-Aid for Scientific Research on Innovative Areas.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Suzuki, S., Kaneko, R., Kodama, T. et al. Comparison of community structures between particle-associated and free-living prokaryotes in tropical and subtropical Pacific Ocean surface waters. J Oceanogr 73, 383–395 (2017). https://doi.org/10.1007/s10872-016-0410-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10872-016-0410-0