Abstract
The white shrimp Penaeus (Litopenaeus) vannamei is the most economically important crustacean species cultivated in the Western Hemisphere. This crustacean shifts its metabolism to survive under extreme environmental conditions such as hypoxia, although for a limited time. Glucose-6-phosphatase (G6Pase) is a key enzyme contributing to maintain blood glucose homeostasis through gluconeogenesis and glycogenolysis. To our knowledge, there are no current detailed studies about cDNA or gene sequences of G6Pase from any crustacean reported. Herein we report the shrimp P. (L.) vannamei cDNA and gene sequences. The gene contains seven exons interrupted by six introns. The deduced amino acid sequence has 35% identity to other homolog proteins, with the catalytic amino acids conserved and phylogenetically close to the corresponding invertebrate homologs. Protein molecular modeling predicted eight transmembrane helices with the catalytic site oriented towards the lumen of the endoplasmic reticulum. G6Pase expression under normoxic conditions was evaluated in hepatopancreas, gills, and muscle and the highest transcript abundance was detected in hepatopancreas. In response to different times of hypoxia, G6Pase mRNA expression did not change in hepatopancreas and became undetectable in muscle; however, in gills, its expression increased after 3 h and 24 h of oxygen limitation, indicating its essential role to maintain glycemic control in these conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glucose-6-phosphatase (G6Pase) (EC 3.1.3.9) is a gluconeogenic and glycogenolytic enzyme that removes phosphate from glucose-6-phosphate (G6P) to release glucose into the bloodstream in the final step of each pathway (van Schaftingen and Gerin 2002). G6Pase is found mainly in liver and kidney of vertebrates and plays the important role of providing glucose during starvation. Unlike most phosphatases acting on water-soluble compounds, it is a membrane-bound enzyme associated with the endoplasmic reticulum. In 1975, W. Arion and co-workers proposed a model for the G6Pase as an unspecific phosphatase with its catalytic site oriented towards the lumen of the endoplasmic reticulum (ER) (Arion et al. 1975), and that the substrate was provided by a translocase specific for G6P, thereby accounting for the specificity of the phosphatase for G6P in intact microsomes. Distinct transporters would allow inorganic phosphate and glucose to leave the vesicles. At variance with this substrate-transport model, other models propose that conformational changes play an important role in the properties of G6Pase. The last 10 years have witnessed important progress in the knowledge of the glucose-6-phosphate hydrolysis system. The genes encoding G6Pase and the glucose-6-phosphate translocase have been cloned and shown to be mutated in glycogen storage disease type Ia and type Ib, respectively. A gene encoding a G6Pase-related protein is specifically expressed in pancreatic islets. Specific potent inhibitors of G6Pase and of the glucose-6-phosphate translocase have been synthesized or isolated from microorganisms. This information as well as other findings support the model initially proposed by Arion. Much progress has also been made about the regulation of the expression of G6Pase by insulin, glucocorticoids, cAMP and glucose (van Schaftingen and Gerin 2002).
G6Pase is necessary to maintain glucose homeostasis and regulates G6P reserves in response to low blood glucose levels or during high energy demands in mammals (Trandaburu 1977; Surholt and Newsholme 1981; Hah et al. 2002; Varga et al. 2019). There is limited information about G6Pase in invertebrates. In the crab Carcinus maenas, the G6Pase enzyme from the digestive gland (hepatopancreas) has high metabolic plasticity and low specificity and plays an important role in systemic glucose homeostasis (Loret and Devos 1992). Changes in G6Pase have been reported in invertebrates. For example, G6Pase activity increased during fasting in hepatopancreas in the crab Menippe mercenaria, without changes in muscle (Scholnick et al. 2006). In contrast, G6Pase decreased in hepatopancreas of WSSV infected shrimp Penaeus monodon (Mathew et al. 2007), while in the chordate amphioxus Branchiostoma japonicum, its expression and activity increased in the digestive diverticulum during fasting (Wang et al. 2015); thus, it is important to understand the specific regulation and G6Pase contribution to maintain cellular processes (Hah et al. 2002).
As mentioned before, the integral membrane G6Pase activity is in the lumen of the ER formed by a complex of the catalytic subunit and a transporter of G6P, producing glucose and inorganic phosphate (Pi) (Marcolongo et al. 2013). Most G6Pases genes have five or six exons with sizes from 800 to 6,000 bp; in humans, the genes are located at 17q21 in the genome (Wang et al. 2015). There are three isoforms of G6Pase in vertebrates and they share structural features, but generally differ in organ expression in specific conditions (Marcolongo et al. 2013).
Low dissolved oxygen (DO) or hypoxia is one of the most essential variables that affect aquatic environments. The DO at which the animals begin to suffer hypoxia varies between species, but generally, adverse effects such as reduced feeding and growth rate, decrease culture yield, and mass mortality of crustaceans may appear when the DO concentration falls below 2 mg/L (Diaz 2001; Wu 2002); however, most organisms have biochemical and physiological mechanisms to respond to oxygen limitations for survival.
The shrimp P.(L.) vannamei is one of the most important crustacean species for global aquaculture, generating high economic resources for shrimp farming countries (Bostock et al. 2010). This shrimp species is an oxyconformer organism that can adjust its metabolism depending on the oxygen available to survive (Wannamaker and Rice 2000) and can withstand DO concentrations as low as 0.2 mg/L for a short time (Pérez-Rostro et al. 2004).
