Abstract
Biomolecular structures at atomic resolution present a valuable resource for the understanding of biology. NMR spectroscopy accounts for 11 % of all structures in the PDB repository. In response to serious problems with the accuracy of some of the NMR-derived structures and in order to facilitate proper analysis of the experimental models, a number of program suites are available. We discuss nine of these tools in this review: PROCHECK-NMR, PSVS, GLM-RMSD, CING, Molprobity, Vivaldi, ResProx, NMR constraints analyzer and QMEAN. We evaluate these programs for their ability to assess the structural quality, restraints and their violations, chemical shifts, peaks and the handling of multi-model NMR ensembles. We document both the input required by the programs and output they generate. To discuss their relative merits we have applied the tools to two representative examples from the PDB: a small, globular monomeric protein (Staphylococcal nuclease from S. aureus, PDB entry 2kq3) and a small, symmetric homodimeric protein (a region of human myosin-X, PDB entry 2lw9).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biomolecular structures at atomic resolution are crucial for interpreting cellular processes in a molecular context. In addition, they serve important roles in drug discovery and functional industrial design, such as the modification of enzyme properties. For all these applications, it is imperative that the biomolecular atomic structures are accurate, precise and truthfully reflect the experimental data on which they were based.
The Protein Data Bank (PDB) (Berman 2008; Bernstein et al. 1977) is the primary repository of the atomic coordinates of three-dimensional (3D) biomolecular structures. It currently contains more than 91,000 entries, which cover proteins, oligonucleotides and their complexes, including small-molecule ligands. The entries solved by Nuclear Magnetic Resonance (NMR) represent approximately 11 % of the total (>10,000 entries). The PDB archive is jointly managed by four partner organisations (RCSB PDB (Berman et al. 2000), PDBe (Velankar et al. 2012), PDBj (Kinjo et al. 2012) and BMRB (Ulrich et al. 2008) under the aegis of the wwPDB (Berman et al. 2007) consortium.
A series of erroneously modelled NMR structures (Clore et al. 1995; Lambert et al. 2004; Nabuurs et al. 2006; Spadaccini et al. 2006) and cases of outright scientific fraud (Borrell 2009) with X-ray derived structures underscore the need for dedicated tools to assess the structural quality of biomolecular structures as well as the agreement with the experimental data. Moreover, the 3D structure and dynamic properties of the biomolecules can change in response to interactions with other molecules and hence it is also imperative to carefully assess the accuracy of the structures.
Structure validation typically encompasses two broad aspects: the agreement of the experimental data with the resulting structure and a geometric validation. In order to calculate the agreement with the experimental data, a theoretical description relating the data to the atomic coordinates is required. These relations are typically also used during the structure calculation procedure to drive the convergence and hence the assessment only conveys the degree to which the structure was calculated properly. If, however, the data are internally inconsistent, this will typically result in statistically poor or unusual distributions of related structural parameters (vide infra). More independent measures are based upon cross-validation methods (Brunger et al. 1993; Clore and Schwieters 2006; Nabuurs et al. 2005; Tjandra et al. 2007) that exclude a fraction of the data in the structure calculation procedures.
Geometric structure validations aim to assess the quality in relation to the chemical and structural knowledge derived from relevant reference structures. Local structural parameters such as bond lengths, bond angles and torsion angles are obtained from X-ray crystallography data of small molecules and ultra-high resolution biomolecular structures (Engh and Huber 1991, 2001), whereas dihedral angle distributions are based upon a set of high-resolution X-ray structures. Clearly, there is an inherent danger that structures are evaluated with respect to an incomplete or biased reference, but it is nowadays generally appreciated that uncommon features flagged by a geometric assessment should be supported by solid experimental data (Vriend 1990; Chen et al. 2010; Bhattacharya et al. 2007; Doreleijers et al. 2012a; Nabuurs et al. 2006; Hooft et al. 1996).
Traditional and still-popular biomolecular NMR structure validation routines have relied on a limited set of tools and metrics. It has been customary to summarise restraint content using a simple count of the number of restraints, whereas it has long been known that these numbers are flawed for multiple reasons (Nabuurs et al. 2003). A recent large-scale analysis also showed great redundancy in the reported number of distance restraints (Doreleijers et al. 2009). It even proved possible to refine NMR-derived structures using random 15N-RDC values to acceptable Q-factors (Bax and Grishaev 2005). PROCHECK-NMR (Laskowski et al. 1996) has been the accepted choice for the assessment of the geometrical quality of NMR ensembles, in spite of it long being out-dated. High percentages of residues in the most favoured Ramachandran plot regions reported by PROCHECK-NMR were commonly regarded as an assurance of a good quality structure, but recent assessments have shown this to be invalid (Doreleijers et al. 2012a, b). Tools designed for X-ray crystallography, such as WHAT IF (Vriend 1990; Hooft et al. 1996) or Molprobity (Davis et al. 2007; Chen et al. 2010), can also be used for NMR-derived structures. However, NMR-specific properties such as the presence of multiple models in one structural ensemble and the potential dynamical aspects represented in this ensemble, often present problems that are not accommodated by these programs. In particular, most of the routines also fail to adequately address the validation of ‘ensembles of ensembles’, where the computational protocols simultaneously aim to treat both the structural model and the available dynamical data (Montalvao et al. 2012; Lindorff-Larsen et al. 2005). Structure validation software dedicated to NMR-derived structures, such as the PSVS suite (Bhattacharya et al. 2007) or CING (Doreleijers et al. 2012a), typically provide solutions for the issues inherently associated with X-ray oriented tools.
Compared to X-ray crystallography, validation of NMR-derived structures is in general more complicated. Not only do the tools need to take into account the aforementioned dynamical effects and the multiple conformers in the structural ensemble, but also the nature of the NMR data, which differs vastly from the X-ray situation. Whereas the latter only concerns the reflections, which are uniform in data content, NMR-derived structures can be based on a large variety of experimental data (Vuister et al. 2011). These can be both local in nature, such as distance and dihedral restraints, global in nature, such as residual dipolar couplings (RDCs) and pseudo-contact shifts (PCS), or describe the overall shape, such as the small angle scattering (SAS) data.
A typical NMR structure calculation protocol involves a customised simulated annealing procedure, typically in torsion-angle space, where restraints are included as pseudo-harmonic potentials (Stein et al. 1997; Güntert 1998). Many additions to the basic protocol have been proposed, such as the use of database potentials (Kuszewski and Clore 2000), radius of gyration (Schwieters and Clore 2008), 15N-T1/T2-relaxation parameters (Tjandra et al. 1997), SAXS-derived potentials (Gabel et al. 2008) or ensembles consistent with S2 order parameters (Best and Vendruscolo 2004). Refinement in explicit water using a more extended force field was shown to significantly improve the structural quality. (Linge et al. 2003; Spronk et al. 2002) Inferential structure determination (Rieping et al. 2005), while computationally expensive, was shown to significantly improve the treatment of dynamical effects and provide for a more unbiased parametrisation of the underlying theoretical models (Bernard et al. 2011).
Assigned chemical shifts are arguably the most important parameters obtained from NMR experiments. They are affected by the immediate chemical environments of the nuclei and can therefore reveal important structural information by themselves, a feature exploited by the recent methods of structure determination from chemical shifts, such as CS-ROSETTA and its derivatives (Shen et al. 2008, 2009b), CHESHIRE (Cavalli et al. 2007) and CS23D (Wishart et al. 2008). However, despite the success of these methods in some cases, they have proven to be not yet fully reliable. The results of the 2010 CASD-NMR competition (Rosato et al. 2009, 2012) showed that occasionally the chemical shift derived structures were up to 12 Å RMSD away from the manually determined reference structures, in spite of their excellent geometric validation scores.
More traditionally, chemical shifts have been used to automatically derive distance restraints primarily from NOE data by programs such as CYANA (Güntert et al. 1997; Herrmann et al. 2002; Lopez-Mendez and Güntert 2006) and Aria (Rieping et al. 2007). The completeness and correctness of chemical shift assignments thus influence the correctness of the distance restraints, the convergence of structure calculation protocols and the accuracy of the final structure ensembles.
Most existing methods for validation of chemical shifts rely on statistical analysis and comparison with databases. Of the methods surveyed for this paper, PSVS uses the Assignment Validation Suite (AVS) (Moseley et al. 2004) to identify outliers, while CING uses VASCO (Rieping and Vranken 2010) for referencing correction and SHIFTX (Neal et al. 2003) for back-calculation. If the difference between the observed and predicted shift values is greater than three standard deviations CING flags the nucleus as a chemical shift outlier. Vivaldi uses VASCO to correct referencing and identify statistical outliers based on amino acid type, secondary structure and accessible surface area. For referencing correction, other existing software includes CheckShift (13C and 15N) (Ginzinger et al. 2007, 2009), LACS (13C and 1H) (Wang et al. 2005; Wang and Markley 2009) and PANAV (Wang et al. 2010a), none of which require structural data, as well as SHIFTCOR (Zhang et al. 2003), which predicts only backbone chemical shifts and requires a structure.
Back-calculation of chemical shifts from structure is a rapidly developing field. Table S1 surveys the different programs used to predict protein chemical shifts. Some of these, i.e. SPARTA (Shen and Bax 2007), SPARTA+ (Shen and Bax 2010b), CamShift (Kohlhoff et al. 2009) and CheShift (Vila et al. 2009) provide chemical shifts predictions only for backbone nuclei, some others, i.e. CH3Shift (Sahakyan et al. 2011a) and ArShift (Sahakyan et al. 2011b) calculate side-chain values, and still others, i.e. SHIFTS (Xu and Case 2001; Moon and Case 2007), SHIFTX2 (Han et al. 2011), PROSHIFT (Meiler 2003), 4DSpot (Lehtivarjo et al. 2009), COSMOS (Möllhoff and Sternberg 2001; Jakovkin et al. 2012) and PPM (Li and Brüschweiler 2012) manage both. While a detailed analysis of these prediction programs is out of scope for this paper, they can be used to identify statistically unusual chemical shifts, by inspecting the differences between the predicted and measured values. However, caution needs to be taken with such an approach for the following reasons: (a) the prediction algorithms strongly depend on the accuracy of the underlying structure, and are therefore only as good as the structures are; (b) an anomalous chemical shift value is not necessarily an error, although it may require some supporting data, such as close vicinity of an aromatic group, unusual local conformation, etc.
Recognising the importance of structure validation, the wwPDB consortium has appointed special validation task forces (VTF) for X-ray crystallography (Read et al. 2011; Gore et al. 2012), electron microscopy (EM) (Henderson et al. 2012), NMR (Montelione et al. submitted) and SAS (Trewhella et al. 2013) methods. These VTFs will define a set of criteria and tools, which will be used at the time of deposition to assess the quality of the structural model, the intrinsic quality of the experimental data, and the fit between both. This paper reviews the tools currently available to NMR spectroscopists for evaluating the quality of their structures. We give an overview of the different checks performed by each package or program and discuss its relative merits using two examples: a small, globular monomeric protein (Staphylococcal nuclease from S. aureus, PDB entry 2kq3 (Wang et al. 2010b), S.Nase) and a small, symmetric homodimer protein [a region of human myosin-X, PDB entry 2lw9 (Lu et al. 2012)]. These two proteins present typical examples in terms of size and experimental data of the systems nowadays studied by NMR spectroscopy, and were solved by conventional triple resonance heteronuclear NMR technology. This review necessarily limits itself to the validation of protein structures and their complexes, as the tools for oligonucleotides and polysaccharides are much less developed. We also limit ourselves to testing ‘NMR-aware’ software, or at least software that does not presume the X-ray crystallographic origin of the structure.
