Abstract
The anisotropic component of the magnetic susceptibility tensor (Δχ tensor) associated with various paramagnetic metal ions can induce pseudocontact shifts (PCSs) and residual dipolar couplings (RDCs) in proteins, yielding valuable restraints in structural studies. In particular, PCSs have successfully been used to study ligands that bind to proteins tagged with a paramagnetic metal ion, which is of great interest in fragment-based drug design. To create easy-to-interpret PCSs, the metal ion must be attached to the protein in a rigid manner. Most of the existing methods for site-specific attachment of a metal tag, however, result in tethers with residual flexibility. Here we present model calculations to quantify the extent, to which mobility of the metal-binding tag can compromise the quality of the Δχ tensor that can be determined from the PCSs observed in the protein. Assuming that the protein can be approximated by a sphere and the tag is attached by a single tether, the results show that a single effective ∆χ tensor can describe the PCSs and RDCs of the protein spins very well even in the presence of substantial tag mobility, implying that PCSs of ligands in binding pockets of the protein can be predicted with similar accuracy. In contrast, the quality of the PCS prediction for nuclear spins positioned above the surface of the protein is significantly poorer, with implications for studies of protein–protein complexes. The simulations probed the sensitivity of the effective Δχ tensor to different parameters, including length of the tether between protein and metal ion, protein size, type and amplitude of tag motion, tensor orientation relative to the protein and direction of tag motion. Tether length and amplitude of motion were identified as two key parameters. It is shown that the amplitude of tag motions cannot be quantified by simple comparisons of the effective Δχ tensor with the alignment tensor determined from RDCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paramagnetic NMR spectroscopy (Bertini et al. 2002) offers unique opportunities in the fields of protein structure determination (Bertini et al. 2001; Arnesano et al. 2005; Schmitz et al. 2012), protein–protein (Pintacuda et al. 2006, Keizers et al. 2010; Saio et al. 2010) and protein–ligand interactions (John et al. 2006; Saio et al. 2011). Among the paramagnetic effects, pseudocontact shifts (PCSs) stand out for the long range of the effect (40 Å and more; Allegrozzi et al. 2000) and the easy way of measurement, namely as the chemical shift difference ΔδPCS observed between NMR spectra of the sample measured with a paramagnetic and a diamagnetic metal ion. PCSs are generated by a paramagnetic metal ion with a non-isotropic magnetic susceptibility. For each nuclear spin, the PCS (measured in ppm) depends on its polar coordinates r, θ and φ with respect to the principal axes of the ∆χ tensor:
where Δχax and Δχrh denote, respectively, the axial and rhombic components of the magnetic susceptibility tensor χ (Bertini et al. 2002), and the Δχ tensor is defined as the χ tensor minus its isotropic component. Equation 1 shows that PCSs can be positive or negative, depending on the position of the nuclear spin with respect to the coordinate system defined by the χ tensor. The orientations of the principal axes of the χ tensor in turn depend on the coordination of the metal ion. To avoid averaging of the PCSs to zero, it is thus important that the metal complex maintains a unique orientation with respect to the protein.
Anisotropic χ tensors also cause weak alignment of the molecules in a magnetic field, resulting in residual dipolar couplings (RDCs). The alignment tensor is simply proportional to the Δχ tensor.
Most biomolecules are naturally devoid of unpaired electrons. Therefore, PCSs require labelling of the biomolecule with a paramagnetic species. Lanthanide ions are the best candidates for this purpose (Otting 2008, 2010) as they afford the largest Δχ tensors, leading to prominent PCSs. In addition, different lanthanide ions have very different χ and Δχ tensor magnitudes, allowing the observation of PCSs at different distances from the paramagnetic centre. Finally, diamagnetic Y3+ is chemically very similar to lanthanide ions, offering an excellent diamagnetic reference.
One of the most popular methods for labelling proteins with paramagnetic tags relies on the attachment of chemically synthesized lanthanide complexes (Rodriguez-Castañeda et al. 2006; Su and Otting 2010, 2011; Keizers and Ubbink 2011; Koehler and Meiler 2011). Most of these tags exploit the reactivity of solvent-exposed cysteine thiol groups for site-selective attachment to the protein, for example by formation of a disulfide bond (Gaponenko et al. 2002; Dvoretsky et al. 2002; Ikegami et al. 2004; Leonov et al. 2005; Haberz et al. 2006; Peters et al. 2011) or by formation of a thioether (Li et al. 2012; Yang et al. 2013), but the resulting tether between protein and tag is almost always, at least to some degree, flexible. This leads to averaging of the PCSs in the protein, usually resulting in smaller-than-expected PCSs. More critically, however, the distance dependence of the PCS effect (Eq. 1) means that the PCSs generated in the protein by a mobile paramagnetic lanthanide can, strictly speaking, no longer be interpreted as arising from a single Δχ tensor. If the tag moves, fitting of the PCSs by a single tensor generates an approximation, which we refer to as the “effective Δχ tensor”.
Different strategies have been developed to circumvent the problems arising from tag mobility, including tag immobilization by attachment via two arms (Prudêncio et al. 2004; Vlasie et al. 2007; Keizers et al. 2007, 2008; Saio et al. 2010; Swarbrick et al. 2011b; Liu et al. 2012), using the shortest tether possible (Jia et al. 2011; Swarbrick et al. 2011a), using bulky tags for which motions are hindered by steric interactions with the protein (Su et al. 2006, 2008; Martin et al. 2007; Häussinger et al. 2009; Graham et al. 2011; Loh et al. 2013) or by integrating a lanthanide-binding peptide into the protein (Barthelmes et al. 2011). In general, however, tags producing single-arm attachments remain attractive. First, suitable attachment sites for single-arm tags can more readily be identified in proteins of unknown structure. Second, a much greater variety of single-arm tags exist. Finally, no double-arm tag exists that can attach a lanthanide to unnatural amino acids. As efficient protocols for the incorporation of unnatural amino acids into proteins have been developed (Liu and Schultz 2010; Young et al. 2010; Loscha et al. 2012; Ozawa et al. 2012), this opens exciting opportunities for the site-specific attachment of lanthanide tags that are independent of sulfur chemistry. We recently demonstrated the first example of a lanthanide tag that can be attached to an unnatural amino acid in the protein by a selective chemical reaction (Loh et al. 2013). While the tag generated significant PCSs in a range of different proteins, it also produced smaller-than-expected effective Δχ tensors, indicating flexibility of the tether between lanthanide complex and protein.