Several studies have reported that shrimp exposure to hypoxia changes the expression of genes encoding enzymes of glycolysis and the Krebs cycle pathways (Ulaje et al. 2019). In addition, the activity of enzymes of anaerobic metabolism is increased as a strategy to maintain the energy balance to survive hypoxic stress (Soñanez-Organis et al. 2010, 2011; Cota-Ruiz et al. 2015; Camacho-Jiménez et al. 2018). Furthermore, higher expression of the genes for the gluconeogenic enzymes fructose-1,6-bisphosphatase (FBP), (Cota-Ruiz et al. 2015, 2016) and phosphoenolpyruvate carboxykinase (PEPCK) (Reyes-Ramos et al. 2018) occurs in hepatopancreas in response to prolonged periods of hypoxia in a organ-specific manner. This is an indication that the gluconeogenic pathway is stimulated. Likewise, the consumption of glycogen reserves in crustaceans due to hypoxia, and its subsequent restoration in post-hypoxic recovery through the recycling of lactate generated in anaerobic metabolism, indicates that these animals are capable of performing both, synthesis and catabolism of glycogen (Ellington 1983; Hervant et al. 1999). However, the metabolic adaptation mechanisms have just started to be unraveled.
Current knowledge about the expression and regulation of G6Pase gene in invertebrates is minimal and, to our knowledge, there are no profound studies about G6Pases genes sequences in crustaceans. In this work, we report the gene structure, coding sequence (CDS), phylogenetic analysis, topology prediction and gene expression in three organs under normoxic conditions and during different periods of hypoxia in hepatopancreas and gills of G6Pase from the white shrimp P. (L). vannamei to contribute to the knowledge of crustacean shrimp energy metabolism and their responses to hypoxia.
Materials and methods
G6Pase cDNA sequence
Total RNA was extracted from gills of healthy juvenile shrimp using TRI® (Sigma-Aldrich, San Luis, Missouri, USA) according to the manufacturer instructions and to remove genomic DNA (gDNA) traces, it was treated with DNase I (Roche, Mannheim, Germany). RNA integrity was confirmed by 1% gel agarose. The first-strand cDNA synthesis from RNA was done using the QuantiTect Reverse transcription system (Qiagen, Hilden, Germany). For this, 1000 ng of total RNA were reverse transcribed with RT Primer Mix. Specific primers (Table 1) were designed for our study for the G6Pase sequence from the partially annotated P. (L.) vannamei genome (Zhang et al. 2019) and transcriptomic data (Ghaffari et al. 2014) using Primer3 (https://primer3.ut.ee/) and Oligo Calc (http://biotools.nubic.northwestern.edu/OligoCalc.html).
The complete coding sequence (CDS) was obtained by PCR using the G6PF2/G6PR2 primers. For a 20 µL reaction, the mix contained 10 µL Hot Start Taq 2 × Master Mix (New England BioLabs, Ipswich, Massachusetts, USA), 500 nM of Fw and Rv primers, 6 µL milliQ water, and 2 µL of cDNA (equivalent to 50 ng of total RNA) and was amplified under the following conditions: 94 °C for 3 min, followed by 94 °C for 30 s, 55 °C for 1 min and 68 °C for 1 min and 40 s per cycle for 42 cycles and finally, 72 °C for 10 min in a PTC 200 Thermal Cycler (Bio-Rad, Hercules, California, USA). Both strands of the PCR products were sequenced in the Laboratory of Genomic Analysis and Technology Core of the University of Arizona (Tucson, Arizona, USA) by the dideoxy chain-termination method. cDNA sequence and deduced protein were compared to nucleotide and protein GenBank databases using the BLAST algorithm (Altschul et al. 1990) at the National Center for Biotechnology Information, Bethesda, MD. CDS analyses were done using ExPASy (http://web.expasy.org/translate/), Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) bioinformatics tools.
G6Pase gene cloning and structure
Genomic DNA was isolated from the muscle of a healthy juvenile shrimp using proteinase K digestion, phenol–chloroform extraction, and ethanol precipitation (Bradfield and Wyatt 1983). The complete G6Pase gene sequence was obtained by overlapping two PCR fragments, the 5’ fragment using G6PF2/qG6PR1 primers and the 3' fragment with qG6PF1/G6PR2 primers (Table 1). The reaction was done using 12.5 µL of Hot Start Taq 2 × Master Mix, 500 nM of each Fw and Rv primer, 25 ng of gDNA, and 9.5 µL of milliQ water to a final volume of 25 µL under the following conditions: a first denaturing step of 75 °C for 15 min and 94 °C for 3 min, followed by 94 °C for 30 s, 59 °C for 1 min and 68 °C for 2 min per cycle for 36 cycles and finally, 72 °C for 10 min in a PTC-200 Thermal Cycler. PCR products were cloned in the pGEM-T Easy Vector System I (Promega, Madison, WI, USA) and used to transform TOP10 Escherichia coli competent cells. Recombinant plasmids were sequenced thoroughly in both DNA strands. The exon–intron identification was done with Spling (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi) and SnapGene (GSL Biotech).
Structural and phylogenetic analysis of protein sequence
Molecular weight and isoelectric point were calculated with ExPASy (https://web.expasy.org/compute_pi/). Search for domains and conserved motifs was done with Pfam 32.0 (https://pfam.xfam.org/). The topological model of the protein was predicted with the HMMTOP transmembrane topology prediction server (Tusnady and Simon 2001) and the representation in two dimensions was constructed with TMRPrest2D (Spyropoulos et al. 2004). The amino acid sequences alignment included 25 G6Pase amino acid sequences obtained from the NCBI database (Table 2) and was performed in CLUSTAL W (Larkin et al. 2007). The phylogenetic analyses were conducted using MEGA version X (Kumar et al. 2018) and the Maximum Likelihood method, based on the Le Gascuel model (Le and Gascuel 2008) with 1000 bootstrap replicates. All positions containing gaps and missing data were eliminated and the tree was rooted with two G6Pase halobacterial sequences from Haloferax gibbonsii and Haloarcula hispanica.