Methods
We will first describe the different programs and tools available for validation of biomolecular NMR structures. An overview of their features is given in Table 1.
PROCHECK-NMR
Historically the most popular NMR-specific validation tool (Laskowski et al. 1996), PROCHECK-NMR is not available via a web-server interface. Instead, a standalone local installation of the program is required. The program is no longer maintained and its underlying scoring database is generally considered out-dated. The program accepts PDB formatted input files and experimental restraints in the Aqua format, also no longer maintained and incapable of handling ambiguous restraints. The output of the program is presented as a collection of postscript-formatted files.
CING
The Common Interface for NMR structure Generation (CING) software package version 1.0 (https://nmr.le.ac.uk) (Doreleijers et al. 2012a) constitutes an integrated framework for the validation of NMR structures. CING assembles a set of experimental and structural data and generates an analysis based on the results of ~ 25 different programs and routines, both internal and external, and dependent upon the supplied input data. CING accommodates a diversity of different experimental data types, as well as handling multi-model ensembles properly in its analysis routines.
The experimental data are tested for internal consistency and agreement with the ensemble. Distance restraints are analysed for duplication, redundancy, completeness (Doreleijers et al. 2005) and information content (Nabuurs et al. 2003). RMSDs and violation analysis is reported. Dihedral restraints are analysed for violations and RMSD. RDC restraints are processed, but currently not validated.
Validation of chemical shift values is based on structural and sequence information, re-referenced using the VASCO routine (Rieping and Vranken 2010) and analysed relative to the BMRB database (Ulrich et al. 2008) and SHIFTX (Neal et al. 2003) back-calculated values. Chemical shifts are also used to assess potential cis/trans-proline errors (Schubert et al. 2002; Shen and Bax 2010a; Siemion et al. 1975), leucine side-chain conformation (Mulder 2009) and to predict ϕ, ψ dihedral angles using the program TALOS+ (Shen et al. 2009a).
The geometric quality of the 3D structure ensembles is assessed in relation to a database of reference structures using WHAT IF (Vriend 1990; Hooft et al. 1996), PROCHECK-NMR (Laskowski et al. 1996) and internal routines. Checks include those for the residue-specific Ramachandran and side-chain rotamer distributions, all dihedral angles, including the ω dihedral, packing, backbone conformations, bumps, bond lengths, bond angles and torsions. The ensemble is also analysed for secondary structure using DSSP (Joosten et al. 2011; Kabsch and Sander 1983) and for solvent accessibility, potential disulphide bridges and salt bridges.
CING uses a circular-variance based algorithm to select for ordered regions. Alternatively, chemical-shift derived S2 order parameters (Berjanskii and Wishart 2005) or user-defined regions can be used for the analysis.
CING generates a hierarchical, comprehensive, interactive HTML/Javascript-based validation report that should be thought of more as a program than as a collection of static HTML pages. The user can interact with the report in several ways using Web 2.0 Javascript functionality. The different pages of the report reflect the natural ordering of either structure or experimental data and are extensively hyperlinked. CING uses a simple Red-Orange-Green (ROG) score that directs the NMR spectroscopist to troublesome areas. The ROG scoring is dependent upon the combined analysis of all results and allows CING to summarise the important issues. A red colouring indicates some potentially serious issues, green denotes the absence of any detected issues and orange (amber) is intermediate between these two situations. In particular, CING’s so-called residue pages display the validation results in direct relation to the relevant experimental data.
The multilingual web server and a web service together are called iCing and allow for anonymous execution of CING validation runs. The iCing server (http://nmr.cmbi.ru.nl/icing/) natively accepts PDB, CYANA (Güntert et al. 1997) and CCPN (Vranken et al. 2005) formatted files for coordinate, restraint, peak list and chemical shift data.
Molprobity
Molprobity (Chen et al. 2010; Davis et al. 2007) is a validation tool evaluating and scoring several structural features. This program is available both as a downloadable stand-alone server and as a web service (http://molprobity.biochem.duke.edu), operating on standard PDB files (or released PDB entries). The latest version was released in February 2013 and is still in alpha testing. For the purpose of this review we used the latest stable version 3. Molprobity is also used by other software reviewed in this paper, all of which used version 3 at the time of writing. Although Molprobity was originally designed to tackle the structural validation of experimental X-ray protein and nucleic acids structures, NMR ensembles require no additional effort from the user. Ensembles are automatically split into single-model PDB files and each model is processed individually. However, at the moment, Molprobity has limited functionality to present combined results from these calculations.
Molprobity starts by analysing any uploaded PDB file, and checks for presence of hydrogen atoms. If needed, Molprobity uses the REDUCE module (Word et al. 1999) to create updated PDB files by introducing and/or removing hydrogen atoms as necessary and propose flips for Asn/Gln/His residues to optimise hydrogen-bonding networks. This feature, although mostly helpful for X-ray structures can be used to confirm the protonation state of modelled histidine side-chains, a common source of errors in structures submitted to the PDB archive.
Molprobity further uses several internal programs to analyse the geometrical quality of the models. Covalent geometry validation of backbone bond lengths and angles is performed by DANGLE and is based on parameters derived by Engh and Huber (1991, 2001) for proteins and Parkinson et al. (1996) for nucleic acids. Protein backbone and side-chain torsion angles are validated using internal routines based on a large set of carefully selected reference data. Backbone angles for the Ramachandran statistics are categorised in four groups, i.e. proline, pre-proline, glycine and a general group covering all other common L-amino acids. Similarly, nucleic acids are evaluated by an internal program SUITENAME to identify improbable ring puckers and unfavourable RNA backbone conformations according to Richardson et al. (2008). One additional score describing the Cβ geometry is calculated based on Lovell et al. (2003).
All-atom contact analysis is a major defining feature of Molprobity. It is performed by the program PROBE, (Word et al. 1999) which generates a list of close contacts for non-covalently bonded atom-pairs that are too close in 3D space, i.e. more than 0.4 Å closer than the sum of Van der Waals radii. An overall close contact score, ‘clash-score’, is calculated as the number of close contacts per 1,000 atoms. Molprobity finally combines the close contact score, percentage of Ramachandran outliers and percentage of bad side-chain rotamers into a highly popular single score per model in the NMR ensemble. The weights of the three scores are chosen such that the single score resembles the crystallographic resolution (in Å) at which such scores are most likely to be observed in X-ray structures.
Results are displayed as interactive web pages consisting of tables, text files containing outliers, Ramachandran plots, etc. Interactive 3D visualisation of validation scores is provided by KiNG (Chen et al. 2009). Scenes can be viewed online using the Java applet version of KiNG. Alternatively, larger scenes can be downloaded and viewed offline.
ResProx
ResProx, Resolution-by-proxy (http://www.resprox.ca), aims at providing a single model-based score that was proposed to function as an accuracy measure similar to the resolution reported for X-ray derived structures in the PDB (Berjanskii et al. 2012). In addition to this generalised resolution parameter, all individual Z-scores used for the calculation are presented in tabular form. For each unsatisfactory score, suggestions are provided on how to increase the overall structure quality and remedy the poor score in particular.
ResProx processes up to 25 measurable protein features, extracted from a multitude of auxiliary programs, in two parallel schemes to calculate two resolution estimates. A ‘decision maker’ will select the most appropriate score to present in the validation report using empirical rules. The first validation scheme uses a machine learning predictor for the resolution based on 25 protein features. This predictor was trained on a set of 2,427 X-ray derived protein structures covering a wide span of reported resolutions, and cross-referenced against a second set of 500 structures. In the second scheme, the ‘Z-mean’ metric is calculated using a linear dependence on a subset of 15 out of 25 criteria using a simple regression scheme.
Five Molprobity scores are used to assess the structure quality: Ramachandran outliers, side-chain rotamer outliers, bond lengths, bond angles and atom clashes. The program VADAR (Willard et al. 2003) contributes 11 scores, covering the validation of hydrogen bond energy through DSSP, χ1 and ω dihedral angles, and general protein packing. VADAR (http://vadar.wishartlab.com/) can also be run separately to get a more detailed view of the derived scores. It generates comprehensive text files containing tables of validation scores for every model as well as summaries for the whole NMR ensemble. Additionally, some validation scores are presented graphically in static images. GeNMR (Berjanskii et al. 2009) provides Ramachandran scores, an atom clash-score and assesses the observed radius of gyration. RosettaHoles2 (Sheffler and Baker 2010) is used to quantify the packing of the protein core. Finally, PROSESS (Berjanskii et al. 2010) is further used to evaluate hydrogen bonding and χ1 dihedral angles.
The stand-alone PROSESS server is also available at http://prosess.ca. Its output consists of multiple HTML pages presenting detailed structural validation scores in tables, graphs and static images of the protein. In addition to structural validation, PROSESS analyses chemical shifts in NMR-STAR (v2.1) format and distance restraints in Xplor format. We tested the geometry and experimental data validation with PROSESS for the two entries (2lw9 and 2kq3), after the supplied restraints files were manually reformatted to comply with the (rather) strict format requirements for PROSESS.
The ResProx web-server requires a PDB-formatted file as input. Alternatively a PDB entry code can be provided to run the ResProx validation on an entry in the PDB archive. Results are presented as simple HTML web pages.
PSVS
Protein Structure Validation Suite (PSVS (Bhattacharya et al. 2007); http://psvs-1_4-dev.nesg.org) is a versatile validation server developed by one of the groups in the Northeast Structural Genomics Consortium (NESG), the only Protein Structure Initiative (PSI) consortium with a substantial NMR component. PSVS is applicable to both X-ray and NMR structures in an effort to be able to compare structural scores directly. It combines the output from a number of programs developed by several groups, i.e. Molprobity (Davis et al. 2007; Chen et al. 2010), Verify3D (Eisenberg et al. 1997), ProsaII (Wiederstein and Sippl 2007), PROCHECK (Laskowski et al. 1993), PDB validation software (http://deposit.rcsb.org/validate), and by the Montelione group itself, i.e. PDBStat (Bhattacharya et al. 2007), FindCore (Snyder and Montelione 2005), AVS (Moseley et al. 2004) and RPF (Huang et al. 2005). PSVS checks both the geometric knowledge-based validation and the fit between the structure and the experimental data, if the latter is available. Many NMR structures feature long disordered termini or loops, which often lack long-range constraints and are not always modelled properly by the structure calculation software. PSVS accounts for this by allowing the users to specify which residues should be subject to the analysis: all, ordered as defined by circular variance (default), core as defined by the FindCore algorithm (Snyder and Montelione 2005), residues forming secondary structure elements or a custom selection. For the purpose of this review we have chosen the default option.
