Abstract
A strategy for the introduction of (1H,13C-methyl)-alanine into perdeuterated proteins is described. Specific protonation of alanine methyl groups to a level of 95% can be achieved by overexpressing proteins in M9/D2O based bacterial growth medium supplemented with 800 mg/l of 2-[2H], 3-[13C] l-alanine. However, though simple, this approach results in undesired, non-specific background labeling due to isotope scrambling via different amino acid metabolic pathways. Following a careful analysis of known metabolic pathways we found that co-addition of perdeuterated forms of α-ketoisovalerate-d7, succinate-d4 and l-isoleucine-d10 with labeled l-alanine, reduces undesired background labeling to <1%. When combined with recently developed methyl TROSY experiments, this methyl-specific labeling protocol permits the acquisition of excellent quality correlation spectra of alanine methyl groups in high molecular weight proteins. Our cost effective strategy offers a significant enhancement in the level of incorporation of methyl-labeled alanine in overexpressed proteins over previously reported methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent progress in NMR spectroscopy of high molecular weight proteins have been strongly connected to the development of new isotopic labeling schemes. High level perdeuteration of proteins combined with optimized experiments has significantly increased the size of proteins for which a NMR study can be realized (Salzmann et al. 2000; Fernandez et al. 2001; Arora et al. 2001; Fiaux et al. 2002; Tugarinov et al. 2005). To date, transverse relaxation optimized spectroscopy (TROSY) has been developed for several atoms in biological molecules including backbone amide groups (Pervushin et al. 1997; Riek et al. 1999), aromatic methines (Pervushin et al. 1998; Brutscher et al. 1998), methylenes (Miclet et al. 2004, 2005) and methyl groups (Tugarinov et al. 2003). Because of their proton multiplicity and fast rotation, methyl groups are characterized by favorable spectroscopic properties. High sensitivity and resolution methyl 1H-13C correlation spectra can be recorded of protein complexes above 100 kDa, provided that these groups are selectively protonated in an otherwise highly perdeuderated protein background (Tugarinov et al. 2003). Methyl groups have been shown to be particularly useful to probe dynamics, interactions and structure properties in large protein assemblies (Gardner and Kay 1998; Tugarinov et al. 2005; Hamel and Dahlquist 2005; Sprangers et al. 2005, 2007; Sprangers and Kay 2007).
Initial protocols for the production of selectively methyl-protonated proteins used 1H,13C-pyruvate as the sole carbon source with growth media prepared in D2O (Rosen et al. 1996). With this method partially protonated Ala, Leu, Val, Ile (Ile-γ2 only) methyl groups were obtained with proton incorporation levels ranging from 40% to 80%. However, due to the partial exchange of protons, the final sample contains four different methyl group isotopomers (CH3, CH2D, CHD2 and CD3), which can significantly complicate subsequent NMR spectra. Later strategies based on the use of selectively protonated α-keto acid precursors in M9/D2O cultures enabled specific incorporation of 13CH3 groups in selected methyl sites. Using the approach developed in the Kay laboratory, the level of incorporation is very high without detectable scrambling of protons (for review see Tugarinov et al. 2006). Protocols using 2-keto, 3,3-[2H2], 4-[13C]-butyrate as the sole proton source produce proteins protonated at isoleucine-δ1 methyl sites only (Gardner and Kay 1997). Similarly, α-ketoisovalerate with specifically protonated methyl sites can be used to produce proteins with only the methyl groups of leucine and valine protonated (Goto et al. 1999; Tugarinov and Kay 2004). Alanine is another methyl site of great interest as the β-methyl group can report directly on the structure and dynamics of the protein backbone. Alanine is also the most abundant amino acid found in proteins (McCaldon and Argos 1988). Unlike isoleucine, leucine and valine residues, which are all synthesized by irreversible metabolic pathways in Escherichia coli, the synthesis of l-alanine is a single step reversible reaction with pyruvate as a precursor. This reaction can be catalyzed by three different transaminases: alanine-oxo acid amino transferase (EC 2.6.1.12), alanine-glycine transaminase (EC 2.6.1.44) and alanine transaminase (EC 2.6.1.2). As pyruvate is a key metabolite involved in a large number of biosynthetic pathways producing proteins with specific protonation of alanine methyl groups and without significant isotopic scrambling is more challenging. The best way to ensure specific labeling is to supplement labeled alanine directly to the culture medium. Isaacson et al. (2007) recently proposed a strategy to specifically incorporate methyl-protonated alanine using rich perdeuterated medium. In rich medium the high background level of perdeuterated amino acids serves to greatly reduce isotopic scrambling. However, this amino acid reservoir also dilutes the methyl-protonated alanine added to the culture medium and consequently reduces the level of incorporation in the target protein. Here we propose an efficient labeling protocol based on M9 culture medium in D2O to accomplish a near complete incorporation of [13C,1H3]-methyl groups in to alanine without detectable scrambling of protons to other sites. This new strategy relies on the introduction of perdeuterated precursors in combination with exogenous labeled alanine to abolish the metabolic scrambling of protons. Particular attention has been paid to the choice of the precursors and carbon sources in order to reduce costs and optimize the level of deuteration.