The potential problems associated with tag mobility prompted us to investigate the degree of mobility that can still generate useful PCSs. In the context of the present work, PCSs are considered useful if they can be fitted by a single effective Δχ tensor and the fit is good enough to use the fitted Δχ tensor for reliable predictions of the PCSs of other nuclear spins in the protein (e.g. for purposes of resonance assignments; Pintacuda et al. 2007; John et al. 2007; de la Cruz et al. 2011; Skinner et al. 2013) or ligand molecules (e.g. for purposes of structure determinations of protein–ligand complexes; John et al. 2006; Saio et al. 2011; Guan et al. 2013).
Finally, we investigated how much information about tag mobility can be gathered from a comparison between a fitted effective Δχ tensor and the alignment tensor derived from RDCs that arise from the paramagnetic alignment of the molecule in the magnetic field.
Materials and methods
Models of proteins and protein–ligand complexes
All simulations were performed using Mathematica (Wolfram Research Inc. 2010). The scripts developed can be downloaded from http://rsc.anu.edu.au/~go/mathematica. Protein nuclei experiencing paramagnetic effects were represented by a Cartesian grid of points (“protein grid”) confined within a sphere (“protein sphere”). We used spheres of 9, 12 and 15 Å radius to represent proteins of three different sizes. The distance between the individual grid points was set to 3 Å.
To model the nuclei of a ligand binding at the surface of the protein, another set of points was selected from the same Cartesian grid, namely the points that were in the layer between two spheres with radii of 2.5 and 3.5 Å larger than that of the protein sphere. The “ligand space” defined in this way was on average 3 Å away from the protein surface and surrounded the entire protein sphere (Fig. 1a).
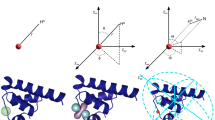
Model of a protein–ligand system with a paramagnetic metal ion bound to the protein. The protein is represented by a magenta sphere. The radius of the sphere was chosen to be 9 Å for a small protein and 15 Å for a large protein. The metal is attached to the surface of the protein sphere via a rigid tether of a constant length (typically 11 Å). The tag is represented by PCS isosurfaces, where blue and red surfaces identify PCS values of +1 and −1 ppm, respectively. In all calculations, the metal was associated with an axially symmetric Δχ tensor with ∆χax = 40 × 10−32 m3. a Grid points inside the protein sphere (3 Å between grid points, points not shown) define the location of the protein spins. Grid points surrounding the protein sphere at an average distance of 3 Å from its surface define the location of the ligand spins (shown as magenta dots). b Three different orientations I–III of the Δχ tensor with respect to the protein. The points inside the protein sphere depict the locations of the protein spins. c Definition of the cone opening angle Ω, which delineates the maximal amplitude of tag movement. The figure uses an equivalent representation, in which the tag is kept in place and the protein moves. Magenta spheres mark the two extreme protein positions. The tether is depicted as a black line. The hinge is positioned at a distance of 2 Å from the surface of the protein sphere to represent the situation of a tag attached to a cysteine side chain. d Cutout sphere, shown in grey, used to select subsets of the spins of the protein (red) and the ligand (blue) grid points for calculating PCS data. In the local fit approach, only the points shown in intense red colour fall within the cutout sphere and contribute to the fit of the effective ∆χ tensor. Similarly, only the points shown in intense blue colour are considered to belong to the ligand spins. PCSs of the protein and ligand spaces are back-calculated only for the selected points, using the effective ∆χ tensor fitted to the protein spins
Model of the mobile metal tag
A paramagnetic centre was positioned at a distance of 11 Å from the surface of the protein sphere to account for the space occupied by a metal-binding tag with tether. To limit the number of variables, an axially symmetric ∆χ tensor, centered on the metal ion, was assumed. ∆χax = 40 × 10−32 m3, being representative of a strongly paramagnetic lanthanide ion, was used in all simulations. Three different mutual orientations of the tensor and protein, denoted as I, II and III (Fig. 1b), were investigated.
The mobility of the tag was modelled by assuming a rigid tether with either one or two hinges. The “one-hinge model” (Fig. 2a) contains a single hinge positioned 2 Å away from the surface of the protein and 9 Å away from the metal. These parameters were chosen to mimic tags tied to the Cβ atom of a cysteine residue. The “two-hinge model” contains an additional hinge at the metal site. In this model, the Δχ tensor is always oriented the same way, like the windscreen wiper on a bus (Fig. 2b). The two-hinge model was motivated by the empirical observation that fitting of a single effective Δχ tensor to PCSs generated by very mobile tags usually leads to metal positions that are further away from the protein surface than expected, whereas the one-hinge model tended to move the paramagnetic centre closer to the protein than expected (see below). The better representation of the experimental situation by the two-hinge model may arise from steric interactions between the metal chelate with the protein and its amino-acid side chains as illustrated in Fig. 2b.
Different structural and motional models of the tag mobility. a “One-hinge model”. A single hinge (shown as a small white circle) is at the protein surface. As a result, the Δχ tensor is always aligned with the tether between protein and metal chelate (represented by a ball with stripes) and therefore changes its orientation with respect to the protein. b Rationalisation for the “two-hinge model”. The metal chelate is a rigid moiety, whereas the tether is flexible. Therefore, as the metal chelate bumps into the protein surface and neighboring amino acid side chains, it tends to reorientate, aligning more closely with its average orientation than in the one-hinge model. The two-hinge model takes this effect into account by assuming a second hinge located at the point of the (dimensionless) metal ion, where the movements around the second hinge always exactly compensate the movement around the first hinge so that the tensor does not change in orientation when the tag moves. c “Line” movement of the tag. The figure shows a representation, in which the tag (depicted by its PCS isosurfaces) stays and the protein moves. (This is equivalent to the tag moving relative to the protein.) The trajectory of the centre of the protein sphere is illustrated by black points and the two end points of the trajectory are marked by protein spheres. d Same as c, except for a “star” movement. The extreme positions of the protein on two of the trajectories are highlighted by magenta spheres
On a technical note, although the final results are presented as if the tag moves relative to the protein, the simulations assumed the protein to move with respect to the tag. This allowed keeping the position and orientation of the ∆χ tensor constant while the protein sphere, along with the grid and ligand space, underwent rotational movements about the hinge. Random reorientational motions at the origin of the Δχ tensor were disregarded, because such motions would conserve the distances between the metal and the protein nuclei and, hence, reduce the size of the average Δχ tensor without affecting the fit of the resulting PCSs by Eq. 1. Only motions that change the distance between the metal and the protein affect the quality of the Δχ-tensor fit.