For geometric validation, PSVS is trained on a set of 252 X-ray structures of globular proteins of maximum 500 residues and with resolution of 1.8 Å or better, sharing at most 50 % sequence identity with each other. Each reported raw score is converted to a Z-score using the mean and standard deviation pre-calculated on the training set. In this implementation, a positive Z-score would indicate that the analysed structure is better than the typical high-resolution X-ray structure. Any negative value for a Z-score would indicate poorer than average quality parameter, a rule of thumb is that Z-scores below −3.5 point to serious problems with modelling, and would require careful analysis of the model and/or the underlying experimental data. For NMR structures, five geometric validation scores are reported as ensemble averages. These five scores are: Molprobity clash-score (Davis et al. 2007; Chen et al. 2010), which gives the number of steric clashes per 1,000 atoms, PROCHECK backbone and all dihedral angle G-factors (Laskowski et al. 1993), Verify3D score (Eisenberg et al. 1997), which gives the likelihood of the observed packing, and ProsaII score (Wiederstein and Sippl 2007), which reports on the likelihood of the observed fold. This allows for a simple and unbiased comparison between NMR and X-ray structures irrespective of the size of the protein. These overall scores are reported in the PSVS summary report, which also identifies secondary structure elements calculated by DSSP (Kabsch and Sander 1983; Joosten et al. 2011), and lists the mean RMSDs of model superposition, a number of per-residue scores, the Ramachandran statistics from both the PROCHECK-NMR (Laskowski et al. 1996) and Molprobity (Chen et al. 2010; Davis et al. 2007) and a figure visualising consistent Ramachandran outlier residues on the structure. Optionally and depending on the types of submitted data, the summary page may also contain statistics on the distance and dihedral angle restraints and their violations, completeness of the chemical shift assignments, list of atoms with unusual chemical shifts, the RPF scores (Huang et al. 2005) describing the goodness of fit between the NOESY peak lists and the ensemble of structures and a generalised RMSD score (GLM-RMSD, vide infra).
The full PSVS report assembles the output from the constituent software packages and allows a keen user to review all of them from one URL or PDF file. The overall grouping is by metric, and the user can drill down within the given page to individual models in the NMR ensemble and individual residues. Viewing all available information about a model or residue is, however, not straightforward and requires manual collation.
PSVS accepts PDB, CYANA (Güntert et al. 1997) and CNS/Xplor (Brunger 2007; Brunger et al. 1998) formatted files as input for the coordinate data. CYANA and CNS/Xplor formatted files are accepted for supplying experimental restraint data. The chemical shifts data can be uploaded as NMR-STAR files (either version 2.1 or 3.1) (Ulrich et al. 2008) whereas the format for peak files is flexible, e.g. tab-delimited, with the possibility for the user to describe the meaning of each column.
GLM-RMSD
GLM-RMSD (Bagaria et al. 2012) is a method to produce an aggregate validation score for a complete structure ensemble from the result of a number of existing programs, which was recently incorporated as part of the PSVS server. The method aims to yield an easily interpretable quality metric representing an estimate of the RMSD from the correct structure. The metric was derived using a generalised linear model based upon a number of well-established parameters: the RPF Discriminatory Power (DP) (Huang et al. 2005), Verify3D (Eisenberg et al. 1997), ProsaII (Wiederstein and Sippl 2007), PROCHECK-ϕ/ψ and all dihedral angle G-factors (Laskowski et al. 1993), Molprobity (Chen et al. 2010; Davis et al. 2007), the Gaussian Network Model (GNM) (Haliloglu et al. 1997), and the molecular size. The initial coefficients and weights for the various inputs were obtained using training data from CASD-NMR (65 structure ensembles for 16 proteins) (Rosato et al. 2009, 2012) and CASP (Moult et al. 1995, 2011). A jack-knifing procedure was used to guard against over-fitting. By successively removing input scores that were redundant or contributed little, a metric was derived that was comprised of a linear combination of only four inputs: the RPF DP score, the PROCHECK-ϕ/ψ score, the Molprobity clash-score, and the molecular size, yielding a correlation coefficient between predicted and actual RMSD values of 0.70 for all test data combined. Interestingly, this suggests that only the PROCHECK-ϕ/ψ and Molprobity scores are sufficient to evaluate the geometric quality of a structure. As 86 % of the structures with a GLM-RMSD < 2 Å were correct and 74 % of the structures with GLM-RMSD > 2 Å were erroneous, a GLM-RMSD of 2 Å was proposed as a quality cut-off. Since the RPF DP score is an important input to the algorithm it requires peak lists to obtain a result, which in turn excluded it from our practical tests for this review (vide infra).
QMEAN
The QMEAN (Benkert et al. 2009) structural quality score is comprised of six individual measures that probe local structure conformation, solvent accessibility and secondary structure. The latter is derived from both the PSIPRED score (McGuffin et al. 2000) and an analysis by DSSP (Kabsch and Sander 1983; Joosten et al. 2011).
The original QMEAN score was protein size dependent as larger proteins received higher absolute scores, which rendered its use somewhat problematic. This measure has now been superseded by a newer, normalised value QMEANnorm, which removes the dependence of the quality score on the size of the model. The QMEANnorm is now routinely reported and all QMEAN scores reported in this manuscript refer to the normalised values.
The QMEAN server (http://swissmodel.expasy.org/qmean) takes PDB-formatted files as input. These have to be supplied as individual files for each model of the ensemble collected into one .zip or .tgz archive. A FASTA sequence describing the protein can also be supplied, but did not change the outcome for the two examples discussed in this paper. QMEAN does not assess the experimental data, nor does it have provisions to determine and accommodate the unstructured regions of the molecule.
QMEAN reports an overview of its results via Email and allows the full set of results to be downloaded as a .tgz formatted archive. The archive provides both an overall QMEAN value and residue specific values for each model, as well as files that contain all the underlying data. The residue-specific QMEAN values are also reported in the Bfac column of a PDB-formatted structure file and graphically displayed as a colour-coded ribbon representation of the protein-backbone. No aggregation over the different models of the ensemble is provided; hence no assessment regarding the disparity in the ensemble is available without further user analysis.
Vivaldi
The Protein Data Bank in Europe PDBe, (Velankar et al. 2012) developed the Vivaldi service [VIsualisation and VALidation DIsplay; http://pdbe.org/vivaldi; (Hendrickx et al. 2013)] to validate NMR structures deposited in the public PDB archive. It combines a variety of validation scores from the external validation package CING (Doreleijers et al. 2012a) with internal routines to validate chemical shifts [VASCO, (Rieping and Vranken 2010)], distance restraints, dihedral restraints and residual dipolar couplings. Furthermore, it uses the OLDERADO (Kelley et al. 1996, 1997; Kelley and Sutcliffe 1997) program to cluster models of the NMR ensemble and to define the ordered core region of the protein. Chemical shifts are obtained from NMR-STAR files processed and archived at BMRB (Ulrich et al. 2008), and experimental restraints are obtained as CCPN projects available at the NMR Restraint Grid (NRG) (Doreleijers et al. 2009) database maintained by BMRB. Thus, Vivaldi does not (yet) provide for uploading and assessment of structural and experimental data by an external user.
Vivaldi utilises an interactive Java applet (OpenAstexViewer) to visualise the validation scores in 3D. In addition, per-residue graphs and textual output aids the user to assess the structural quality of an NMR ensemble.
NMR constraints analyser
The NMR Constraints Analyser (Heller and Giorgetti 2010) was explicitly designed for constraint analysis only. It is available as a web server at http://molsim.sci.univr.it/bioinfo/web/, complete with detailed documentation. The contents of the NRG FRED database maintained by BMRB (Doreleijers et al. 2009) are available to the program and can be selected easily by entering the appropriate PDB entry code. External user data can be uploaded as PDB formatted files (.pdb and .mr) with restraints in either CNS (Brunger 2007; Brunger et al. 1998) or CYANA/DYANA (Güntert et al. 1997) format. The accepted formats are well documented, but lack detailed feedback for incorrect input. In addition to the constraint analysis, the program calculates distance restraint completeness, according to procedure described by Doreleijers et al. (1999).
The output of the programs is reported as an interactive webpage that consists of three parts: a graph of the sequence with the number of constraints, the number of violations, an indicator for the presence of torsion angle restraints and the calculated completeness. In addition to these sequence dependent results, a set of tables are reported showing the restraints for one or more selected atoms and a Jmol viewer showing a ribbon diagram of the backbone, colour-coded according to the number of restraints.
Other publicly available servers
While we aimed to cover as exhaustive a list of validation software as possible, the scope of this paper necessarily limits us to testing and describing those servers that are ‘NMR-aware’ and/or aggregate scores of multiple sources. The software listed below can be adapted for NMR-structures, but that often requires specifying a model and chain identifier when running the validation task and sometimes even separating the ensemble into separate files with one model in each file. For these reasons, we did not attempt to test these servers extensively and limit ourselves to brief descriptions.
PDB Validation software (http://deposit.rcsb.org/validate/) performs basic geometry and nomenclature checks for NMR entries. Currently, it is applied to all depositions of NMR structures in the PDB archive. It is also included in PSVS and Quality Control Check.
PROSA (https://prosa.services.came.sbg.ac.at) (Wiederstein and Sippl 2007) is included in the PSVS server, which prepares the input files and averages the output from PROSA over the ensemble. As a standalone server, PROSA accepts ensembles of structures but analyses only one model at a time.
SAVES (Structure Analysis and Verification Server; http://services.mbi.ucla.edu/SAVES) combines 6 structural validation programs, with one of them X-ray specific. Only one model from the NMR ensemble is allowed at a time. Results are presented on simple web pages using colour-coding to indicate possible issues (yellow) and errors (red) with links to graphs an images.
Quality Control Check (http://smb.slac.stanford.edu/jcsg/QC) is a validation server developed by the Joint Center for Structural Genomics (JCSG) is also X-ray centric. It requires an upload of a separate file for each model of the NMR ensemble. It includes 9 validation programs, but only a subset of these (e.g. Molprobity and the PDB Validation software) are relevant to NMR structures.
WHAT IF [http://swift.cmbi.ru.nl/servers/html/index.html; (Vriend 1990)] server is not very NMR aware. Although it can handle an NMR ensemble, it does not produce any aggregated scores. Moreover, the extensive textual output produced for each member of the ensemble is difficult to analyse manually. For this reason, WHAT IF is used by both CING and Vivaldi to derive structural parameters, which are subsequently processed and analysed for the full ensemble and properly presented.
Harmony [http://caps.ncbs.res.in/harmony; (Pugalenthi et al. 2006)] server uses multiple sequence alignment to assess the local structural environment. The information from amino acid substitutions among homologous sequences (in the form of environment-dependent amino acid substitution tables) is then used as a tool for identifying errors that may be present in the protein structure. The server is directed toward X-ray structures, but accepts a PDB file containing an NMR ensemble. The results, however, do not indicate how the individual conformers are scored. Separate outputs are returned for each chain.
Results
We tested the performance of the different packages using two recently solved protein structures as examples. PDB entry 2kq3 (Wang et al. 2010b) was also used in the recent description of the CING package (Doreleijers et al. 2012a) and was now also subjected to the other analyses. PDB entry 2lw9 (Lu et al. 2012) represents the structural ensemble of a relatively small dimeric protein. It was solved using conventional protocols with distance restraints and backbone dihedral angle restraints only, as is still the practice for the majority of entries. Of particular interest is the assessment of a symmetric dimer, as this class of molecules pose specific issues with respect to the experimental procedures by which the intermolecular restraints were derived. For each entry, the data used in the analyses below were obtained as CCPN projects from the NMR Restraints Grid (Doreleijers et al. 2009) database maintained by BMRB (Ulrich et al. 2008), and if necessary exported into other formats with the help of FormatConverter. The chemical shifts files were taken directly from BMRB.