Materials and methods
Preparation of 2-[2H] l-alanine
2-[2H], 3-[13C] l-alanine and 2-[2H], 2,3-[13C2] l-alanine were synthesized using a modification of the procedure described by Goss and Newill for l-halotryptophan synthesis (Goss and Newill 2006). The exchange of the α-proton was achieved using a tryptophan synthase-rich cell lysate prepared from an LB culture of E. coli transformed with pSTB7, a high copy number plasmid that constitutively expresses Salmonella enterica tryptophan synthase (commercially available ATCC 37845; Goss and Newill 2006). For the proton to deuterium exchange, 1 g 13C-labeled l-alanine was dissolved in 60 ml of a 50 mM TRIS solution in D2O at pH 7.6, in presence of 1 mg of pyridoxal phosphate (PLP or vitamin B6). Lyophilized lysate, corresponding to 1 l of E. coli/pSTB7 culture (Goss and Newill 2006), was added and the mixture incubated at 35°C for 24 h. The extent of proton-deuterium exchange was monitored by NMR. After 24 h the solution was lyophilized. The resulting lysate was resuspended in 60 ml D2O and incubated for a further 24 h to complete the exchange. The reaction was stopped and tryptophan synthase precipitated by heating the solution to 80°C for 30 min. Following centrifugation, the supernatant containing labeled alanine was collected and used without further purification. The level of deuteriation was estimated to be over 98% from the analysis of the α proton NMR signal.
Optimization of the incorporation of alanine in overexpressed ubiquitin
Initial experiments to determine the level of protonated alanine incorporation into ubiquitin were performed using 3-[13C] l-alanine (CortecNet) to reduce costs. Escherichia coli BL21(DE3) cells were transformed with a pET41c plasmid carrying the human His-tagged ubiquitin (pET41c-His-Ubi) gene and transformants were grown in 70 ml unlabeled M9/H2O media containing 1 g/l 15NH4Cl, and 5 g/l glycerol (unlabeled) as the sole carbon source. When the optical density (OD) at 600 nm reached 0.8, a 30 ml solution containing 3-[13C] l-alanine was added. After an additional 1 h, protein expression was induced by the addition of IPTG to a final concentration of 1 mM. Induction was performed for 3 h at 37°C. Ubiquitin was purified by Ni-NTA (Qiagen) chromatography in a single step.
The optimal quantity of alanine required to achieve near complete incorporation in the overexpressed protein was assessed in a series of cultures (100 ml each) in which different amounts of 3-[13C] l-alanine were added 1 h prior induction (final concentration ranging from 0 to 800 mg/l). To quantify the reduction of isotope scrambling, several samples of ubiquitin were produced in the presence of 800 mg/l of 3-[13C] l-alanine with the addition of different metabolites: α-ketoisovalerate 200 mg/l; succinate (2.5 g/l); pyruvate (3 g/l) and acetate (3 g/l); and l-isoleucine with concentrations ranging from 0 to 100 mg/l.
Production of U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β proteins
Escherichia coli BL21(DE3) carrying the plasmid of the overexpressed protein (ubiquitin or MSG) were progressively adapted, in three stages over 24 h to M9/D2O media containing 1 g/l 15ND4Cl, 2.5 g/l glycerol-d5 (Cambridge Isotope Laboratories, Inc.) and 1 g/l d-glucose-d7 (Isotec). In the final culture, the bacteria were grown at 37°C in M9 media prepared with 99.85% D2O (Euriso-top). When the OD (600 nm) reached 0.8, a solution containing methyl-protonated alanine (prepared with the protocol described above) and different perdeuterated precursors in D2O was added. After addition the concentrations in culture medium were: 800 mg/l of 2-[2H], 3-[13C] l-alanine; 2.5 g/l of succinate-d4 (Cambridge Isotope Laboratories, Inc.); 200 mg/l for α-ketoisovalerate-d7 (CDN Isotopes Inc); and 60 mg/l for isoleucine-d10 (Cambridge Isotope Laboratories, Inc.). Protein expression was induced by the addition of IPTG to a final concentration of 1 mM 1 h later. Ubiquitin was purified by Ni-NTA chromatography a single step. Malate Synthase G (MSG) was purified in initially by Chelating Sepharose chromatography (GE Healthcare) followed by gel filtration chromatography (Superdex 200 pg GE Healthcare). All the perdeuterated precursors and carbon sources used in this protocol were chosen based on their availability from standard suppliers at relatively low cost. [13C,1H3] labeling of the Cβ position of alanine in U-[2H], U-[15N]-enriched proteins using the approach described here increases the isotope cost by ~400 € per liter of culture compared to standard protocols for the production of U-[2H], U-[15N]-proteins, (estimation based on the price of isotope purchased for this study in 2007 and 2008).