The hinge in the one-hinge model was permitted to undergo rotational movements in different directions. Viewed from the coordinate system of the tag, the centre of the protein sphere was assumed to move between equidistant points arranged along either a line of points (Fig. 2c) or a star-like set of trajectories (Fig. 2d). The line model assumes a one-dimensional movement, where the protein centre is confined within a single plane. There are several possible orientations of this plane with respect to the tensor orientation II and III (further explored in Fig. S1). The star model approximates a movement-in-a-cone model, where the protein centre stays within a cone with opening angle Ω (Fig. 1c). Ω is determined by the end points of any one of the trajectory lines shown in Fig. 2c and d. Data were simulated for opening angles between 60o and 120o. The protein centre was moved along the trajectories at closely spaced equidistant points, with equal population of each point, resulting in an overall higher population near the centre of the cone.
PCS calculation and Δχ-tensor fits
The movement of the tag with respect to the protein was assumed to be fast on the NMR time scale, so that the PCS values computed for each position and orientation of the tag would average to a single PCS value for each nuclear spin of the protein. We refer to the average PCS as “experimental PCS” or “observable PCS”. The PCSs of the ligand space were computed in the same way. Fits of ∆χ tensors to the simulated PCSs were performed by a Mathematica script using the same algorithm as the program Numbat (Schmitz et al. 2008). Only PCSs in the range between −3 and +3 ppm were used for the tensor fit, to eliminate the potentially strong influence of large PCSs on the overall ∆χ tensor. The reason for omitting larger PCSs is two-fold. First, large PCSs are experimentally more difficult to assign. Second, large PCSs arise from spins in the vicinity of the paramagnetic metal ion, where signals are easily broadened beyond detection due to paramagnetic relaxation enhancements (PRE). The resulting ∆χ tensor is referred to as the “effective ∆χ tensor”, and a PCS value back-calculated using the effective Δχ tensor is referred to as “back-calculated PCS”.
The quality factor is conventionally calculated as the root-mean-square deviation between the observed and back-calculated PCS, RMSDPCS, divided by the root-mean-square of the observed PCSs, RMSPCSobs: Q = RMSDPCS/RMSPCSobs. In the present work, we used RMSDPCS values as the quality criterion of the ∆χ-tensor fits, because these RMSD values have the same unit as PCSs (ppm) and thus can readily be compared with the magnitude of experimental errors of PCS measurements. In the following, we drop the subscript of RMSDPCS.
Global and local tensor fit
We tested two different approaches for ∆χ-tensor fitting and PCS back-calculation. In both cases, the same protein spins were used to fit the tensor and back-calculate PCSs. The “global tensor” fit included the PCSs from all protein nuclei and the tensor was used to back-calculate the PCSs of all protein and ligand spins. In contrast, the “local tensor” fit approach considered PCSs only from the region of the protein and ligand space which fell within a “cutout sphere”. The cutout sphere was chosen to have a radius equal to that of the protein and centered at the surface of the protein as shown in Fig. 1d. Different orientations of the cutout sphere with respect to the tensor are possible (Fig. S2). To limit the number of variables, we always placed the cutout sphere at the side of the protein, i.e. on a line perpendicular to the tether of the tag.
RDC calculation
The following equation was used to describe the RDC 1 D HN (in Hz) between 1H and 15N spins (Bertini et al. 2002)
where γH and γN denote the magnetogyric ratios of the nuclear spins H and N, respectively, r HN is the internuclear distance, h is Planck’s constant, k B the Boltzmann constant, T the temperature, and the angles Θ and Φ are the polar angles describing the orientation of the bond vector with respect to the principal axes of the Δχ tensor.
“Experimental RDC” values were calculated in the same way as the experimental PCSs, namely as the average of the RDC calculated for each orientation of the tag with respect to the protein. Only one-bond RDCs between 1H and 15N spins, 1 D(1HN,15N), were considered. A uniform distribution of bond orientations was generated. The resulting array of vectors pointing in different directions was rotated along with the protein grid for each motion of the tag with respect to the protein. The simulation was performed for a single point of the protein grid, as RDCs do not depend on the position of the nucleus with respect to the paramagnetic centre.
Rotamer library
Libraries of possible tag conformations were calculated using the program PyParaTools (Stanton-Cook et al. 2010). The program randomly alters the dihedral angles of every rotatable bond in the tether between protein and metal-chelate of the tag, accepting only conformations free of steric clashes with the protein.
Results
Parameters from rotamer libraries
To establish model parameters in agreement with experimentally available tags, we used data from the dengue virus NS2B-NS3 protease with the cyclen tag C1 (Graham et al. 2011), for which single-cysteine mutants at position 57 in NS2B and at positions 34 and 68 in NS3 (denoted as mutant A, B and C, respectively) have been made and PCS data recorded with the C1 tag (de la Cruz et al. 2011). As the 3D structure of this protein is available, the conformational space accessible to the C1 tag could readily be explored by generating a library of tag conformations by randomly altering the dihedral angles of every rotatable bond in the tether between protein and metal-binding site.
The rotamer libraries of mutants A–C revealed average distances of the metal from the cysteine Cβ atom of 9.3, 8.3 and 8.5 Å, respectively. We therefore set the length of the tether in the models (Fig. 2a, b) to 9 Å. Furthermore, we measured the angle ω between the vector connecting the average metal position with the tag attachment site (assumed to be the cysteine Cβ atom) and the vector connecting the actual metal position with the tag attachment site (Fig. 3a). The angle capturing more than 90 % of the ω values (ω90 %; Fig. 3b) was used as the opening angle of the cone in our model (note that the total opening angle is Ω = 2.ω90 %). ω90 % values were calculated to be 35o, 66o and 55o for the C1 tag attached at sites A, B and C, respectively. Based on these results, we investigated the effect of tag motions in our model for opening angles Ω ranging from 60o to 120º.