CING
The analyses of the entries 2kq3 and 2lw9 proceeded with all checks applied. The full reports can be examined via the NRG-CING website at http://nmr.cmbi.ru.nl/NRG-CING/data/kq/2kq3/2kq3.cing/2kq3/HTML/index.html, and http://nmr.cmbi.ru.nl/NRG-CING/data/lw/2lw9/2lw9.cing/2lw9/HTML/index.html, respectively. Automated analysis of the ordered regions using the circular variance criteria shows that the ordered sections of PDB entry 2kq3 include 122 out of 140 residues whereas for entry 2lw9 this amounts to 86 out of the 102 total residues in chains A and B (84 %) (Tables 2 and S2).
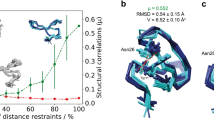
The overall ROG scores, i.e. 0.17/0.65/0.17, for the ordered residues of entry 2lw9 are indicative of problems. Figure 1a shows the residue-specific ROG scores mapped upon the ribbon diagram of the 2lw9 protein. The orange- or red-labelled residues nearly encompass the complete protein, suggesting a general problem. The overall WHAT IF χ1χ2 rotamer normality score of −8.2 ± 0.3, as reported in the CING summary pages, suggests a problem with the side-chain conformations. Indeed, examination of the residue-specific pages of the CING report clearly indicates that the side-chain conformation of many residues is problematic. An example is shown in Fig. 1d for residue Leu9 of chain A of 2lw9, which displays the χ1χ2 plot (the so-called Janin plot). All 20 conformers in the ensemble cluster in a relatively narrow range and exhibit a consistently staggered χ1-rotamer. The problematic side-chain conformations are also flagged by the residue-specific Janin Z-scores (cf. Fig. 1b, bottom panel). The consistent low values of this parameter are also one of the main causes of the orange or red ROG scores of the corresponding residues.
CING results for PDB entry 2lw9. The full CING report can be accessed from http://nmr.cmbi.ru.nl/NRG-CING/data/lw/2lw9/2lw9.cing/2lw9/HTML/index.html. a Ribbon diagram of the first model of 2lw9 colour-coded according to the CING ROG scores. b Residue specific scores for chain A. Top to bottom: secondary structure as derived by DSSP, residue-specific WHAT IF accessibility, Ramachandran, backbone normality, rotamer and Janin scores as function of residue number of chain A. c Ramachandran plot for Leu9. Individual conformers are displayed as green plus signs. Experimental ϕ, ψ dihedral restraints are shown as a transparent light-orange box. Leucine-specific distribution of the accessible conformational ϕ, ψ space, as derived from the WHAT IF reference database of high-resolution crystal structures, is shown in green/yellow/grey. d χ1χ2 plot (also referred to as Janin plot) for Leu9. Labelling as in c). e Overview of the distance restraints page, selected for critiqued restraints only
The 2lw9 protein folds into a simple structure comprised of only two helices per monomer. Indeed, most of the backbone adopts this helical arrangement and the CING DSSP-based analysis (Fig. 1b) confirms their presence. For most of the backbone conformation CING does not signal problems (cf. Fig. 1b, c). One notable residue at the C-terminal end of helix 1 (Thr30), however, displays poor packing, Ramachandran and backbone normality scores (Fig. 1b), resulting in a red residue ROG score. The 2lw9 protein is a symmetric dimer and the analysis results for the corresponding residues in the two different chains are generally similar.
Crucial to a proper validation assessment is the analysis of the experimental data. CING assembles report pages for all experimental data made available to the program. The pages are interactive, as they allow for sorting and selection. Figure 1e shows the report page for the distance restraints of the 2lw9 entry, displaying only the critiqued restraints, i.e. those for which CING detected problems. The results display a series of disturbing lower bounds violations. In particular, it highlights the surprising distance restraints with lower bounds of 4.8–5.0 Å and upper bounds of 7.5–10.4 Å. CING also performs an analysis of the chemical shift assignments if such data are supplied. For 2lw9, the program flags six illogical missing stereo-specific assignments.
Supplementary Fig. S1 displays similar panels to Fig. 1 with the CING analysis results for 2kq3. Figure S1a displays backbone traces of the first member of the 2kq3 NMR ensemble, superposed with the trace of the S.Nase X-ray structure [PDB entry 1ey0 (Chen et al. 2000)]. Residue-specific backbone RMSD values for the ordered regions typically are in the 0.4–1.1 Å range. Regions significantly surpassing these values, e.g. Ile18-Gly20, are often flagged for suspect conformations. Inspection of the side-chain conformations often also yielded unusual results. For example, the Janin-plot of Lys9 reveals a bifurcated distribution of staggered conformers (Fig. S1d). Comparison with the crystal structure clearly reveals the differences in conformation (Fig. S1a). A detailed analysis on the basis of the full CING report was also presented before (Doreleijers et al. 2012a).
Molprobity
Analysis of PDB entries 2lw9 and 2kq3 was initiated from the main Molprobity website (http://molprobity.biochem.duke.edu/) (Davis et al. 2007; Chen et al. 2010) using the built-in feature to retrieve coordinate files from the public PDB archive. All validation scores relevant to NMR protein structures were calculated and analysed.
At the time of writing, Molprobity is undergoing a major version upgrade (V3.19 to V4.00a), which is mainly focussed on improving the calculation of clash-scores. Both versions are available from the website. In this paper, Molprobity V3.19 was used for analysing PDB entries 2lw9 and 2kq3 in order to maintain consistency with other validation packages, which at the time of writing were not yet updated to use the newest version of Molprobity.
The summary statistics Table (Fig. 2a) for entry 2lw9 shows perfect quality scores for bond-angles and bond-lengths (0 % outliers) and good geometry for the Cβ atom (no deviations above 0.25 Å). Molprobity’s assessment of both backbone and side-chain torsion angles is rather poor, displaying 2.8 ± 1.4 % Ramachandran outliers and 35.8 ± 3.9 % unfavoured side-chain rotamers, a result in line with the analysis by CING. Furthermore, clash-scores of 17.3 ± 2.9 are observed indicating an overall problem in protein packing. This results in an overall Molprobity score of 3.4 ± 0.1 Å. Analysis of per-residue tabular output (Fig. 2b) and KiNG images (Chen et al. 2009) (Fig. 2c) for the first model of the NMR ensemble shows that atom clashes are spread throughout the whole interface between the main α-helices of chains A and B, whereas side-chain rotamer outliers are found over the full length of the protein.
Molprobity output for PDB entry 2lw9. a Overview HTML page of the Molprobity analysis containing colour-coded overview scores. b Overview table showing outliers per residue for model 1. c KiNG (Chen et al. 2009) image for model 1 showing small and large overlaps as yellow and red dots, respectively
Geometric validation of PDB entry 2kq3 (cf. Supplementary Fig. S2) shows similar scores as obtained for entry 2lw9. No outliers in bond lengths and angles were observed in any of the 20 models of the NMR ensemble, yet many atom clashes and unfavourable dihedral angles are observed throughout the structures, with 4.8 ± 1.5 % Ramachandran outliers and 31.0 ± 3.0 % bad side-chain rotamers. Furthermore, Molprobity reports very high clash-scores (i.e. 35.7 ± 2.5 serious clashes per 1,000 atoms) spread over the entire protein core.
ResProx
Analysis of PDB entries 2lw9 and 2kq3 was initiated from the main ResProx website (http://www.resprox.ca/) (Berjanskii et al. 2012) using the built-in feature to retrieve coordinate files from the public PDB archive.
The average ‘resolution-by-proxy’ score over the ensemble for PDB entry 2lw9 is 2.9 ± 0.1 Å and is classified as ‘bad’ (>2.5 Å). A breakdown of this score is provided in the Z-score report, showing all 15 measured scores that contribute to the overall resolution. Eight scores are annotated ‘good’ and two ‘bad’ across the whole NMR ensemble, while the remaining five scores have both good and bad models. Interestingly, Molprobity Ramachandran score is considered good in 13 out of 20 models (Z = 1.4 ± 0.8), whereas Molprobity itself reported this score as worrisome (vide supra). Using different cut-offs and/or reference structures could be the underlying cause of this. Furthermore, the Ramachandran score calculated by GeNMR (Berjanskii et al. 2009) (Z = 2.1 ± 0.5) is considered borderline ‘bad’. Another discrepancy exists between χ1 angle scores obtained from VADAR (Willard et al. 2003) and PROSESS (Berjanskii et al. 2010), where the former score is considered ‘bad’ (Z = 3.2 ± 0.2), whereas the latter is considered ‘good’ (Z = 1.6 ± 0.4) and between the Molprobity clash-score (bad; Z = 2.4 ± 0.2) and GeNMR bump score (good; Z = 0.2 ± 0.2). Other ‘bad’ scores include RosettaHoles2 and the GeNMR radius of gyration score. PROSESS summarises a great number of scores for each category by a set of “overall” scores and a global quality score on a scale from 0 (worst) to 10 (best). Only these are reported in Table 3, although individual global and per/residue scores are also available for inspection on the detailed results pages. They convey the same information as discussed above for ResProx, but also include scores for the quality of backbone chemical shifts (poor for entry 2lw9) and distance restraints (good for chain A, only 1 restraint violation > 0.5 Å, but poor for chain B, 5 restraint violations). It is unclear how the summary PROSESS table reports the number of restraint violations, as for individual models this number varies from 1 to 5. All of PROSESS scores are reported separately for each chain.
The reported resolution for PDB entry 2kq3 is 3.2 ± 0.1 Å and thus classified as ‘bad’. Both Ramachandran scores (Molprobity 2.5 ± 0.8 and GeNMR 3.8 ± 0.2), χ1 angle scores (VADAR 3.9 ± 0.2 and PROSESS 2.2 ± 0.3), clash-scores (Molprobity 3.5 ± 0.1 and GeNMR 2.3 ± 0.3) and Θ-hydrogen bond angle score (PROSESS 2.4 ± 0.4) are all beyond two standard deviations of the expected values and thus considered bad. Experimental data validation from PROSESS indicates that chemical shifts are within expected ranges, while the fit to distance restraints is bad with 16 violations (Supplementary Table S3).
PSVS
Figure 3 and Supplementary Figure S3 show the results of the PSVS (Bhattacharya et al. 2007) analysis for entries 2lw9 and 2kq3, respectively. These results are mostly consistent with the assessments from other validation servers described above. The smaller differences in global scores (e.g. Molprobity Ramachandran statistics) arise from the selection of residues submitted for analysis: e.g. ordered residues (cf. Tables 2 and S2) when running PSVS and all residues when running Molprobity itself.