Hypoxia bioassay
Juvenile shrimp obtained from a local farm were acclimated in 100 L tanks connected to a closed circulation filtration system with seawater under normoxic conditions [5 mg DO/L], constant salinity and temperature (35 ppt and 28 °C, respectively), in the Laboratory of Marine Invertebrate Physiology. The shrimp were transported to the laboratory as postlavae and maintained in quarantine for approximately two months until they reached the appropriate size. In this period, the animals were analyzed by molecular and microbiological tests (PCR and bacteriological count) to discard White Spot Syndrome Virus (WSSV), Taura Syndrome Virus (TSV), Infectious Hypodermal Hematopoietic Necrosis Virus (IHHNV), Vibrio parahaemolyticus and Hepatobacter penaei infected animals. Shrimp were fed with commercial feed equivalent to 3% of their dry weight in three portions per day with daily removal of residual food and feces and constant monitoring of temperature, salinity, ammonia, and dissolved oxygen. Then, the shrimp (13.8 ± 1.4 g) were randomly distributed in 4 aquaria (11–12 shrimp/tank) and kept for three days in these tanks. Before the test, the shrimp were fed only once, and afterward, a total exchange of seawater was done in each aquarium. The molting stage of the shrimp was not identified but clearly recently molted specimens were discarded. The following treatments were performed: 0 h normoxia (control at the beginning of the test) [4.99 ± 0.16 mg DO/L, 28 ± 0.1 °C], 3 h hypoxia [1.5 ± 0.2 mg DO/L, 27.4 ± 0.2 °C], 24 h hypoxia [1.5 ± 0.2 mg DO/L, 27.2 ± 0.2 °C], and 48 h hypoxia [1.5 ± 0.3 mg DO/L, 27.1 ± 0.3 °C] in separate tanks. Normoxia was maintained by air bubbling with air diffuser stones, while hypoxia was produced by bubbling nitrogen decreasing the oxygen concentration progressively for 1 h. Dissolved oxygen and temperature were monitored every hour with a portable oximeter (model YSI proODO, Yellow Spring, OH, USA) and adjusted when necessary. Ammonia was monitored daily, and if necessary, water exchanges were done to keep levels below 3.0 mg/L. After time elapsed, 4–5 shrimp were sampled per treatment starting with the control group. The organs of each shrimp were dissected and placed in a cold microtube containing 10 volumes of TRI reagent (w/v), immediately frozen in a dry ice/96% ethanol mixture, and then stored at -80 °C until the analysis.
G6Pase mRNA quantification
G6Pase expression in normoxia was evaluated in hepatopancreas, gills, and muscle, while hepatopancreas and gills were used to analyze the effect of hypoxia by RT-qPCR (see below). Total RNA was isolated from 50 mg of each organ and treated with DNase I to remove gDNA remnants. The qG6PF2/qG6PR2 primers were used to amplify the G6Pase transcript to generate a 307 bp amplicon. One qPCR product per organ was sequenced in both directions to confirm the specificity of the quantification. The ribosomal protein L8 (GenBank accession no. DQ316258.1), that has been previously used as reference gene to normalize expression data for hypoxia studies in shrimp (Cota-Ruiz et al. 2015), was used with the primers L8F2/L8R2 previously designed in our laboratory to amplify a 166 bp fragment (Trasviña-Arenas et al. 2013) (Table 1). The G6Pase standard curve was performed using as a template a 765 bp purified PCR product previously obtained with the qG6PF2/G6PR2 primers with 1:10 serial dilutions (from 8 × 10–5 to 8 × 10–9 ng/µL) while the L8 standard curve was performed with serial dilutions 1:5 (from 25 to 4 × 10–2 ng/µL derived from total RNA) of gills cDNA to calculate amplification efficiency. As mentioned before, a 307 bp fragment was used to quantitate expression for G6Pase and a 166 bp fragment for ribosomal protein L8. cDNAs were synthesized for each organ using 500–750 ng of total RNA as a template with the QuantiTect Reverse Transcription Kit. Four technical replicates (two cDNA reactions for each RNA and two PCR reactions per cDNA) were analyzed in 4–5 shrimp per treatment. Due to the low abundance of G6Pase mRNA in hepatopancreas in hypoxia, the expression experiments were done in two stages. For normoxia expression in hepatopancreas, gills and muscle, and hypoxia in gills in the first stage, each qPCR reaction included 3 µL of 5 × HOT FIREPol® EvaGreen® (Solis BioDyne, Tartu, Estonia), 500 or 700 nM of each primer for G6Pase or L8, respectively, 1 µL of cDNA (equivalent to 25 ng of total RNA) and nuclease-free water for a final volume of 15 µL. For hypoxia expression in hepatopancreas in the second stage, each qPCR reaction included 7.5 uL of 2 × Sso Advanced universal SYBR® Green supermix (Bio-Rad, Hercules, CA, USA), 500 or 600 nM of each primer for G6Pase or L8, respectively, 2 µL of cDNA (equivalent to 100 ng of total RNA) and nuclease-free water for a final volume of 15 µL; a non-template control reaction was included in each run to check PCR contamination. The reactions were performed in a CFX96 Touch Real-Time PCR Detection System thermal cycler (Bio-Rad) with an initial cycle of 95 °C for 12 min, followed by 43 cycles of 95 °C for 15 s, 60.2 °C for 20 s and 72 °C for 20 s in the first stage, and the protocol for the second stage was an initial cycle of 94 °C for 3 min, followed by 45 cycles of 95 °C for 15 s, 60.2 °C for 35 s and 72 °C for 55 s. Finally, a melting curve analysis was done with a gradual increase of temperature (0.5 °C/3 s) from 65 °C to 95 °C to examine the presence of primers dimers or non-specific amplifications. The expression of G6Pase relative to L8 was calculated with the 2−ΔCt method (Schmittgen and Livak 2008).