Selection of model parameters based on tag mobility in a real protein. a Possible tag positions in the dengue virus NS2B-NS3 protease labelled with the C1 tag at residue 57 in NS2B (mutant A), residue 34 in NS3 (mutant B) or residue 68 in NS3 (mutant C). The rotatable bonds in the tether of the C1 tag were allowed to assume staggered or nearly staggered conformations in random combinations that are compatible with the sterically allowed confines of the tagged protein. The figure shows the positions of the metal in the resulting tag rotamer library (cloud of blue dots). The average metal position is marked by a black point. The red point marks the effective tag attachment site and the vertex of the angle ω, which defines the angular deviation of actual metal positions (blue) from the average position (black). b Deviation of the metal position from its average. All ω angle values in the tag rotamer library of the mutant A are depicted as blue points. ω90 % (value indicated by the red line) denotes the value that captures 90 % of the ω angle values determined from the rotamer library
Global Δχ-tensor fit
Using the models described above, we monitored the quality of the tensor fits, systematically changing different parameters of the simulations. The back-calculated PCS data, PCScalc, using the effective Δχ tensor for both protein and ligand spins, were plotted against the experimental PCS data, PCSexp. The effective Δχ tensor was always determined by fits that used the PCSs from the protein sphere only. The quality of the fits was assessed by correlation plots and RMSD values between the back-calculated and experimental PCSs.
Initially, three different orientations of the Δχ tensor relative to the protein sphere were investigated (Fig. 1b) for three different protein sizes (simulated by changing the radius of the protein sphere) and four different movement models: “line one-hinge”, “line two-hinge”, “star one-hinge”, “star two-hinge” (Fig. 2).
The results show that excellent correlations can be obtained for the protein for all opening angles with small RMSD values between the experimental and back-calculated PCS values, of a magnitude comparable to typical uncertainties in PCS measurements (less than 0.05 ppm; Figs. 4–6). As expected, the quality of the correlations decreased for larger opening angles (Fig. 4). In all simulations, the quality of the correlations was significantly worse for the ligand space than for the protein sphere, although the correlations were still clearly recognizable. The fits were particularly good for a smaller protein radius (Fig. 5a). The mutual orientation of protein and tag had a lesser influence on the quality of the tensor fits (Fig. 5b). In summary, mobility of the tag hardly compromises the predictive value of the effective Δχ tensors for NMR resonances of the protein sphere, whereas the PCScalc values predicted for the ligand space are more strongly affected by the tag mobility.
Example of simulations performed to assess the impact of tag mobility on the effective Δχ tensor fitted to the PCSs observed in the protein. In the model used, the protein sphere has a radius of 9 Å, the Δχ tensor assumes orientation I, all protein spins are used to fit the effective Δχ tensor (“global fit”) and the tag moves according to the “star two-hinge” model. Three different cone opening angles Ω are considered. The Δχax and Δχrh values of the effective Δχ tensors are listed, including the coordinates of their centres. A negative z-coordinate indicates that the centre of the effective Δχ tensor is further away from the protein surface than in the absence of tag mobility. The quality of the fits by a single effective Δχ tensor is illustrated by the correlation between observable PCSs (on the horizontal axis, in ppm, calculated by averaging between the PCSs predicted for the different tag conformations) and back-calculated PCSs (on the vertical axis, in ppm, calculated using the single effective Δχ tensor). The PCSs of the ligand spins were not used to fit the effective Δχ tensors. PCSs greater than ±3 ppm were excluded from the fits. The RMSD values (in ppm) report the deviation between observable and back-calculated PCSs
Impact of protein size and tensor orientation on the quality of the effective Δχ tensor fits. The quality of the fits is expressed by the RMSD values (in ppm) between observable and back-calculated PCSs for the protein PCSs (magenta) and ligand PCSs (blue). Only the protein PCSs were used for fitting Δχ tensors. The results are shown for the “star two-hinge” movement model with an opening angle Ω = 120°, using the global fit approach. a Three different protein sizes are considered (9, 12 and 15 Å in radius) using tensor orientation I. b Three different tensor orientations (I, II and III, see Fig. 1b) are considered, using a protein radius of 9 Å
Quite generally, the effective Δχ tensors were a poor reflection of the actual Δχ tensor of the tag. For example, the effective Δχ tensors invariably displayed a significant Δχrh component for the “line” movement models, although the actual Δχ tensor was axially symmetric (Fig. 6). Figure 6 also highlights that the overall magnitudes of the effective Δχ tensors were remarkably variable, including switches of sign. The two-hinge model produced larger-than-expected Δχ tensors centered at a position further away from the protein than the average metal position, which is in agreement with common experience for tags with flexible tethers. The one-hinge model, however, produced effective Δχ tensors that tended to be centered at a position closer to the protein surface. In this model, the magnitude of the effective Δχ tensor was more comparable to that of the actual Δχ tensor. One must keep in mind, however, that the one-hinge model underestimates the degree of reorientational motions of the metal chelate that would reduce the Δχ tensor by averaging even without translocation of the metal ion.
Local Δχ-tensor fit
The lesser quality of the correlations between experimental and back-calculated PCSs for the ligand space (Figs. 4–6) arises from the difficulty to predict PCSs for nuclear spins that are located far from those that have been used to fit the effective Δχ tensor. On the other hand, a real ligand is not able to cover the entire protein surface as specified by our model (Fig. 1a). Instead, it would be confined to a single site on the protein surface. An improved correlation may thus be obtained by using an effective Δχ tensor that is determined by fitting the PCSs of only those nuclear spins of the protein that are in close proximity of the ligand. The improvement obtained by such a “local effective Δχ tensor” would be at the expense of accurate PCS predictions for those nuclear spins in the protein sphere that had not been used in the Δχ-tensor fit. We selected the protein spins to be included in the fit of the local effective Δχ tensor by defining a “cutout sphere”, as described in Fig. 1d.