Selection from the PSVS report on PDB entry 2lw9. Sequence numbering in chain B is automatically adjusted to 62–112 for clarity of presentation. a Residues with consistent Ramachandran outliers. b PROCHECK G-factors for backbone dihedral angles. c PROCHECK G-factors for all dihedral angles. d Verify3D scores. e ProsaII energy. f Number of steric clashes. Values in panels (b–f) are plotted as a function of protein sequence. Scores in panels (d–e) for each position i are averaged over a window of [i − 3, i + 3], and are plotted for each model in the ensemble
The global PSVS scores for entry 2lw9 indicate that the packing of the structure is not likely (Z-score for Verify3D of −6.6), and that there are more than the usual number of clashes (Molprobity Z-score of −4.9 for steric clashes). The Ramachandran statistics also indicates that 2 % of the residues over all models are in disallowed regions, with two consistent outliers Thr30 and Asn2 on both chains (Fig. 3a) reported by both Molprobity and PROCHECK-NMR. The other global parameters indicate that the backbone is modelled mostly correctly (Z-score for ProsaII is 1.41 and for PROCHECK-ϕ/ψ angles 2.48). While the side-chain dihedral angles are poor, they are generally within the range commonly observed in NMR structures (PROCHECK all-dihedral-angle Z-score of −2.4) (Lemak et al. 2011). However, all of these global scores may mask individual outliers, and thus inspection on the residue level is necessary (Fig. 3b–f). This analysis confirms that there is a problematic spot around residue Thr30 involving both backbone and side-chain dihedral angles (Fig. 3b, c), while steric clashes are quite numerous, but spread throughout the protein (Fig. 3f). The highest numbers of van der Waals violations (up to 15) are observed for residues Ile15 and Gln35. The AVS analysis of 2lw9 reported an assignment completeness of 36 %; however this low number is due to the fact that the entry is a dimer and the real assignment completeness is therefore closer to 73 %. Only one outlier, Cδ of Arg41, is identified. The analysis of distance and dihedral angle restraints indicates that there were very few restricting long-range restraints (0.2 per residue), and that there were 2 violations per model, which were larger than 0.5 Å.
The results for PDB entry 2kq3 from PSVS indicate that while the protein fold is likely overall correct (Verify3D, ProsaII and PROCHECK-ϕ/ψ Z-scores only moderately negative), the Ramachandran analysis by both Molprobity and PROCHECK-NMR flags some local problems with respect to the backbone. However, the side-chains are most likely modelled incorrectly, resulting in the PROCHECK all-dihedral-angle Z-score of −5.6 and the Molprobity clash Z-score of −8.5. Such values are typically observed in structures that were not refined in explicit water, a procedure known to significantly improve the side-chain packing and side-chain conformations (Nabuurs et al. 2004; Linge et al. 2003; Spronk et al. 2002), a conclusion also supported by CING analysis (Doreleijers et al. 2012b). PSVS also identified more than 40 distance restraint violations per model, with 36 of them greater than 0.5 Å, which may indicate that the data from which the restraints were derived, may have been contradictory or the calibration procedure during the conversion of NOE peaks to restraints inappropriate. Seventeen chemical shift outliers are reported for this entry by the AVS module. The completeness of side-chain resonance assignments is 82 %, although for the aromatic rings, it drops to only 45 %.
QMEAN
The QMEAN analysis was run using its server (http://swissmodel.expasy.org/qmean) (Benkert et al. 2009). The use of an additional FASTA-formatted file with a description of the protein sequence did not alter the results. Figure 4 displays the results obtained for the first model of the 2lw9 ensemble. Manual averaging of the QMEAN scores for all 20 models yielded 0.64 ± 0.03 (Z-score −1.1 ± 0.3). As indicated by the red cross in Fig. 4a, the 2lw9 ensemble scores below average for proteins of comparable size. The QMEAN score is composed of six underlying metrics and their scores are displayed in Fig. 4c. In particular, the Cβ interaction parameter, which is a secondary structure-specific measure, and the torsion parameter, which encodes for a three residue extended torsion, display significant negative values indicative of problems with this structure. QMEAN neither discriminates in its scoring for the unstructured regions of the protein nor examines the underlying experimental NMR data.
Output generated by the QMEAN server for the first model of the ensemble of PDB entry 2lw9. a QMEAN scores as function of the protein sequence length. The score for the first model of the 2lw9 ensemble is indicated by the red cross. b Ribbon diagram colour-coded blue to red (good to bad) according to the residue-specific QMEAN predicted local error. c QMEAN and underlying metrics Z-scores (red indicates below average, blue above average scores)
Figure 4b shows the ribbon diagram of 2lw9 with each residue colour-coded according to the predicted local error. Notably and as was also found by CING and PSVS (vide supra), the C-terminal ends of the two helices are clearly flagged, as are the unstructured C-terminal ends of the protein.
Supplementary Fig. S4 shows the results of the QMEAN analysis for the first model of the 2kq3 ensemble. As was the case for the 2lw9 ensemble, the 2kq3 ensemble scores below average with a QMEAN score for the 20 models of 0.71 ± 0.03 (Z-score −0.4 ± 0.4). Interestingly and in line with the analyses of the other program suites (cf. Supplementary Figs. S1-3), the first β-strand is flagged as a region of predicted local error.
Vivaldi
The validation report of PDB entry 2lw9 is available through the Vivaldi web service at http://www.pdbe.org/vivaldi/2lw9 (Hendrickx et al. 2013). Representative output for PDBe entries 2lw9 and 2kq3 is shown in Fig. 5 and Supplementary Figure S5, respectively. Figure 5a shows a very tight bundle of 20 structures for entry 2lw9, mostly in a helical conformation. Multiple stable domains are obtained from the analysis by the program OLDERADO (Kelley et al. 1996, 1997; Kelley and Sutcliffe 1997) (Tables 2 and S2) and comprise the whole protein except for the N-terminal residues A:1 (and symmetry related B:53) and the C-terminal residues A:45–51 (B:96–103). Since Vivaldi obtains the ROG and geometric validation scores from the NRG-CING web service, the information it presents is already described in the section on CING results, although there are differences in terms of what cut-offs are used to draw the users’ attention to problematic spots (e.g. Ramachandran outliers).
Representative graphical output of the Vivaldi web service at PDBe for PDB entry 2lw9 a Superimposed NMR ensemble coloured by rigid-body domains (OLDERADO). b Per-residue CING scores plotted on the most representative model (model 14) of the NMR ensemble as calculated by OLDERADO. c Distance restraint violations plotted on the most representative model of the ensemble. Yellow lines represent violations below 0.3 Å. Amber lines for violations between 0.3 and 0.5 Å and red lines for violations above 0.5 Å. d Chemical shift outliers identified using VASCO presented as Van der Waals spheres and coloured by a continuous gradient based on the absolute Z-score (3.5 yellow, 5.0 and above red). e Per-residue graph showing the absolute sum of violations in Å. Coloured bars indicate the averaged value over the NMR ensemble. Black dots show violations for the most representative model. Continuous gradient colouring (0.0 Å green, 0.1 Å yellow, >0.2 Å red). f Chemical shift Z-scores on a per-atom basis. Continuous gradient colouring of the absolute Z-score (<2.0 green, 3.5 yellow, >5.0 red)
The Vivaldi analysis of the deposited restraint data shows thirty-three residues with a high number (>50) of distance restraints with relatively few restraints violations (Fig. 5c, e). Taken together with numerous atom clashes and poor packing, this suggests an over-fitting to experimental restraints during structure determination and refinement calculations.
Chemical shift analysis using VASCO (Rieping and Vranken 2010) (Fig. 5d, f) shows a good agreement between the experimental data and the structure. Seven carbon atoms were flagged as chemical shift outliers (Z-score > 3). These outliers have no direct electrostatic interactions with other residues or aromatic side-chains in close proximity to explain their unexpected chemical shifts. As the chemical shift validation routines used by CING and by Vivaldi use different underlying statistics, the referencing corrections are different, which may explain the differences in chemical shift analysis.
Analysis of PDB entry 2kq3 can be obtained from Vivaldi at http://www.pdbe.org/vivaldi/2kq3. Supplementary Figure S5a shows a tightly bundled core region (amber), a flexible N-terminal tail (Thr2-His8) and a flexible loop (Glu43-Ala58).
CING ROG scores (Figure S5b) are predominantly red indicating general problems with the modelled structure. WHAT IF scores indicate moderate Ramachandran, bond length or χ1 angle problems, and atom clashes are reported throughout the protein. Bond angle outliers are reported for His8 and His121 due to the non-planarity of Nε2. This is a commonly observed problem in NMR structures throughout the PDB archive. Atom clashes are reported throughout the core domain and amount to over 0.2 Å for 21 residues and over 0.4 Å for 4 additional residues.
Unusual chemical shift values are identified for 72 atoms from 40 different residues (supplementary Figs S5d, f) and are mostly concentrated on lysine residues. As VASCO does not take aromatic interactions into account, manual inspection of these outliers is advised. Six chemical shift outliers are identified for Lys9, all with negative Z-scores (i.e. the experimental chemical shift is smaller than the expected shift), thus suggesting a substantial ring-current effect induced by an aromatic side-chain. Inspection of the structure, however, does not yield a likely candidate. The other 39 residues with chemical shift outliers are scattered throughout the molecule.
Vivaldi analysed 2,091 distance restraints, which mainly cover residue ranges 7–41 and 61–140. The molecule has approximately 20 distance restraints per residue on average. Restraint violations are shown in Supplementary Fig. S5c, e.
NMR constraints analyser
Figure 6 and Supplementary Figure S6 show representative output of the NMR Constraints Analyser (Heller and Giorgetti 2010) web server for PDB entries 2lw9 and 2kq3, respectively. The program was tested in December 2012. Tabulation of the restraint content and completeness as function of residue is displayed in Fig. 6b for 2lw9. The program displays restraints for a single chain at a time, also in the case of dimers. Clicking the bar graph selects the corresponding residue for display in the viewer (not shown), and restraints selected in the table (cf. Supplementary Fig. 6c) can be displayed in the viewer as well. Regions of the molecule that are well or badly defined by restraints appear clearly, but there are no reference values to indicate local or global structure quality as such. Compared to dedicated analysis programs, such CcpNmr Analysis (Vranken et al. 2005), the NMR Constraints Analyser is neat and easy to use, but clearly lacking in detail. The restraint tables give the upper distance limit and the number of violated models, but lack information about lower limits, or actual distances or violation values. Also, restraints involving pseudo-atoms, such as methyl groups, cannot be visualised on the Jmol viewer.
Representative output of the NMR Constraints Analyser web server, showing the results of the analysis of PDB entry 2lw9. a Structure of 2lw9, colored by constraint density (red: many; blue: few), with Lys22 shown. b Constraint completeness (red line), number of constraints (dark bars) and number of violations (red triangles) along chain B of 2lw9. Although chains A and B are identical, the program only gives results for one chain of one structure model. c Constraints for Lys22 Hδ1 and HN
Discussion
Over the past decades, NMR has proved itself as a very versatile technique for structure determination of biomacromolecules and as a credible complement to X-ray crystallography. However, it is prone to serious errors particularly when misinterpreted, conflicting or over interpreted data are used (Bhattacharya et al. 2007; Doreleijers et al. 2012a; Lemak et al. 2011; Mao et al. 2011). Hence, the validation of input data, the resulting structures and the fit between the structural models and the experimental data is an absolute necessity for assessing and using NMR-derived structures in other biological applications. This need was also recognised by the wwPDB consortium (Berman et al. 2007) who appointed an NMR validation taskforce (NMR-VTF). The primary task for the NMR VTF was to define commonly accepted procedures and guidelines for validation of NMR structures. The NMR-VTF has now put forward its recommendations (Montelione et al. submitted), which will ultimately result in a set of tools that will be applied to all NMR entries deposited in the PDB archive. The authors of this review paper are directly involved in the implementation of these tools, most of which will be based upon the programs discussed in this review. At present, a regularly updated archive of CING validation reports of nearly all NMR entries of the PDB archive, called NRG-CING (Doreleijers et al. 2012b), is available for inspection at http://nmr.cmbi.ru.nl/NRG-CING.