Statistical analysis
Data are presented as means ± SD or medians and ranges as indicated. The distribution of data was tested with the Shapiro–Wilk normality test. Homogeneity of variance was verified with Levene or Brown-Forsythe test. Data of G6Pase mRNA expression in different shrimp organs in normoxia were analyzed using one-way ANOVA and differences between means were detected with Tukey test. Because data of hypoxia effect in G6Pase mRNA expression in hepatopancreas and gills fail in normality assumption, Kruskal Wallis and Dunn tests were performed to assess differences between medians. All the analyses were done in the NCSS 2020 software (NCSS LLC, Kaysville, Utah, USA) with a significance level of p < 0.05. Graphics were made in GraphPad Prism version 9.
Results
G6Pase gene, CDS, and deduced protein
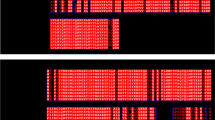
The gene and cDNA sequences of G6Pase obtained from the white shrimp were deposited in GenBank with the accession number MW246958 (Fig. 1). The gene nucleotide sequence we obtained is 99.07% identical to the genome sequence deposited in GenBank by Zhang et al. (2019), and 99.8% identical to the transcript included in the transcriptomic data of Ghaffari et al. (2014). The gene sequence starting from the initial codon for MET, ATG, is 2823 bp long. The genomic sequence was obtained with two overlapping fragments from genomic DNA. It comprises seven exons of 86, 165, 207, 131, 252, 119, and 117 bp each, interrupted by six introns with five split codons. All the introns contain the consensus 5′-GT-AG-3′ splice junctions necessary for correct RNA splicing of most eukaryotic genes (Breathnach et al. 1978) and the number and size of exons and introns correspond with the ones reported in other species (Wang et al. 2015). The average base composition A/T and G/C of the exons are 56.43% and 43.57%, respectively, whereas for the introns are 73% and 27%, respectively. The CDS was initially identified from gills cDNA and is 1,077 bp long encoding 358 amino acids residues from the initial methionine (ATG) to the stop codon (TAA) at position 1075, resulting in calculated values for the isoelectric point and molecular weight of 9.25 and 40.74 kDa, respectively. The catalytic amino acids were identified by sequence alignments. Analysis with the Pfam 32.0 tool revealed that shrimp G6Pase has the PAP2-glucose-6-phosphatase domain characteristic of all G6Pase isoforms and no signal peptide presence was predicted with signalP, which is typical of known G6Pases.
Nucleotide and amino acid sequences of the G6Pase gene of shrimp. Only the coding sequence is numbered. Primer sequences are underlined, split codons are in bold and underlined, initial Met and stop codons are in gray. The catalytic amino acids are between asterisk ⁕ and the PAP2-G6Pase domain is between brackets []. Exons are presented in capital letters and introns in lowercase letters
Homology and phylogenetic analysis of P. (L). vannamei G6Pase protein sequence
BLAST analysis of the deduced protein indicated an identity of 23.7 to 35.9% with the G6Pase sequences from the fruit fly (Drosophila melanogaster, GenBank accession no. NP_001097063), human (Homo sapiens, GenBank accession no. P35575), zebrafish (Danio rerio, GenBank accession no. NP_001157278.1) and eastern river shrimp (Macrobrachium nipponense, GenBank accession no. ALK82315.1). The highest homology of the white shrimp G6Pase was found with M. nipponense and D. rerio with the catalytic amino acids contained in highly conserved regions (Fig. 2). The phylogenetic analysis of 25 G6Pase sequences (Table 2) clearly showed two well-differentiated clusters between vertebrates and invertebrates locating the white shrimp sequence in the latter, within the same clade with the shrimp M. nipponense (35.9% identity) from other arthropods (Fig. 3).
G6Pase topological model
The 2D hypothetical model predicts seven transmembrane helices in the white shrimp G6Pase (Fig. 4). According to the topological analysis, three loops (in 177–190, 237–268, 319–328 positions), the catalytic amino acids (RHRH), and the N-terminal are oriented towards the ER lumen, and three loops (in 146–159, 207–218, 285–300 positions) and the C-terminal towards the cytoplasm, thus the G6Pase characteristic orientation of the active site is conserved.
Differential organ-expression of G6Pase in normoxia and hypoxia
G6Pase expression in hepatopancreas, gills and muscle was evaluated by RT-qPCR in normoxia (Fig. 5). Expression of the shrimp G6Pase gene was detected in the three evaluated organs with the highest expression in hepatopancreas (p < 0.05), with 6.1 and 3.5-fold than in gills and muscle, respectively. Similar results were reported for the amphioxus B. japonicum that had a higher G6Pase expression in digestive diverticulum than in muscle and gills under developmental conditions (Wang et al. 2015), B. japonicum occupies a nodal evolutionary position from vertebrates and invertebrates. Likewise, G6Pase activity was 20 times higher in hepatopancreas than in claw muscle under feeding conditions in the stone crab M. mercenaria (Scholnick et al. 2006).
Relative gene expression of G6Pase in white shrimp organs under normoxia. Data were normalized with the mRNA expression of the ribosomal protein L8 per the 2−ΔCt method. Bars represent means ± SD. The differences between the treatments are represented by literals (p < 0.05). The n values are included for each group
The effect of hypoxia on G6Pase relative gene expression was evaluated in hepatopancreas and gills by RT-qPCR (Fig. 6) and initially also evaluated in muscle, but expression in hypoxic conditions was undetectable. The PCR of amplification efficiencies and correlation coefficient in real time were 91.1%, 0.99 and 90.5%, 0.99 for G6Pase and L8, respectively. No significant differences were detected within the treatments in hepatopancreas due to the high variability among the shrimp, although the means for the hypoxia groups are lower by approximately 0.5-fold compared to the normoxia control. In gills, there was a significant increase (p < 0.05) of 11.3 and 5.2-fold at 3 h and 24 h respectively compared to normoxia and no significant changes were detected at 48 h, but with a tendency to increase (2.1-fold) with respect to normoxia. The effect of hypoxia on the expression of G6Pase has not been studied in crustaceans or other invertebrates, but it is known that in vertebrates species such as the fish Sparus aurata (Metón et al. 2004; Wang et al. 2015; Zhang et al. 2020) and in the stone crab M. mercenaria (Scholnick et al. 2006), starvation stress results in increased activity of the enzyme in the liver and hepatopancreas, respectively.