The local effective Δχ tensor obtained in this way proved to produce much more reliable PCS predictions for the ligand which was assumed to be confined to the ligand space that is also within the cutout sphere. Figure 7a and b illustrates the superior quality of the PCS prediction for the reduced ligand space defined in this manner in the worst-case-scenario when the cone opening angle is 120º and the protein radius is 15 Å. Testing this approach for three different protein sizes (with radii of 9, 12 and 15 Å) and the three different tensor orientations I–III with respect to the protein, we observed similar improvements in the quality of the PCS correlations. Figure S3 contains a compilation of simulations performed with systematically varied parameters. It shows that, for the tensor magnitude chosen in our calculations, excellent correlations between back-calculated and experimental PCSs are characterized by RMSD values < 0.05 ppm, whereas an RMSD of 0.1 ppm can be associated with significant outliers.
Comparison of local fits versus global fits and effect of tether length and opening angle. The histograms report the RMSD values (in ppm) between observable and back-calculated PCSs for the protein and ligand, using the “star two-hinge” movement model for the three different tensor orientations I–III. a Using a protein radius of 15 Å, a 9 Å tether, opening angle Ω = 120°, and the global tensor fit approach. b Same as a, but using the local tensor fit approach. c Effect of a shorter tether. The tag was tied to the protein sphere (9 Å radius) via a 5 Å tether. The tag moved with an opening angle Ω = 120°. The calculations used the local fit approach with a cutout sphere (Fig. 1d). Different from all other simulations in this work, nuclear spins coming within 12 Å of the metal during the simulation were assumed to be unobservable due to PRE. d Same as c, except Ω = 90°
Tags with short tethers
To limit the number of variables, the simulations described above used a single tether length of 9 Å, based on the rotamer libraries of the dengue virus NS2B-NS3 protease established with the C1 tag. Notably, however, the rotamer libraries showed that the distance between the metal and cysteine Cβ-atom could vary between 3.4 and 10.5 Å. We therefore also investigated the effect of a shorter tether length (5 Å). This turned out to produce markedly larger deviations between back-calculated and observable PCSs, which can be explained by the larger PCS gradients obtained for the same opening angles. In contrast to all other simulations performed in this work, this simulation also took PREs into account by removing the PCSs from all nuclear spins that were getting closer than 10 Å of the metal ion and only the local fit approach with a cutout sphere was explored. Systematic variation of the protein radius, movement model, opening angle and tensor orientation revealed the largest RMSD for a tag with a 5 Å tether, if it was tied to a protein sphere of 9 Å radius, moved according to the “star two-hinge” model and had the tensor in orientation II. As illustrated in Fig. 7c, this worst-case scenario led to RMSD values of above 0.5 ppm for the ligand spins, when the opening angle was 120°. The situation was better for other tensor orientations and improved particularly for a smaller opening angle Ω (Fig. 7d). As before, the fit to the protein spins was markedly better than for the ligand spins.
Comparison with rotamer library
The large effect arising from the tether length prompted us to investigate the full rotamer library of C1 tags attached to mutants A-C in the dengue virus NS2B-NS3 protease (de la Cruz et al. 2011). For best comparison with the model calculations above, we grafted the rotamer libraries of mutants A-C onto protein spheres of 9, 12 and 15 Å radius. Recognizing that each rotamer in the library has a specific location and orientation of the bound metal with respect to the protein nuclei, the average PCS expected for each nuclear spin of the protein can readily be calculated to simulate experimental PCSs. We assumed an axially symmetric Δχ tensor with ∆χax = 40 × 10−32 m3 and that the tensor axis is orthogonal to the plane of the cyclen ring of the C1 tag.
For each of the mutants A-C, experimental and back-calculated PCSs were calculated for the protein and ligand spins using both global and local tensor fit approaches. Figure 8a and b shows that the agreement between back-calculated and experimental PCSs is excellent for the protein space, but not as good for the ligand spins, especially in the case of the mutants B and C, for which the effective opening angle is greater (Fig. 3a). Notably, the data shown in Fig. 8b for mutant B were the worst-case scenario, as lower RMSD values were obtained for larger protein radii. Figure 8c illustrates the correlations between the back-calculated and experimental PCSs for the protein sphere obtained in the local fits leading to Fig. 8b. Clearly, the correlations are excellent for mutants A and B, and very good for mutant C. The corresponding correlations for the ligand spins (Fig. 8d), however, are good only for mutant A and reasonable for mutant C. The correlation is spoilt for mutant B by a single outlier. This outlier corresponds to a ligand spin that is relatively close to the metal (within 17 Å of the average metal position, whereas the average ligand spin is about 26 Å from the average metal position). This distance is sufficiently long that the ligand spin would not be affected by excessive PREs (in contrast to some of the ligand spins in the global fit scenario of Fig. 8a). These data are another illustration of the result that reliable PCS predictions can be expected only for RMSD values < 0.05 ppm.
Correlations between back-calculated and experimental PCSs calculated for rotamer libraries of the C1 tag grafted onto a protein sphere of 12 Å radius as in Fig. 1a. The rotamer libraries were derived from mutants A-C of the dengue virus NS2B-NS3 protease (de la Cruz et al. 2011). a Histograms reporting the RMSDs between back-calculated and experimental PCSs, if all protein atoms are included in the Δχ tensor fit (“global fit”) and the ligand layer includes all atoms (located on average 3 Å from the surface of the protein). b Same as a, but using the local Δχ tensor fit, where the protein and ligand spins are selected by a cutout sphere as defined in Fig. 1d. c Correlations between the back-calculated and experimental PCSs for the protein spins, where the experimental PCSs were calculated using the rotamer libraries of the C1 tag for the mutants A–C of the dengue virus NS2B-NS3 protease. The RMSD values of these correlations are reported by the magenta bars in b. For improved plot resolution, only PSCs in the range between −0.7 and 0.7 ppm are displayed, although PCSs as large as −2.5 ppm were calculated for the mutants A and B. The correlations for the large PCSs were of similar quality as those in the range shown. d Same as c, but for the ligand spins. The RMSD values of these correlations are reported by the blue bars in b
Alignment tensor fit
Any molecule producing PCSs also aligns weakly in a magnetic field and therefore results in RDCs. In the absence of tag mobility, the magnetically induced alignment tensor is simply proportional to the Δχ tensor. In the presence of tag motions, however, the Δχ tensor and the alignment tensor average in different ways, as RDCs are independent of the distance from the metal ion (Eq. 2). Therefore, RDC data generated by a flexible tag can always be described perfectly by a single average alignment tensor irrespective of tag mobility and metal position, whereas the quality of an effective Δχ tensor is affected by any tag motions that relocate the metal ion relative to the protein.