NMR-derived structures typically encompass both structured and less-structured regions. The latter typically score worse on parameters used to characterise ordered structure. As a near complete set of NMR data, especially the chemical shifts, is required for the proper analysis of the structure, it is desirable to still include the full molecule in the validation analysis. Dedicated NMR validation programs, such as PSVS (Bhattacharya et al. 2007) or CING (Doreleijers et al. 2012a), routinely report on both the structured and full-length molecule. Table 2 lists the structured regions defined for entry 2lw9 as obtained by the different programs (Table S2 contains the corresponding information for entry 2kq3). The FindCore algorithm (Snyder and Montelione 2005) is clearly more restrictive when compared to the methods based on dihedral order parameter. For the latter, all algorithms yield almost the same results, differing only slightly for residues 94–96 at the C-terminal end of chain B.
Table 3 lists a summary of the structural and data assessment by the different programs for PDB entry 2lw9 (Table S3 reports on entry 2kq3). Overall, all programs indicate substantial problems with entry 2lw9. Whereas the overall fold is likely correct, conformational parameters related to the backbone and side-chain conformation and packing indicate significant problems. In particular, the C- and N-termini of the two helices are specifically flagged by multiple programs. At the level of restraints, the different programs all signal problems, i.e. violations, with respect to the agreement between the structural and experimental data. Disturbing lower-bound restraint violations and odd distance restraints are flagged by CING. The structural and data analysis together suggests errors in the modelling protocol used to derive the structural ensemble.
PDB entry 2kq3 (Wang et al. 2010b) has been used before as an example for the description of the CING program. Like CING, all other programs identify similar problems related to conformation and packing for this entry (cf. Supplementary Figures S2-6). We previously indicated a number of specific problematic areas, such as the first β-strand, which are also flagged by the other programs. Refinement in explicit solvent remedies these issues to some extent, and for this entry we previously showed that we could improve upon both the backbone and side-chain conformations (Doreleijers et al. 2012a).
The tested methods for the validation of chemical shift assignments do not produce a consistent picture for the two entries (cf. Tables 3 and S3). Currently VASCO produces the longest list of unusual chemical shifts, but it can only examine entries already present in the PDB and BMRB databases, making it less useful during the structure determination process. PSVS includes the AVS method, and can identify at least some outliers, but does not correct referencing. CING does correct the referencing but is more lenient towards declaring a shift value an outlier. In our opinion, it appears sensible to perform at least these two analyses and to confirm that any outliers are genuine, rather than due to clerical errors or wrong assignments.
This review was not aimed at discussing the two entries per se; rather they served as examples for the procedures implemented in the different programs. Many of these, i.e. CING, ResProx (Berjanskii et al. 2012), PSVS (Bhattacharya et al. 2007), QMEAN (Benkert et al. 2009) and Vivaldi (Hendrickx et al. 2013), are in effect based (in part) upon the results of a number of other underlying programs, that are sometimes partially overlapping. For example, PSVS and ResProx both use scores from the Molprobity program (Davis et al. 2007; Chen et al. 2010) and Vivaldi is heavily based upon the CING/WHAT IF (Vriend 1990; Hooft et al. 1996) assessments. Careful comparison of the different results yields some notable features. Whereas both PSVS and ResProx use the Molprobity Ramachandran score, the results for 2lw9 are qualified as ‘bad’ and ‘good’ by the two programs, respectively (cf. Table 3). The ResProx scores for side-chain assessment [VADAR χ1 (Willard et al. 2003) and PROSESS χ1 (Berjanskii et al. 2010)] also receive conflicting labels, as do its scores for packing [Molprobity clash-score and GeNMR bumps (Berjanskii et al. 2009)], suggesting that potentially the rescaling of the original scores to generalised Z–scores requires revisiting. Alternatively, these differences may be genuine and reflect the different sensitivities of the parameters to the problems present in entry 2lw9. Overall, our analysis of entries 2lw9 and 2kq3 by the different validation program suites suggests that, in addition to aggregated or transformed scores, it is beneficial for the user to also have access to original values of the parameters as obtained from the underlying program. This allows for a more straightforward comparison of the results obtained by the different validation suites. For the PSVS and CING programs, the original results are already directly accessible.
The comparison also raises a more fundamental question related to the significance of the different parameters; i.e. what validates the validators? Here, we would suggest the notion of ‘usual suspects’: patterns of poor indicators typically signal problems and only in exceptional cases are there genuine reasons to discard the overall conclusions. As many of the tools are based upon prior knowledge derived from the PDB database, features not yet present or under-represented may potentially be flagged unnecessarily. However, given the now extensive coverage of structural motifs in the PDB archive, such occurrences are very rare and should be treated with extreme caution. Examples of these are the chemical modification of residues, or the inclusion of D-enantiomers or other unusual amino acids.
The assessment of the structural quality on the basis of a combined set of different parameters has also proven to be a viable approach for the identification of the serious cases of outright incorrect structures (Bhattacharya et al. 2007; Doreleijers et al. 2012a). In fact, the developments of both the PSVS and CING suites were prompted by the erroneous structure 1tgq (discussed by Nabuurs et al., 2005), now replaced by PDB entry 2b95. A subsequent PSVS analysis of entry 1tgq clearly marked it as highly suspect (Bhattacharya et al. 2007), while its CING ROG scores, i.e. 0.54/0.30/0.16, also flagged it as highly problematic. In contrast, the revised entry 2b95 yields the much more acceptable ROG scores 0.37/0.35/0.28 and the highly homologous entry 1y4o yields 0.16/0.15/0.69 (Doreleijers et al. 2012b), characteristic of a well-modelled structure.
With the exception of Vivaldi, which operates directly upon the data deposited in the PDB and BMRB archives, the other programs that are currently maintained all feature an on-line server (cf. Table 1) for user submission of data. The file formats for the input to these different validation programs vary considerably: all programs accept PDB version 3 formatted files for the structural data, although some can handle only one model at a time (e.g. QMEAN). At present, none of the programs appear capable of using the much more modern mmCIF or PDBML/XML formats for the structural data. The CING program also accepts the CCPN format for structural data. Only a subset of the validation programs, i.e. CING, PSVS, PROSESS and Vivaldi, also validate the restraint and other experimental data. Formats for these are much more diverse, with either CYANA (PSVS, CING), Xplor/CNS (PSVS, PROSESS) or CCPN (CING) formatted data being accepted. Vivaldi and NRG-CING use the experimental restraints data remediated by BMRB and available from the NRG database.
Not only the input, but also the output generated by the different programs varies greatly. In certain cases (e.g. QMEAN, WHAT IF, Molprobity) the results for the different models in the ensemble are presented as separate entities and hence require manual averaging, a generally cumbersome procedure. PSVS features a summary page with key metrics summarised at a glance and collates detailed validation information in drill down pages (HTML or PDF). CING features hyperlinked and interactive webpages, which facilitate directed examination of the results. Particular emphasis is placed on the relation between experimental data and structural results. Vivaldi is a visualisation tool featuring interactive 3D viewer and graphs, exposing the validation information to non-expert users of the PDB archive. A notable feature of ResProx is the extensive list of suggestions that potentially could improve the different validation scores.
The development of validation software is a continuous process that has to keep pace with the development of NMR methodology. In our opinion, the community of NMR software developers could consider the following points to make the structure and experimental data validation more widespread and results more easily compared: (1) agree on a test set of macromolecular structures with known ‘good’ and ‘bad’ features to benchmark and compare their tools; (2) agree on standardised input formats for experimental NMR data, as conversion between the numerous existing formats is not trivial and most servers accept only a small subset of formats; moreover, there is some variation even within a given format, making the experience of a non-expert user quite frustrating; (3) if constituent validation scores are converted into Z-scores, the raw scores should still be made available; (4) the validation servers should state versions of constituent software used to obtain the scores; (5) they should provide APIs or machine-readable output. We have also found a number of features in the surveyed servers very useful that perhaps can be emulated by other developers: (1) suggestions on how to address a problematic structural feature (as done by ResProx); (2) ability to directly compare X-ray and NMR structures (e.g. PSVS, ResProx, Molprobity); (3) easy navigation in the results and functionality to present all relevant scores for individual residues or even atoms (CING) rather than grouping by scores only; (4) detailed analysis of peak lists (PSVS, CING).
Conclusions
Structural quality and the agreement between experimental data and structural results can be greatly improved by the application of validation routines. Nowadays, several packages taken together already supply ample tools to avoid trivial and hence unnecessary errors. By consistently using these tools as an integrated part of the structure determination process, the resulting outcome will not only be better in terms of quality, but also more confidently address the biological problem. Fortunately, the (anonymous) CING user statistics suggest an increasing number of regular returning users that submit jobs likely to represent different stages of their structure determination process. It is our recommendation that the assessment of the structural quality should be done in relation to the experimental data. We often find that regions of poor structural quality also display poorer agreement with the experimental data, such as NOE restraints (Nabuurs et al. 2005), or peaks (RPF scores; Huang et al. 2005; Bhattacharya et al. 2007) and back-calculated chemical shifts. Programs like PSVS and CING perform an integrated analysis that provides this information relatively easily, thus allowing for improvement of the structure calculations. Ultimately, this could result in the most optimal structures being deposited to the PDB archive.
The next stage for NMR structure validation also should include cross-validation with independent data (Brunger et al. 1993; Clore and Schwieters 2006; Nabuurs et al. 2005; Tjandra et al. 2007) as a standard procedure. Although significantly more complicated for NMR-derived biomolecular structures compared to X-ray structures, because of the diversity in the NMR data, the much increased data content of the average experimental NMR data set relative to the situation 10–15 years ago renders the cross validation procedures quite feasible.
The proper analysis of NMR-derived structures containing oligonucleotides or small-molecule ligands is currently still incomplete. Although most programs, e.g. CING and PSVS, will readily accept these and perform basic assessments, few dedicated tools are currently available for these non-protein macromolecules. Proper validation also requires provisions for NMR-specific phenomena, most prominently dynamics. All current analysis routines fail when confronted with ensembles of ensembles, generated to model the different dynamical states in concert with the structure. All these much-required developments are topics of on-going research and implementation.