Effect of hypoxia on G6Pase gene expression in hepatopancreas (A) and gills (B) of white shrimp. Data were normalized with the mRNA expression of the ribosomal protein L8 per the 2−ΔCt method. N: control normoxia, H3: 3 h in hypoxia, H24: 24 h in hypoxia, H48: 48 h in hypoxia. Bars denote medians and ranges. The differences between the treatments are represented by literals (p < 0.05). The n values are included for each group
Discussion
The analysis of enzymes involved in carbohydrate metabolism is essential to understand their regulation and physiological significance in different species. The enzymes that catalyze irreversible reactions in central metabolism are particularly interesting to comprehend the pathways and their regulators that provide vital physiological responses for the cells. In the white shrimp, the genes of four enzymes that catalyze irreversible reactions of glycolysis and the corresponding for gluconeogenesis have been identified: pyruvate carboxylase (PC) (our unpublished data), PEPCK (Reyes-Ramos et al. 2018), FBP (Cota-Ruiz et al. 2015) and herein we report the G6Pase sequence, a key enzyme involved in glucose homeostasis. To our best knowledge, this is the first detailed report of this gene in crustaceans.
There are three isozymes for G6Pase in vertebrates: G6Pase-I, G6Pase-II, and G6Pase-III (Hutton and O’Brien 2009); however, information about characterization, expression, and regulation of G6Pase genes is rather limited in invertebrates. The cDNA coding sequence was initially obtained from gills in our laboratory, and the complete gene sequence was experimentally determined from genomic DNA. When the genome sequence of P. (L.) vannamei was uploaded to the NCBI database (but not fully annotated), the cDNA nucleotide sequence that we had determined was carefully compared to the genomic data and found the interrupted G6Pase gene sequence. Analysis of G6Pases genes from different species included in GenBank (not fully complete or annotated) showed that the exon–intron structure of the shrimp G6Pase is more similar to arthropods such as in the moth Ostrinia furnacalis (Gene ID: 114,364,740) and the silk moth Bombyx mori (Gene ID: 101,742,080) which contain seven exons interrupted by six introns, while most of the genes in mammals are composed of five or six coding exons. The shrimp gene shares the same size and position of the second and fourth coding exons of the above mentioned arthopods, likely due to their phylogenetic proximity, although the length of the introns and complete genes are different.
Even though the overall identity between the G6Pase proteins is not very high, the regions containing the catalytic amino acids are conserved (Ghosh et al. 2002; Metón et al. 2004). The G6Pase protein from B. japonicum has 20–40% identity with other G6Pases of different species, and this enzyme plays a key role in glucose homeostasis (Wang et al. 2015). The 358 amino acid shrimp G6Pase has the conserved phosphatases sequence motif PAP2 (KXXXXXXRP-(X12-54)-PSGH-(X31-54)-SRXXXXX HXXXD) characteristic of all G6Pases (Stukey and Carman 1997). Additionally, the prediction of secondary structure based on hydropathy analysis using HMMTOP indicates the presence of transmembrane helices in G6Pase and coincides with homologs from vertebrates (Marcolongo et al. 2013), but the number of helices differs. The presence of seven TM helices separated by six hydrophilic loops in white shrimp G6Pase is presented in the model (Fig. 4).
In the phylogenetic analysis, the shrimp G6Pase is placed in the invertebrate group, with proximity only to the other shrimp species including the M. nipponense G6Pase and separated from the vertebrate sequences. It is worth noting that while isoforms in vertebrates can be grouped into different clades, in invertebrates there is no notable difference, indicating the higher proximity of G6Pases in this group. However, although the overall percentage of identity is low, there are highly conserved regions and the shrimp sequence is more similar to isoforms 1 and 2 of the fishes Astyanax mexicanus (30.72%), and Chanos chanos (30.52%) respectively.
In vertebrates, the liver is the central organ responsible for glucose homeostasis by de novo synthesis from non-carbohydrate precursors such as lactate or by hydrolysis of glycogen (Ghosh et al. 2002). In both metabolic pathways, the final reaction is catalyzed by G6Pase; therefore, this enzyme has been widely studied as an indicator of energy flow regulation, especially under stressful conditions such as starvation or under periods of high energy demand (Hah et al. 2002; Melkonian and Schury 2019). Under optimal development conditions, little has been studied about G6Pase at a transcriptional level in invertebrates. However, in vertebrates as spotted gar, zebrafish, medaka, and rainbow trout, the liver is the main organ with the highest G6Pase expression compared to gills, muscle, kidney, demonstrating that it has a similar organ-specific expression pattern (Marandel et al. 2017). The same behavior was found in shrimp, indicating that hepatopancreas could be responsible for glycemic control in normoxia, coinciding with previous reports (Cervellione et al. 2017). Hence, it is possible that G6Pase functions as a good indicator of shrimp metabolic and physiological state.
In contrast, the expression of the gluconeogenic enzymes FBP and PEPCK is lower in the hepatopancreas compared to the muscle and the gills (Cota-Ruiz et al. 2015; Reyes-Ramos et al. 2018), which could indicate the activation of glycogenolysis instead of gluconeogenesis in this organ as a result of the fasting period to which the organisms were subjected (24 h at the sampling of this experiment). Consequently, the lack of energy substrates could induce the breakdown of the stored glycogen aiding to distribute glucose through the hemolymph to the target tissues for its use in the ATP rapid generation pathways as previously described, linking oxidative metabolism with the use and mobilization of carbohydrates between organs in shrimp (Sánchez-Paz et al. 2007; Ulaje et al. 2019). On the other hand, muscle and gills could play a secondary role in this metabolic process during normoxia, but it has been reported that G6Pase activity in starvation increases in both organs (Thabrew et al. 1971; Mathew et al. 2007), which suggests a condition and organ-specific regulation of the glycemic control to compensate the stress suffered by the animals.