Is it possible to take the deviation between the effective Δχ tensor and the associated magnetically induced alignment tensor as a measure of tag mobility? To test this question, we simulated experimental 1 D(1HN,15N) RDCs, using our model with different opening angles Ω of the tag motions (Fig. 1c) and fitted the average alignment tensors. Next, each alignment tensor A was converted into a Δχ tensor using
where B 0 is the magnetic field strength, μ0 the induction constant, k the Boltzmann constant, and T the temperature (Bertini et al. 2002). Figure 9 compares the Δχax values obtained from RDCs and PCSs for different opening angles of the cone that represents the amplitude of tag mobility. As expected, the tensors are identical in the absence of tag mobility (i.e. zero opening angle) and the RDCs decrease with the averaging that comes with increasing opening angle. In contrast, the effective Δχ tensor determined from PCSs becomes larger for greater opening angles. Importantly, the magnitude of both effects is sensitive to the orientation of the original Δχ tensor with respect to the protein. Furthermore, using the “star one-hinge” model produced data, where the Δχax values determined from PCSs and RDCs were the same within 20 % even for an opening angle of Ω = 120o (data not shown). Therefore, the discrepancy between the two tensors is not a reliable measure of the amplitude of the tag motions.
Dependence of the ∆χax parameter of the effective ∆χ tensor calculated from the fits of observable PCS (magenta) and RDC (blue) values on the cone opening angle Ω for the three different orientations I–III of the ∆χ tensor (Fig. 1b). The model represented the protein as a sphere with a radius of 9 Å, using the global fit approach and the “star two-hinge” model
Discussion
The simulations presented in this work validate the use of one-arm lanthanide tags for structural studies by PCS, but the results also highlight the importance of considering the parameters pertaining to the specific system under investigation. Firstly, it is important to remember that, owing to the distance dependence of the PCS effect (Eq. 1), a single effective Δχ tensor is only an approximation to the real situation, which, if the tag motions translocate the metal ion with respect to the protein, in principle must be described by a multitude of Δχ tensors and metal positions. Therefore, completely accurate fits of the experimental PCSs cannot be achieved in the presence of tag mobility. Since effective Δχ tensors are obtained by minimizing the RMSD between back-calculated and experimental PCSs, large PCSs can contribute disproportionately to the RMSD value. Large PCSs arise only in the vicinity of the metal ion and therefore stem more likely from tag conformations at extreme positions of the tag motions. This explains why the omission of the largest PCSs from the Δχ tensor fit yields effective Δχ tensors that predict PCSs of additional spins with better accuracy. Similarly, the accuracy of the predictions increases, if the opening angle of the tag movements is relatively small and the tether between protein and metal chelate is relatively long.
Secondly, the effective Δχ tensor, obtained by fitting the PCSs observed for protein spins, can display quite different Δχax and Δχrh parameters compared with the real Δχ tensor associated with the metal chelate, if the lanthanide tag moves with respect to the protein. Nonetheless, the effective Δχ tensor can predict the PCSs of nuclear spins that were not used in the tensor fit with high accuracy, even if the tether linking the paramagnetic metal with the protein is mobile within a cone with an opening angle of 120o, which is representative of the situation encountered with real proteins and tags. It is useful to distinguish two situations. (1) If a small ligand molecule binds to a pocket in the surface of the target protein, its coordinates can be considered to be located within the grid of spins referred to as the protein sphere in the present simulations. In this situation, the fit between back-calculated and experimental PCSs is expected to be excellent for both protein and ligand molecules, especially if the fit of the effective Δχ tensor involves only protein spins near the ligand-binding site. This also explains the remarkably good correlations obtained between back-calculated and experimentally measured PCSs for three different proteins labelled with the C1 tag (Graham et al. 2011; de la Cruz et al. 2011). (2) If the PCS predictions are made for nuclear spins of ligands that are located further away from the target protein, such as encountered in studies of protein–protein interactions, the quality of the predictions for the ligand rapidly deteriorates with increasing distance from the spins used for fitting the effective Δχ tensor. Some fortuitous combinations of orientation and trajectory of the tag relative to the protein may still result in high-quality predictions of the PCSs for the ligand space but, in general, outliers must be expected. For single-arm tags with short tethers, it is thus important to limit the amplitude of metal movement, for example by additional coordination to a carboxyl group of the protein (Swarbrick et al. 2011a; Yagi et al. 2013). If the aim is to obtain a good model of a protein–protein complex from PCSs generated by a mobile lanthanide tag, it is likely necessary to enhance the available experimental information by preparing several samples, in which the tag is positioned at different sites.
Thirdly, our simulations show that local effective Δχ tensors provide significantly more accurate predictions of PCSs of ligand molecules than Δχ-tensor fits that include all available protein spins. Since the binding site of the ligand is often known from chemical shift perturbations or can be predicted from biological information, it is straightforward to select the protein part that should be used for the Δχ tensor fit. Many different protocols for selecting the PCSs for inclusion in the fit can be conceived. The simple approach proposed here (selecting PCSs only of protein spins located within the protein radius from the binding site) attempts to include an adequate number of proteins spins for the fit. Note that, in order to prevent uncertainties of the PCS measurements affecting the fitted Δχ tensors, it is advisable to use a much larger number of PCSs in the fit than the absolute minimum (8, as Δχax, Δχrh, the three metal coordinates and three Euler angles of the tensor need to be fitted). Quite generally, the local effective Δχ-tensor approach will work best, if the paramagnetic tag is not too close to the ligand to avoid generating overly large PCSs, yet not too far so that the PCSs can be measured with acceptable relative experimental errors.