References
Bagaria A, Jaravine V, Huang YPJ, Montelione GT, Güntert P (2012) Protein structure validation by generalized linear model root-mean-square deviation prediction. Protein Sci 21:229–238. doi:10.1002/Pro.2007
Bax A, Grishaev A (2005) Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struc Biol 15:563–570. doi:10.1016/J.Sbi.2005.08.006
Benkert P, Künzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37:W510–W514. doi:10.1093/Nar/Gkp322
Berjanskii MV, Wishart DS (2005) A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc 127:14970–14971. doi:10.1021/Ja054842f
Berjanskii M, Tang P, Liang J, Cruz JA, Zhou JJ, Zhou Y, Bassett E, MacDonell C, Lu P, Lin GH, Wishart DS (2009) GeNMR: a web server for rapid NMR-based protein structure determination. Nucleic Acids Res 37:W670–W677. doi:10.1093/Nar/Gkp280
Berjanskii M, Liang YJ, Zhou JJ, Tang P, Stothard P, Zhou Y, Cruz J, MacDonell C, Lin GH, Lu P, Wishart DS (2010) PROSESS: a protein structure evaluation suite and server. Nucleic Acids Res 38:W633–W640. doi:10.1093/Nar/Gkq375
Berjanskii M, Zhou JJ, Liang YJ, Lin GH, Wishart DS (2012) Resolution-by-proxy: a simple measure for assessing and comparing the overall quality of NMR protein structures. J Biomol NMR 53:167–180. doi:10.1007/S10858-012-9637-2
Berman HM (2008) The Protein Data Bank: a historical perspective. Acta Crystallogr A 64:88–95. doi:10.1107/S0108767307035623
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. doi:10.1093/Nar/28.1.235
Berman H, Henrick K, Nakamura H, Markley JL (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35:D301–D303. doi:10.1093/Nar/Gkl971
Bernard A, Vranken WF, Bardiaux B, Nilges M, Malliavin TE (2011) Bayesian estimation of NMR restraint potential and weight: a validation on a representative set of protein structures. Proteins 79:1525–1537. doi:10.1002/Prot.22980
Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) Protein Data Bank—computer-based archival file for macromolecular structures. J Mol Biol 112:535–542
Best RB, Vendruscolo M (2004) Determination of protein structures consistent with NMR order parameters. J Am Chem Soc 126:8090–8091. doi:10.1021/Ja0396955
Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66:778–795. doi:10.1002/Prot.21165
Borrell B (2009) Fraud rocks protein community. Nature 462:970. doi:10.1038/462970a
Brunger AT (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2:2728–2733. doi:10.1038/Nprot.2007.406
Brunger AT, Clore GM, Gronenborn AM, Saffrich R, Nilges M (1993) Assessing the quality of solution nuclear-magnetic-resonance structures by complete cross-validation. Science 261:328–331. doi:10.1126/Science.8332897
Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54:905–921. doi:10.1107/S0907444998003254
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A 104:9615–9620. doi:10.1073/pnas.0610313104
Chen JM, Lu ZQ, Sakon J, Stites WE (2000) Increasing the thermostability of staphylococcal nuclease: implications for the origin of protein thermostability. J Mol Biol 303:125–130. doi:10.1006/Jmbi.2000.4140
Chen VB, Davis IW, Richardson DC (2009) KiNG (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci 18:2403–2409. doi:10.1002/Pro.250
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21. doi:10.1107/S0907444909042073
Clore GM, Schwieters CD (2006) Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol 355:879–886. doi:10.1016/J.Jmb.2005.11.042
Clore GM, Omichinski JG, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn AM (1995) Interhelical angles in the solution structure of the oligomerization domain of P53. Science 267:1515–1516. doi:10.1126/Science.7878474
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi:10.1093/Nar/Gkm216
Doreleijers JF, Raves ML, Rullmann T, Kaptein R (1999) Completeness of NOEs in protein structures: a statistical analysis of NMR data. J Biomol NMR 14:123–132. doi:10.1023/A:1008335423527
Doreleijers JF, Nederveen AJ, Vranken W, Lin JD, Bonvin AMJJ, Kaptein R, Markley JL, Ulrich EL (2005) BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J Biomol NMR 32:1–12. doi:10.1007/S10858-005-2195-0
Doreleijers JF, Vranken WF, Schulte C, Lin JD, Wedell JR, Penkett CJ, Vuister GW, Vriend G, Markley JL, Ulrich EL (2009) The NMR restraints grid at BMRB for 5,266 protein and nucleic acid PDB entries. J Biomol NMR 45:389–396. doi:10.1007/S10858-009-9378-Z
Doreleijers JF, da Silva AWS, Krieger E, Nabuurs SB, Spronk CAEM, Stevens TJ, Vranken WF, Vriend G, Vuister GW (2012a) CING: an integrated residue-based structure validation program suite. J Biomol NMR 54:267–283. doi:10.1007/S10858-012-9669-7
Doreleijers JF, Vranken WF, Schulte C, Markley JL, Ulrich EL, Vriend G, Vuister GW (2012b) NRG-CING: integrated validation reports of remediated experimental biomolecular NMR data and coordinates in wwPDB. Nucleic Acids Res 40:D519–D524. doi:10.1093/Nar/Gkr1134
Eisenberg D, Luthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Method Enzymol 277:396–404
Engh RA, Huber R (1991) Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr A 47:392–400. doi:10.1107/S0108767391001071
Engh RA, Huber R (2001) International Tables for Crystallography. In: Rossmann MG, Arnold E (eds) International Tables for Crystallography, vol F. Kluwer Academic Publishers, Dordrecht, pp 382–392
Gabel F, Simon B, Nilges M, Petoukhov M, Svergun D, Sattler M (2008) A structure refinement protocol combining NMR residual dipolar couplings and small angle scattering restraints. J Biomol NMR 41:199–208. doi:10.1007/S10858-008-9258-Y
Ginzinger SW, Gerick F, Coles M, Heun V (2007) CheckShift: automatic correction of inconsistent chemical shift referencing. J Biomol NMR 39:223–227. doi:10.1007/S10858-007-9191-5
Ginzinger SW, Skocibusic M, Heun V (2009) CheckShift improved: fast chemical shift reference correction with high accuracy. J Biomol NMR 44:207–211. doi:10.1007/S10858-009-9330-2
Gore S, Velankar S, Kleywegt GJ (2012) Implementing an X-ray validation pipeline for the Protein Data Bank. Acta Crystallogr D 68:478–483. doi:10.1107/S0907444911050359
Güntert P (1998) Structure calculation of biological macromolecules from NMR data. Q Rev Biophys 31:145–237. doi:10.1017/S0033583598003436
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298. doi:10.1006/Jmbi.1997.1284
Haliloglu T, Bahar I, Erman B (1997) Gaussian dynamics of folded proteins. Phys Rev Lett 79:3090–3093. doi:10.1103/Physrevlett.79.3090
Han B, Liu YF, Ginzinger SW, Wishart DS (2011) SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR 50:43–57. doi:10.1007/S10858-011-9478-4
Heller DM, Giorgetti A (2010) NMR constraints analyser: a web-server for the graphical analysis of NMR experimental constraints. Nucleic Acids Res 38:W628–W632. doi:10.1093/Nar/Gkq484
Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng ZK, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang HW, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL (2012) Outcome of the first electron microscopy validation task force meeting. Structure 20:205–214. doi:10.1016/J.Str.2011.12.014
Hendrickx PMS, Gutmanas A, Kleywegt GJ (2013) Vivaldi: visualisation and validation of biomacromolecular NMR structures from the PDB. Proteins 81:583–591. doi:10.1002/prot.24213
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227. doi:10.1016/S0022-2836(02)00241-3
Hooft RWW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381:272
Huang YJ, Powers R, Montelione GT (2005) Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc 127:1665–1674. doi:10.1021/Ja047109h
Jakovkin I, Klipfel M, Muhle-Goll C, Ulrich AS, Luy B, Sternberg U (2012) Rapid calculation of protein chemical shifts using bond polarization theory and its application to protein structure refinement. Phys Chem Chem Phys 14(35):12263–12276. doi:10.1039/C2cp41726j
Joosten RP, Beek TAHT, Krieger E, Hekkelman ML, Hooft RWW, Schneider R, Sander C, Vriend G (2011) A series of PDB related databases for everyday needs. Nucleic Acids Res 39:D411–D419. doi:10.1093/Nar/Gkq1105
Kabsch W, Sander C (1983) Dictionary of protein secondary structure—pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637. doi:10.1002/Bip.360221211
Kelley LA, Sutcliffe MJ (1997) OLDERADO: on-line database of ensemble representatives and domains. Protein Sci 6:2628–2630
Kelley LA, Gardner SP, Sutcliffe MJ (1996) An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng 9:1063–1065. doi:10.1093/Protein/9.11.1063
Kelley LA, Gardner SP, Sutcliffe MJ (1997) An automated approach for defining core atoms and domains in an ensemble of NMR-derived protein structures. Protein Eng 10:737–741. doi:10.1093/Protein/10.6.737
Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, Kengaku Y, Cho H, Standley DM, Nakagawa A, Nakamura H (2012) Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res 40:D453–D460. doi:10.1093/Nar/Gkr811
Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M (2009) Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc 131:13894–13895. doi:10.1021/Ja903772t
Kuszewski J, Clore GM (2000) Sources of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J Magn Reson 146:249–254. doi:10.1006/Jmre.2000.2142
Lambert LJ, Schirf V, Demeler B, Cadene M, Werner MH (2004) Flipping a genetic switch by subunit exchange (vol 20, pg 7149, 2001). EMBO J 23:3186. doi:10.1038/Sj.Emboj.7600313
Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi:10.1107/S0021889892009944
Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. doi:10.1007/Bf00228148
Lehtivarjo J, Hassinen T, Korhonen SP, Peräkylä M, Laatikainen R (2009) 4D prediction of protein H-1 chemical shifts. J Biomol NMR 45:413–426. doi:10.1007/S10858-009-9384-1
Lemak A, Gutmanas A, Chitayat S, Karra M, Farès C, Sunnerhagen M, Arrowsmith CH (2011) A novel strategy for NMR resonance assignment and protein structure determination. J Biomol NMR 49:27–38. doi:10.1007/S10858-010-9458-0
Li DW, Brüschweiler R (2012) PPM: a side-chain and backbone chemical shift predictor for the assessment of protein conformational ensembles. J Biomol NMR 54(3):257–265. doi:10.1007/s10858-012-9668-8
Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M (2005) Simultaneous determination of protein structure and dynamics. Nature 433(7022):128–132. doi:10.1038/Nature03199
Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M (2003) Refinement of protein structures in explicit solvent. Proteins 50:496–506. doi:10.1002/Prot.10299
Lopez-Mendez B, Güntert P (2006) Automated protein structure determination from NMR spectra. J Am Chem Soc 128:13112–13122. doi:10.1021/Ja061136l
Lovell SC, Davis IW, Arendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins Struct Funct Genet 50:437–450. doi:10.1002/Prot.10286
Lu Q, Ye F, Wei ZY, Wen ZL, Zhang MJ (2012) Antiparallel coiled-coil-mediated dimerization of myosin X. Proc Natl Acad Sci USA 109:17388–17393. doi:10.1073/Pnas.1208642109
Mao BC, Guan RJ, Montelione GT (2011) Improved technologies now routinely provide protein NMR structures useful for molecular replacement. Structure 19:757–766. doi:10.1016/J.Str.2011.04.005
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi:10.1093/Bioinformatics/16.4.404
Meiler J (2003) PROSHIFT: protein chemical shift prediction using artificial neural networks. J Biomol NMR 26:25–37. doi:10.1023/A:1023060720156