The studies about shrimp have shown a complex metabolic regulation system to resist different stressors. Until now, hypoxia has received ample interest since climate change and anthropogenic activities are affecting marine and shrimp farm waters. Carbohydrate metabolism enzymes and their genes appear to respond in a specific organ-condition manner as mentioned before. Hypoxia does not seem to affect G6Pase expression in the hepatopancreas in this study conditions. This could indicate that glucose synthesis in this organ is independent of oxygen deficiency-induced stress. Nevertheless, gluconeogenesis appears to be activated in crustaceans by prolonged hypoxia (Cota-Ruiz et al. 2015; Reyes-Ramos et al. 2018) or post-hypoxic recovery to synthesize glucose using the end products of anaerobic metabolism such as lactate as precursors (Hervant et al. 1999). Thus, we cannot discard the participation of other G6Pase isoforms involved in glucose homeostasis, as is known in mammals (Lord-Dufour et al. 2009). The expression of G6Pase in gills increased after 3 h and 24 h of hypoxia, suggesting that glucose is generated de novo; in contrast, the expression of FBP and PEPCK decreased (Cota-Ruiz et al. 2015; Reyes-Ramos et al. 2018), while the concentration of glucose and relative expression of its transporter increased (Martínez-Quintana et al. 2014, 2016) under similar stressful conditions, which could again indicate catabolism of glycogen. Since it is known that crustaceans with different tolerance to hypoxia increase their hemolymph glucose and decrease their glycogen stores in the organs (Taylor and Spicer 1987), there is likely glycogenolysis activation in the white shrimp. It might be possible that gills exposed to low oxygen levels at the first exposed hours require additional energy vs other organs to perform essential processes such as respiration, detoxification, and osmotic regulation (Henry et al. 2012; Ulaje et al. 2019). Hence, G6Pase through glycogenolysis is generating enough glucose for the processes mentioned above.
Additionally, at 48 h of hypoxia in gills, the expression of G6Pase decreases, and it is reset at the level found in normoxia. Likewise, in this organ, the expression of FBP and PEPCK also decreases, which likely indicates that gluconeogenesis is not occurring, in contrast to the activation of gluconeogenesis in hepatopancreas that might occur at 48 h of hypoxia (Cota-Ruiz et al. 2015; Reyes-Ramos et al. 2018). This is probably the result of depression of metabolism (or hypometabolism) (Gorr et al. 2006) as a physiological strategy for the survival of shrimp to hypoxia to conserve energy through a coordinated decrease of the functions that mainly use ATP in the cell (for example, the synthesis and degradation of proteins) and prioritize essential and survival processes as mentioned above, to prevents lethal drops in cellular ATP levels. This could also be an indication of the synergistic regulation between gluconeogenesis and glycogenolysis not only as a glucose generation pathway to obtain fuel, but also as a strategy to take advantage of the carbon skeletons generated as lactate during anaerobic glycolysis.
Conclusions
This work reinforces the important role of the use and mobilization of carbohydrates in shrimp to survive and resist the low concentration of dissolved oxygen in different periods. The white shrimp G6Pase appears to contain all the necessary components of an active enzyme and the expression patterns in the three studied organs in normoxic and hypoxic conditions demonstrate its important role in glucose regulation probably mainly in gills by a glycogenolytic pathway in oxygen limited condition. Further research considering analysis of the promoter sequence, enzyme activity, and the implication of other G6Pase isoforms in response to different stressors in aquaculture will be helpful to the understanding of the physiological implications of this gene in these marine organisms.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Arion WJ, Wallin BK, Lange AJ, Ballas LM (1975) On the involvement of a glucose 6-phosphate transport system in the function of microsomal glucose 6-phosphatase. Mol Cell Biochem 6:75–83
Bostock J, McAndrew B, Richards R et al (2010) Aquaculture: Global status and trends. Philos Trans r Soc B Biol Sci 365:2897–2912
Bradfield JY, Wyatt GR (1983) X-linkage of a vitellogenin gene in Locusta migratoria. Chromosoma 88:190–193. https://doi.org/10.1007/BF00285619
Breathnach R, Benoist C, O’Hare K et al (1978) Ovalbumin gene: Evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA 75:4853-4857. https://doi.org/10.1073/pnas.75.10.4853
Camacho-Jiménez L, Peregrino-Uriarte AB, Martínez-Quintana JA, Yepiz-Plascencia G (2018) The glyceraldehyde-3-phosphate dehydrogenase of the shrimp Litopenaeus vannamei: Molecular cloning, characterization and expression during hypoxia. Mar Environ Res 138:65–75. https://doi.org/10.1016/j.marenvres.2018.04.003
Cervellione F, McGurk C, Van den Broeck W (2017) “Perigastric organ”: a replacement name for the “hepatopancreas” of Decapoda. J Crustac Biol 37:353–355. https://doi.org/10.1093/jcbiol/rux020
Cota-Ruiz K, Leyva-Carrillo L, Peregrino-Uriarte AB et al (2016) Role of HIF-1 on phosphofructokinase and fructose 1, 6-bisphosphatase expression during hypoxia in the white shrimp Litopenaeus vannamei. Comp Biochem Physiol -Part A Mol Integr Physiol 198:1–7. https://doi.org/10.1016/j.cbpa.2016.03.015