Finally, the discrepancies between average alignment tensors determined from RDCs and effective Δχ tensors determined from PCSs contain no easily recoverable information about the size of the conformational space accessible to the metal tag. Depending on the parameters used to simulate the tag motions, we found that the two tensors can be very similar for large opening angles Ω or quite different for relatively small Ω values. When the tensors are different, this makes it more difficult to correct the PCSs of nuclear spins with large CSAs for the residual CSA effects associated with the alignment tensor (John et al. 2005), as the correction should be based on the actual alignment tensor rather than making the assumption that the alignment tensor is simply proportional to the effective Δχ tensor. Conversely, the tensors determined from RDCs and PCSs are expected to differ even in the complete absence of tag motions, because the order parameter of the bonds is less than 1 or because the structure of the protein in solution is known with insufficient accuracy. The structural uncertainties could be eliminated by identifying the largest RDC value, which most likely arises from a bond closely aligned with the longest axis of the alignment tensor and therefore is proportional to Δχax. The remaining uncertainties concern the value of the order parameter and whether an RDC can be measured for a bond that is closely aligned with the long principal axis of the alignment tensor.
In conclusion, single-arm attachments of lanthanide ions to proteins present a valid strategy for obtaining valuable long-range structural information in proteins and protein–ligand complexes from PCS data, provided that caveats arising from tag mobility are kept in mind. Even in the case of large tag motions that produce difficult-to-interpret PCSs, however, lanthanide tags can usefully be deployed to generate alignment tensor orientations that cannot be achieved by conventional alignment media. Any tag that generates large PCSs necessarily also produces significant RDCs, and RDCs can be perfectly interpreted by a single average alignment tensor.
References
Allegrozzi M, Bertini I, Janik MBL, Lee YM, Liu G, Luchinat C (2000) Lanthanide-induced pseudocontact shifts for solution structure refinements of macromolecules in shells up to 40 Å from the metal ion. J Am Chem Soc 122:4154–4161
Arnesano F, Banci L, Piccioli M (2005) NMR structures of paramagnetic metalloproteins. Q Rev Biophys 38:167–219
Barthelmes K, Reynolds AM, Peisach E, Jonker HR, DeNunzio NJ, Allen KN, Imperiali B, Schwalbe H (2011) Engineering encodable lanthanide-binding tags into loop regions of proteins. J Am Chem Soc 133:808–819
Bertini I, Donaire B, Jiménez B, Luchinat G, Parigi G, Piccioli M, Poggi L (2001) Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k. J Biomol NMR 21:85–98
Bertini I, Luchinat C, Parigi G (2002) Magnetic susceptibility in paramagnetic NMR. Prog NMR Spectrosc 40:249–273
de la Cruz L, Ngyuen THD, Ozawa K, Shin J, Graham B, Huber T, Otting G (2011) Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: fold analysis by pseudocontact shifts. J Am Chem Soc 133:19205–19215
Dvoretsky A, Gaponenko V, Rosevear PR (2002) Derivation of structural restraints using a thiol-reactive chelator. FEBS Lett 528:189–192
Gaponenko V, Altieri AS, Li J, Byrd A (2002) Breaking symmetry in the structure determination of (large) symmetric protein dimers. J Biomol NMR 24:143–148
Graham B, Loh CT, Swarbrick JD, Ung P, Shin J, Yagi H, Jia X, Chhabra S, Barlow N, Pintacuda G, Huber T, Otting G (2011) DOTA-amide lanthanide tag for reliable generation of pseudocontact shifts in protein NMR spectra. Bioconjugate Chem 22:2118–2125
Guan JY, Keizers PHJ, Liu WM, Löhr F, Skinner SP, Heeneman EA, Schwalbe H, Ubbink M, Siegal G (2013) Small-molecule binding sites on proteins established by paramagnetic NMR spectroscopy. J Am Chem Soc. doi:10.1021/ja401323m
Haberz P, Rodriguez-Castañeda F, Junker J, Becker S, Leonov A, Griesinger C (2006) Two new chiral EDTA-based metal chelates for weak alignment of proteins in solution. Org Lett 8:1275–1278
Häussinger D, Huang JR, Grzesiek S (2009) DOTA-M8: an extremely rigid, high-affinity lanthanide chelating tag for PCS NMR spectroscopy. J Am Chem Soc 131:14761–14767
Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C (2004) Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR 29:339–349
Jia X, Maleckis A, Huber T, Otting G (2011) 4,4′-dithiobisdipicolinic acid: a small and convenient lanthanide binding tag for protein NMR spectroscopy. Chem Eur J 17:6830–6836
John M, Park AY, Pintacuda G, Dixon NE, Otting G (2005) Weak alignment of paramagnetic proteins warrants correction for residual CSA effects in measurements of pseudocontact shifts. J Am Chem Soc 127:17190–17191
John M, Pintacuda G, Park AY, Dixon NE, Otting G (2006) Structure determination of protein-ligand complexes by transferred paramagnetic shifts. J Am Chem Soc 128:12910–12916
John M, Schmitz C, Park AY, Dixon NE, Huber T, Otting G (2007) Sequence- and stereospecific assignment of methyl groups using paramagnetic lanthanides. J Am Chem Soc 129:13749–13757
Keizers PHJ, Ubbink M (2011) Paramagnetic tagging for protein structure and dynamics analysis. Prog Nucl Magn Reson Spectrosc 58:88–96
Keizers PHJ, Desreux JF, Overhand M, Ubbink M (2007) Increased paramagnetic effect of a lanthanide protein probe by two-point attachment. J Am Chem Soc 129:9292–9293
Keizers PHJ, Saragliadis A, Hiruma Y, Overhand M, Ubbink M (2008) Design, synthesis, and evaluation of a lanthanide chelating protein probe: CLaNP-5 yields predictable paramagnetic effects independent of environment. J Am Chem Soc 130:14802–14812
Keizers PHJ, Mersinli B, Reinle W, Donauer J, Hiruma Y, Hannemann F, Overhand M, Bernhart R, Ubbink M (2010) A solution model of the complex formed by adrenodoxin and adrenodoxin reductase determined by paramagnetic NMR spectroscopy. Biochemistry 49:6846–6855