Möllhoff M, Sternberg U (2001) Molecular mechanics with fluctuating atomic charges—a new force field with a semi-empirical charge calculation. J Mol Model 7(4):90–102. doi:10.1007/s008940100008
Montalvao RW, De Simone A, Vendruscolo M (2012) Determination of structural fluctuations of proteins from structure-based calculations of residual dipolar couplings. J Biomol NMR 53(4):281–292. doi:10.1007/S10858-012-9644-3
Montelione GT, Berman H, Nilges M, Bax A, Güntert P, Herrmann T, Kleywegt GJ, Markley JL, Richardson JS, Schwieters CD, Vuister GW, Vranken W, Wishart DS (submitted) Recommendations of the wwPDB NMR validation task force. Structure
Moon S, Case DA (2007) A new model for chemical shifts of amide hydrogens in proteins. J Biomol NMR 38:139–150. doi:10.1007/S10858-007-9156-8
Moseley HNB, Sahota G, Montelione GT (2004) Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR 28:341–355
Moult J, Pedersen JT, Judson R, Fidelis K (1995) A large-scale experiment to assess protein-structure prediction methods. Proteins Struct Funct Genet 23:R2–R4. doi:10.1002/Prot.340230303
Moult J, Fidelis K, Kryshtafovych A, Tramontano A (2011) Critical assessment of methods of protein structure prediction (CASP)- Round IX. Proteins 79:1–5. doi:10.1002/Prot.23200
Mulder FAA (2009) Leucine side-chain conformation and dynamics in proteins from C-13 NMR chemical shifts. ChemBioChem 10:1477–1479. doi:10.1002/Cbic.200900086
Nabuurs SB, Spronk CAEM, Krieger E, Maassen H, Vriend G, Vuister GW (2003) Quantitative evaluation of experimental NMR restraints. J Am Chem Soc 125:12026–12034. doi:10.1021/Ja035440f
Nabuurs SB, Nederveen AJ, Vranken W, Doreleijers JF, Bonvin AMJJ, Vuister GW, Vriend G, Spronk CAEM (2004) DRESS: a database of REfined solution NMR structures. Proteins 55:483–486. doi:10.1002/Prot.20118
Nabuurs SB, Krieger E, Spronk CAEM, Nederveen AJ, Vriend G, Vuister GW (2005) Definition of a new information-based per-residue quality parameter. J Biomol NMR 33:123–134. doi:10.1007/S10858-005-2826-5
Nabuurs SB, Spronk CAEM, Vuister GW, Vriend G (2006) Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. PLoS Comput Biol 2:71–79. doi:10.1371/journal.pcbi.0020009
Neal S, Nip AM, Zhang HY, Wishart DS (2003) Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. J Biomol NMR 26:215–240
Parkinson G, Vojtechovsky J, Clowney L, Brunger AT, Berman HM (1996) New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr D 52:57–64. doi:10.1107/S0907444995011115
Pugalenthi G, Shameer K, Srinivasan N, Sowdhamini R (2006) HARMONY: a server for the assessment of protein structures. Nucleic Acids Res 34:W231–W234. doi:10.1093/Nar/Gkl314
Read RJ, Adams PD, Arendall WB, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lütteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH (2011) A new generation of crystallographic validation tools for the Protein Data Bank. Structure 19:1395–1412. doi:10.1016/J.Str.2011.08.006
Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, Keating KS, Pyle AM, Micallef D, Westbrook J, Berman HM (2008) RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA 14:465–481. doi:10.1261/Rna.657708
Rieping W, Vranken WF (2010) Validation of archived chemical shifts through atomic coordinates. Proteins 78:2482–2489. doi:10.1002/Prot.22756
Rieping W, Habeck M, Nilges M (2005) Inferential structure determination. Science 309:303–306. doi:10.1126/Science.1110428
Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M (2007) ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23:381–382. doi:10.1093/Bioinformatics/Btl589
Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Güntert P, Herrmann T, Huang YJ, Jonker HRA, Mao B, Malliavin TE, Montelione GT, Nilges M, Raman S, van der Schot G, Vranken WF, Vuister GW, Bonvin AMJJ (2009) CASD-NMR: critical assessment of automated structure determination by NMR. Nat Methods 6:625–626. doi:10.1038/Nmeth0909-625
Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Güntert P, He YF, Herrmann T, Huang YPJ, Jaravine V, Jonker HRA, Kennedy MA, Lange OF, Liu GH, Malliavin TE, Mani R, Mao BC, Montelione GT, Nilges M, Rossi P, van der Schot G, Schwalbe H, Szyperski TA, Vendruscolo M, Vernon R, Vranken WF, de Vries S, Vuister GW, Wu B, Yang YH, Bonvin AMJJ (2012) Blind testing of routine, fully automated determination of protein structures from NMR data. Structure 20:227–236. doi:10.1016/J.Str.2012.01.002
Sahakyan AB, Vranken WF, Cavalli A, Vendruscolo M (2011a) Structure-based prediction of methyl chemical shifts in proteins. J Biomol NMR 50:331–346. doi:10.1007/S10858-011-9524-2
Sahakyan AB, Vranken WF, Cavalli A, Vendruscolo M (2011b) Using side-chain aromatic proton chemical shifts for a quantitative analysis of protein structures. Angew Chem Int Edit 50:9620–9623. doi:10.1002/Anie.201101641
Schubert M, Labudde D, Oschkinat H, Schmieder P (2002) A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on C-13 chemical shift statistics. J Biomol NMR 24:149–154. doi:10.1023/A:1020997118364
Schwieters CD, Clore GM (2008) A pseudopotential for improving the packing of ellipsoidal protein structures determined from NMR data. J Phys Chem B 112:6070–6073. doi:10.1021/Jp076244o
Sheffler W, Baker D (2010) RosettaHoles2: a volumetric packing measure for protein structure refinement and validation. Protein Sci 19:1991–1995. doi:10.1002/Pro.458
Shen Y, Bax A (2007) Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR 38:289–302. doi:10.1007/S10858-007-9166-6
Shen Y, Bax A (2010a) Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR 46:199–204. doi:10.1007/S10858-009-9395-Y
Shen Y, Bax A (2010b) SPARTA plus: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR 48:13–22. doi:10.1007/S10858-010-9433-9
Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu GH, Eletsky A, Wu YB, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA 105:4685–4690. doi:10.1073/Pnas.0800256105
Shen Y, Delaglio F, Cornilescu G, Bax A (2009a) TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223. doi:10.1007/S10858-009-9333-Z
Shen Y, Vernon R, Baker D, Bax A (2009b) De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR 43:63–78. doi:10.1007/S10858-008-9288-5
Siemion IZ, Wieland T, Pook KH (1975) Influence of distance of proline carbonyl from beta and gamma carbon on C-13 chemical-shifts. Angew Chem Int Ed Engl 14:702–703. doi:10.1002/Anie.197507021
Snyder DA, Montelione GT (2005) Clustering algorithms for identifying core atom sets and for assessing the precision of protein structure ensembles. Proteins 59:673–686. doi:10.1002/Prot.20402
Spadaccini R, Perrin H, Bottomley MJ, Ansieau S, Sattler M (2006) Structure and functional analysis of the MYND domain (Retracted article. See vol 376, pp. 1523, 2008). J Mol Biol 358:498–508. doi:10.1016/J.Jmb.2006.01.087
Spronk CAEM, Linge JP, Hilbers CW, Vuister GW (2002) Improving the quality of protein structures derived by NMR spectroscopy. J Biomol NMR 22:281–289. doi:10.1023/A:1014971029663
Stein EG, Rice LM, Brunger AT (1997) Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J Magn Reson 124:154–164. doi:10.1006/Jmre.1996.1027
Tjandra N, Garrett DS, Gronenborn AM, Bax A, Clore GM (1997) Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy. Nat Struct Biol 4:443–449. doi:10.1038/Nsb0697-443
Tjandra N, Suzuki M, Chang SL (2007) Refinement of protein structure against non-redundant carbonyl C-13 NMR relaxation. J Biomol NMR 38:243–253. doi:10.1007/S10858-007-9165-7
Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM (2013) Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure 21:875–881. doi:10.1016/j.str.2013.04.020
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL (2008) BioMagResBank. Nucleic Acids Res 36:D402–D408. doi:10.1093/Nar/Gkm957
Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, Fernandez Montecelo MA, van Ginkel G, Golovin A, Gore SP, Gutmanas A, Haslam P, Hendrickx PMS, Heuson E, Hirshberg M, John M, Lagerstedt I, Mir S, Newman LE, Oldfield TJ, Patwardhan A, Rinaldi L, Sahni G, Sanz-Garcia E, Sen S, Slowley R, Suarez-Uruena A, Swaminathan GJ, Symmons MF, Vranken WF, Wainwright M, Kleywegt GJ (2012) PDBe: Protein Data Bank in Europe. Nucleic Acids Res 40:D445–D452. doi:10.1093/Nar/Gkr998
Vila JA, Arnautova YA, Martin OA, Scheraga HA (2009) Quantum-mechanics-derived C-13(alpha) chemical shift server (CheShift) for protein structure validation. P Natl Acad Sci USA 106 (40):16972–16977. doi:10.1073/Pnas.0908833106
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas P, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696. doi:10.1002/Prot.20449
Vriend G (1990) WHAT IF—a molecular modeling and drug design program. J Mol Graphics 8:52–56
Vuister GW, Tjandra N, Shen Y, Grishaev A, Grzesiek S (2011) Measurement of structural constraints. In: Lian L-Y, Robers G (eds) Protein NMR Spectroscopy: Principle Techniques and Applications. Wiley & Sons Ltd, West Sussex (UK), pp 83–158
Wang LY, Markley JL (2009) Empirical correlation between protein backbone N-15 and C-13 secondary chemical shifts and its application to nitrogen chemical shift re-referencing. J Biomol NMR 44:95–99. doi:10.1007/S10858-009-9324-0
Wang LY, Eghbalnia HR, Bahrami A, Markley JL (2005) Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR 32:13–22. doi:10.1007/S10858-005-1717-0
Wang BW, Wang YJ, Wishart DS (2010a) A probabilistic approach for validating protein NMR chemical shift assignments. J Biomol NMR 47:85–99. doi:10.1007/S10858-010-9407-Y
Wang M, Feng YA, Yao HW, Wang JF (2010b) Importance of the C-terminal loop L137–S141 for the folding and folding stability of staphylococcal nuclease. Biochemistry-Us 49:4318–4326. doi:10.1021/Bi100118k
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410. doi:10.1093/Nar/Gkm290
Willard L, Ranjan A, Zhang HY, Monzavi H, Boyko RF, Sykes BD, Wishart DS (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 31:3316–3319. doi:10.1093/Nar/Gkg565
Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G (2008) CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res 36:W496–W502. doi:10.1093/Nar/Gkn305
Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC (1999) Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol 285:1711–1733. doi:10.1006/Jmbi.1998.2400
Xu XP, Case DA (2001) Automated prediction of (15)N, (13)C(alpha), (13)C(beta) and (13)C ‘ chemical shifts in proteins using a density functional database. J Biomol NMR 21:321–333. doi:10.1023/A:1013324104681
Zhang HY, Neal S, Wishart DS (2003) RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25:173–195. doi:10.1023/A:1022836027055
Acknowledgments
We thank Profs Laue, Vriend and Kleywegt and members of the wwPDB NMR VTF for continuing discussions. We are especially grateful to all the structural biologists, who deposit their structures and experimental data in the public archives. This work was supported by BBSRC grants BB/J007471/1, BB/J007897/1 and BB/K021249/1 and European Community FP7 e-Infrastructure “WeNMR” project (Grant 261572). The NMR-related work at PDBe is further supported by EMBL-EBI and the Wellcome Trust (grant 088944).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vuister, G.W., Fogh, R.H., Hendrickx, P.M.S. et al. An overview of tools for the validation of protein NMR structures. J Biomol NMR 58, 259–285 (2014). https://doi.org/10.1007/s10858-013-9750-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9750-x