Cota-Ruiz K, Peregrino-Uriarte AB, Felix-Portillo M et al (2015) Expression of fructose 1,6-bisphosphatase and phosphofructokinase is induced in hepatopancreas of the white shrimp Litopenaeus vannamei by hypoxia. Mar Environ Res 106:1–9. https://doi.org/10.1016/j.marenvres.2015.02.003
Diaz RJ (2001) Overview of hypoxia around the world. J Environ Qual 30:275–281. https://doi.org/10.2134/jeq2001.302275x
Ellington WR (1983) The recovery from anaerobic metabolism in invertebrates. J Exp Zool 228:431–444. https://doi.org/10.1002/jez.1402280305
Ghaffari N, Sanchez-Flores A, Doan R et al (2014) Novel transcriptome assembly and improved annotation of the whiteleg shrimp (Litopenaeus vannamei), a dominant crustacean in global seafood mariculture. Sci Rep 4:1–10. https://doi.org/10.1038/srep07081
Ghosh A, Shieh JJ, Pan CJ et al (2002) The catalytic center of glucose-6-phosphatase. HIS176 is the nucleophile forming the phosphohistidine-enzyme intermediate during catalysis. J Biol Chem 277:32837–32842. https://doi.org/10.1074/jbc.M201853200
Gorr TA, Gassmann M, Wappner P (2006) Sensing and responding to hypoxia via HIF in model invertebrates. J Insect Physiol 52:349–364
Hah JS, Ryu J, Lee W et al (2002) The hepatocyte glucose-6-phosphatase subcomponent T3: Its relationship to GLUT2. Biochim Biophys Acta - Biomembr 1564:198–206. https://doi.org/10.1016/S0005-2736(02)00450-9
Henry RP, Lucu Č, Onken H, Weihrauch D (2012) Multiple functions of the crustacean gill: Osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front Physiol 3 NOV:431. https://doi.org/10.3389/fphys.2012.00431
Hervant F, Garin D, Mathieu J, Freminet A (1999) Lactate metabolism and glucose turnover in the subterranean crustacean Niphargus virei during post-hypoxic recovery. J Exp Biol 202:579–592. https://doi.org/10.1242/jeb.202.5.579
Hutton JC, O’Brien RM (2009) Glucose-6-phosphatase catalytic subunit gene family. J Biol Chem 284:29241–29245
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. https://doi.org/10.1093/molbev/msn067
Lord-Dufour S, Copland IB, Levros LC et al (2009) Evidence for transcriptional regulation of the glucose-6-phosphate transporter by HIF-1α: Targeting G6PT with mumbaistatin analogs in hypoxic mesenchymal stromal cells. Stem Cells 27:489–497. https://doi.org/10.1634/stemcells.2008-0855
Loret SM, Devos PE (1992) Hydrolysis of G6P by a microsomal aspecific phosphatase and glucose phosphorylation by a low Km hexokinase in the digestive gland of the crab Carcinus maenas: variations during the moult cycle. J Comp Physiol B 162:651–657. https://doi.org/10.1007/BF00296647
Marandel L, Panserat S, Plagnes-Juan E et al (2017) Evolutionary history of glucose-6-phosphatase encoding genes in vertebrate lineages: Towards a better understanding of the functions of multiple duplicates. BMC Genomics 18:342. https://doi.org/10.1186/s12864-017-3727-1
Marcolongo P, Fulceri R, Gamberucci A et al (2013) Multiple roles of glucose-6-phosphatases in pathophysiology: State of the art and future trends. Biochim Biophys Acta - Gen Subj 1830:2608–2618
Martínez-Quintana JA, Peregrino-Uriarte AB, Gollas-Galván T et al (2014) The glucose transporter 1 -GLUT1- from the white shrimp Litopenaeus vannamei is up-regulated during hypoxia. Mol Biol Rep 41:7885–7898. https://doi.org/10.1007/s11033-014-3682-8
Martínez-Quintana JA, Valenzuela-Soto EM, Martínez-Téllez MA et al (2016) Functionality of the white shrimp glucose transporter 1: Expression in Xenopus oocytes and gene silencing during hypoxia. J Exp Mar Bio Ecol 474:109–115. https://doi.org/10.1016/j.jembe.2015.10.004
Mathew S, Nair AKK, Anandan R et al (2007) Biochemical studies on changes associated with enzymes of glucose metabolism in white spot syndrome virus (WSSV) infected with Penaeus monodon (Fabricius). African J Biotechnol 6:1944–1948. https://doi.org/10.5897/ajb2007.000-2282
Melkonian EA, Schury MP (2019) Physiology. StatPearls Publishing, Gluconeogenesis
Metón I, Caseras A, Fernández F, Baanante IV (2004) Molecular cloning of hepatic glucose-6-phosphatase catalytic subunit from gilthead sea bream (Sparus aurata): Response of its mRNA levels and glucokinase expression to refeeding and diet composition. Comp Biochem Physiol - B Biochem Mol Biol 138:145–153. https://doi.org/10.1016/j.cbpc.2004.03.004
Pérez-Rostro CI, Racotta IS, Ibarra AM (2004) Decreased genetic variation in metabolic variables of Litopenaeus vannamei shrimp after exposure to acute hypoxia. J Exp Mar Bio Ecol 302:189–200. https://doi.org/10.1016/j.jembe.2003.10.010
Reyes-Ramos CA, Peregrino-Uriarte AB, Cota-Ruiz K et al (2018) Phosphoenolpyruvate carboxykinase cytosolic and mitochondrial isoforms are expressed and active during hypoxia in the white shrimp Litopenaeus vannamei. Comp Biochem Physiol Part - B Biochem Mol Biol 226:1–9. https://doi.org/10.1016/j.cbpb.2018.08.001
Sánchez-Paz A, García-Carreño F, Hernández-López J et al (2007) Effect of short-term starvation on hepatopancreas and plasma energy reserves of the Pacific white shrimp (Litopenaeus vannamei). J Exp Mar Bio Ecol 340:184–193. https://doi.org/10.1016/j.jembe.2006.09.006