Koehler J, Meiler J (2011) Expanding the utility of NMR restraints with paramagnetic compounds: background and practical aspects. Prog NMR Spectrosc 59:360–389
Leonov A, Voigt B, Rodriguez-Castañeda F, Sakhaii P, Griesinger C (2005) Convenient synthesis of multifunctional EDTA-based chiral metal chelates substituted with an S-mesylcysteine. Chem Eur J 11:3342–3348
Li QF, Yang Y, Maleckis A, Otting G, Su XC (2012) Thiol-ene reaction: a versatile tool in site-specific labelling of proteins with chemically inert tags for paramagnetic NMR. Chem Commun 48:2704–2706
Liu CC, Schultz PG (2010) Adding new chemistries to the genetic code. Annu Rev Biochem 79:413–444
Liu WM, Keizers PHJ, Hass MAS, Blok A, Timmer M, Sarris AJC, Overhand M, Ubbink M (2012) A pH-sensitive, colorful, lanthanide-chelating paramagnetic NMR probe. J Am Chem Soc 134:17306–17313
Loh CT, Ozawa K, Tuck KL, Barlow N, Huber T, Otting G, Graham B (2013) Lanthanide tags for site-specific ligation to an unnatural amino acid and generation of pseudocontact shifts in proteins. Bioconj Chem 24:260–268
Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G (2012) Multi-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed 51:2243–2246
Martin LJ, Hähnke MJ, Nitz M, Wöhnert J, Silvaggi NR, Allen KN, Schwalbe H, Imperiali B (2007) Double-lanthanide-binding tags: design, photophysical properties, and NMR applications. J Am Chem Soc 129:7106–7113
Otting G (2008) Prospects for lanthanides in structural biology by NMR (2008). J Biomol NMR 42:1–9
Otting G (2010) Protein NMR using paramagnetic ions. Annu Rev Biophys 39:387–405
Ozawa K, Loscha KV, Kuppan KV, Loh CT, Dixon NE, Otting G (2012) High-yield cell-free protein synthesis for site-specific incorporation of unnatural amino acids at two sites. Biochem Biophys Res Commun 418:652–656
Peters F, Maestre-Martinez M, Leonov A, Kovačič L, Becker S, Boelens R, Griesinger C (2011) Cys-Ph-TAHA: a lanthanide binding tag for RDC and PCS enhanced protein NMR. J Biomol NMR 51:329–337
Pintacuda G, Park AY, Keniry MA, Dixon NE, Otting G (2006) Lanthanide labeling offers fast NMR approach to 3D structure determination of protein–protein complexes. J Am Chem Soc 128:3696–3702
Pintacuda G, John M, Su XC, Otting G (2007) NMR structure determination of protein-ligand complexes by lanthanide labeling. Acc Chem Res 40:206–212
Prudêncio M, Rohovec J, Peters JA, Tocheva E, Boulanger MJ, Murphy MEP, Hupkes HJ, Koster W, Impagliazzo A, Ubbink M (2004) A caged lanthanide complex as a paramagnetic shift agent for protein NMR. Chem Eur J 10:3252–3260
Rodriguez-Castañeda F, Haberz P, Leonov A, Griesinger C (2006) Paramagnetic tagging of diamagnetic proteins for solution NMR. Mag Res Chem 44:10–16
Saio T, Yokochi M, Kumeta H, Inagaki F (2010) PCS-based structure determination of protein–protein complexes. J Biomol NMR 46:271–280
Saio T, Ogura K, Shimizu K, Yokochi M, Burke TR, Inagaki F (2011) An NMR strategy for fragment-based ligand screening utilizing a paramagnetic lanthanide probe. J Biomol NMR 51:395–408
Schmitz C, Stanton-Cook MJ, Su XC, Otting G, Huber T (2008) Numbat: an interactive software tool for fitting Δχ-tensors to molecular coordinates using pseudocontact shifts. J Biomol NMR 41:179–189
Schmitz C, Vernon R, Otting G, Baker D, Huber T (2012) Protein structure determination from pseudocontact shifts using ROSETTA. J Mol Biol 416:668–677
Skinner SP, Moshev M, Hass MAS, Ubbink M (2013) PARAssign—paramagnetic NMR assignments of protein nuclei on the basis of pseudocontact shifts. J Biomol NMR 55:379–389
Stanton-Cook M, Su XC, Otting G, Huber T (2010) http://com-bio.anu.edu.au/mscook/PPT
Su XC, Otting G (2010) Paramagnetic labelling of proteins and oligonucleotides for NMR. J Biomol NMR 46:101–112
Su XC, Otting G (2011) Erratum to: paramagnetic labelling of proteins and oligonucleotides for NMR. J Biomol NMR 50:99–100
Su XC, Huber T, Dixon NE, Otting G (2006) Site-specific labelling of proteins with a lanthanide-binding tag. ChemBioChem 7:1469–1474
Su XC, McAndrew K, Huber T, Otting G (2008) Lanthanide-binding peptides for NMR measurements of residual dipolar couplings and paramagnetic effects from multiple angles. J Am Chem Soc 130:1681–1687
Swarbrick JD, Ung P, Chhabra S, Graham B (2011a) An iminodiacetic acid based lanthanide binding tag for paramagnetic exchange NMR spectroscopy. Angew Chem Int Ed 50:4403–4406
Swarbrick JD, Ung P, Su XC, Maleckis A, Chhabra S, Huber T, Otting G, Graham B (2011b) Engineering of a bis-chelator motif into a protein α-helix for rigid lanthanide binding and paramagnetic NMR spectroscopy. Chem Commun 47:7368–7370
Vlasie MD, Comuzzi C, van den Nieuwendijk AMCH, Prudêncio M, Overhand M, Ubbink M (2007) Long-range-distance NMR effects in a protein labeled with a lanthanide-DOTA chelate. Chem Eur J 13:1715–1723
Wolfram Research Inc (2010) Mathematica, Version 8.0
Yagi H, Maleckis A, Otting G (2013) A systematic study of labelling an α-helix in a protein with a lanthanide using IDA-SH or NTA-SH tags. J Biomol NMR 55:157–166
Yang Y, Li QF, Cao C, Huang F, Su XC (2013) Site-specific labeling of proteins with a chemically stable, high-affinity tag for protein study. Chem Eur J 19:1097–1103
Young TS, Ahmad I, Yin JA, Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395:361–374
Acknowledgments
We thank Mr. Kala Bharath Pilla and Dr. Thomas Huber for stimulating discussions. Financial support by the Australian Research Council is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shishmarev, D., Otting, G. How reliable are pseudocontact shifts induced in proteins and ligands by mobile paramagnetic metal tags? A modelling study. J Biomol NMR 56, 203–216 (2013). https://doi.org/10.1007/s10858-013-9738-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9738-6