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Scholnick DA, Barabas AE, Cowan SS (2006) The influence of chloride on glucose export in marine crabs: Sensitivity of glucose-6-phosphatase to chloride ion. J Crustac Biol 26:510–514. https://doi.org/10.1651/S-2669a.1
Soñanez-Organis JG, Peregrino-Uriarte AB, Sotelo-Mundo RR et al (2011) Hexokinase from the white shrimp Litopenaeus vannamei: cDNA sequence, structural protein model and regulation via HIF-1 in response to hypoxia. Comp Biochem Physiol - B Biochem Mol Biol 158:242–249. https://doi.org/10.1016/j.cbpb.2010.12.006
Soñanez-Organis JG, Racotta IS, Yepiz-Plascencia G (2010) Silencing of the hypoxia inducible factor 1 -HIF-1- obliterates the effects of hypoxia on glucose and lactate concentrations in a tissue-specific manner in the shrimp Litopenaeus vannamei. J Exp Mar Bio Ecol 393:51–58. https://doi.org/10.1016/j.jembe.2010.06.031
Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ (2004) TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20:3258–3260. https://doi.org/10.1093/bioinformatics/bth358
Stukey J, Carman GM (1997) Identification of a novel phosphatase sequence motif. Protein Sci 6:469–472. https://doi.org/10.1002/pro.5560060226
Surholt B, Newsholme EA (1981) Maximum activities and properties of glucose 6-phosphatase in muscles from vertebrates and invertebrates. Biochem J 198:621–629. https://doi.org/10.1042/bj1980621
Taylor AC, Spicer JI (1987) Metabolic responses of the prawns Palaemon elegans and P. serratus (Crustacea: Decapoda) to acute hypoxia and anoxia. Mar Biol 95:521–530. https://doi.org/10.1007/BF00393095
Thabrew MI, Poat PC, Munday KA (1971) Carbohydrate metabolism in Carcinus maenas gill tissue. Comp Biochem Physiol - Part B Biochem 40:531–541. https://doi.org/10.1016/0305-0491(71)90238-0
Trandaburu T (1977) Fine structural localization of glucose-6-phosphatase activity in the pancreatic islets of two amphibian species (Salamandra salamandra L. and Rana esculenta L.). Acta Histochem 59:246–253. https://doi.org/10.1016/S0065-1281(77)80046-9
Trasviña-Arenas CH, Garcia-Triana A, Peregrino-Uriarte AB, Yepiz-Plascencia G (2013) White shrimp Litopenaeus vannamei catalase: Gene structure, expression and activity under hypoxia and reoxygenation. Comp Biochem Physiol Part - B Biochem Mol Biol 164:44–52. https://doi.org/10.1016/j.cbpb.2012.10.004
Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. https://doi.org/10.1093/bioinformatics/17.9.849
Ulaje SA, Rojo‐Arreola L, Lluch‐Cota SE, et al (2019) Gene expression and energetic metabolism changes in the whiteleg shrimp (Litopenaeus vannamei) in response to short‐term hypoxia. Aquac Res 50:are.13960. https://doi.org/10.1111/are.13960
van Schaftingen E, Gerin I (2002) The glucose-6-phosphatase system. Biochem J 362:513–532. https://doi.org/10.1042/bj3620513
Varga V, Murányi Z, Kurucz A et al (2019) Species-specific glucose-6-phosphatase activity in the small intestine—Studies in three different mammalian models. Int J Mol Sci 20:5039. https://doi.org/10.3390/ijms20205039
Wang Y, Wang H, Li M et al (2015) Identification, expression and regulation of amphioxus G6Pase gene with an emphasis on origin of liver. Gen Comp Endocrinol 214:9–16. https://doi.org/10.1016/j.ygcen.2014.12.021
Wannamaker CM, Rice JA (2000) Effects of hypoxia on movements and behavior of selected estuarine organisms from the southeastern United States. J Exp Mar Bio Ecol 249:145–163. https://doi.org/10.1016/S0022-0981(00)00160-X
Wu RSS (2002) Hypoxia: From molecular responses to ecosystem responses. Mar Pollut Bull 45:35–45. https://doi.org/10.1016/S0025-326X(02)00061-9
Zhang X, Yang Z, Deng D et al (2020) Effects of dietary starch levels on the growth, plasma metabolites and expression of genes involved in glucose transport and metabolism in Pelodiscus sinensis juveniles. Aquac Res 51:738–750. https://doi.org/10.1111/are.14424
Zhang X, Yuan J, Sun Y et al (2019) Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun 10:1–14. https://doi.org/10.1038/s41467-018-08197-4
Acknowledgements
We are grateful for funding to CONACyT, Ciencia Básica grant A1-S-24557 to GYP. We thank Dr. Jorge L. Duarte-Gutiérrez, Dr. Dahlia M. Nuñez-Hernandez, Biol. Adrian Gamez-Alejo, for technical help provided in the hypoxia bioassays.
Funding
This study was funded by the National Science and Technology Council, Mexico (CONACyT), grant Ciencia Básica A1-S-24557 to GYP and a scholarship for Ph.D studies to LEHA.
Author information
Authors and Affiliations
Contributions
LEHA performed experiments, data analysis and writing of original draft. KCR performed initial experiments and data analysis. SGJ, provided shrimp and bioassay facilities. ABPU supervised experiments and methodology, gave helpful suggestions, critically revised and approved the final manuscript. GYP conceived, designed and supervised the experiments, critically revised, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing or financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hernández-Aguirre, L.E., Cota-Ruiz, K., Peregrino-Uriarte, A.B. et al. The gluconeogenic glucose-6-phosphatase gene is expressed during oxygen-limited conditions in the white shrimp Penaeus (Litopenaeus) vannamei: Molecular cloning, membrane protein modeling and transcript modulation in gills and hepatopancreas. J Bioenerg Biomembr 53, 449–461 (2021). https://doi.org/10.1007/s10863-021-09903-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-021-09903-6