NMR spectroscopy
All spectra of ubiquitin were recorded on a Varian DirectDrive spectrometer operating at a proton frequency of 600 MHz equipped with a cryogenic triple resonance pulsed field gradient probehead. Data were acquired at 25°C in 100% D2O buffer containing 100 mM sodium phosphate (pH = 6.7, uncorrected). Protein concentrations were within the range of 0.5 to 2.0 mM. The 13C enrichment level at the Cβ position of alanine was determined by analyzing the multiplet structure of cross-peaks between alanine Hα and Hβ protons observed in a 2D-TOCSY experiments recorded of ubiquitin overexpressed in M9/H2O medium in presence of 3-[13C] l-alanine. The intensity of the central line of the multiplet corresponds to the proportion of Hβ bound to 12Cβ, while the two outer-lines (separated by 125 Hz) correspond to the proportion of 13C incorporated at the β position of alanines. The level of 13C labeling on other methyl, methylene and methine positions was determined by comparing the intensities of the resonances in 13C-HSQC spectra acquired in constant time (CT) mode. Signal intensities were corrected to account for different sample concentrations, and each cross peak normalized, as a percentage, against a comparable reference spectrum acquired of U-[13C]-ubiquitin.
All spectra of MSG were recorded on a Varian Inova spectrometer operating at a proton frequency of 800 MHz equipped with a cryogenic triple resonance pulsed field gradient probehead. Data were acquired on a 0.8 mM U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β MSG sample at 37°C in 100% D2O buffer containing 20 mM MES (pH = 7.0 uncorrected), 20 mM MgCl2, 5 mM DTT. The 1H-13C HMQC was recorded with 2576 complex data points in direct dimension (maximum t2 = 198 ms) and 160 points in carbon dimension (maximum t1 = 80 ms), with two scans by increment. In order to facilitate assignment of the alanine methyl resonances a 3D HCC experiment was recorded of 1 mM U-[2H], U-[15N], U-[12C], U-[13C2H]-Ala-α, U-[13C1H3]-Ala-βMSG sample prepared using the protocol described here. The experiment was collected with 192 points in the Cα dimension (maximum t1 = 30 ms), 104 points in the Cβ dimension (maximum t2 = 16 ms), and 1820 points in the direct dimension (maximum t3 = 140 ms) and 8 scans per increment, for a total acquisition time of 2.5 days. All data were processed and analyzed with the nmrPipe/nmrDraw suite of programs (Delaglio et al. 1995).
Results and discussion
Alanine incorporation
The first parameter optimized was the amount of exogenous alanine required to ensure complete incorporation of labeled alanine in proteins overexpressed in E. coli culture. To minimize isotope scrambling, labeled alanine was added to the culture one hour prior to induction. The level of incorporation of alanine into ubiquitin was followed by adding varying amounts of 3-[13C] l-alanine (concentrations from 50 to 800 mg/l of M9/H2O culture media). Figure 1 shows the level of 13C incorporation at the Cβ position of alanine in ubiquitin as a function of the amount of labeled alanine added to the culture medium. The incorporation level reaches a plateau above 90% for a concentration of exogenous l-alanine in excess of 600-mg/l. This result is in agreement with level of inhibition previously reported in literature (Muchmore et al. 1989).
Level of incorporation of 13C at the Cβ-alanine position in overexpressed ubiquitin as a function of the amount of exogeneous 3-[13C] l-alanine added. The 13C enrichment level at the Cβ position of alanine was determined by analyzing the multiplet structure of cross-peaks between alanine Hα and Hβ protons observed in 2D-TOCSY experiments
Figure 2A represents an extract of a 13C-HSQC spectrum of ubiquitin produced in E. coli in M9/H2O medium supplemented by 800 mg/l of 3-[13C] l-alanine one hour prior to induction. As expected, intense signals for the two alanine residues of ubiquitin are observed, but several additional signals could also be detected for other methyl, methylene and methine sites. Comparison of signal intensities with a 13C-HSQC spectrum recorded on a uniformly 13C-labeled ubiquitin sample confirms that the Cβ atoms of alanine are labeled at more than 95% (data not shown). The level of 13C labeling of leucine, valine and isoleucine (Ile-γ2 only) methyl groups is approximately 25%. The incorporation of 13C in all other methyl (Ile-δ1, Met and Thr), methylene and methine sites ranges from 5% to 15%.
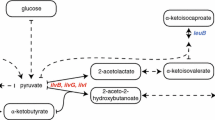
A 2D 1H-13C HSQC spectrum of ubiquitin overexpressed in presence of 13C-alanine. The spectrum was recorded at 25°C in D2O on a NMR spectrometer operating at a proton frequency of 600 MHz. The first contour level corresponds to 5 % of the maximal intensities of Cβ-alanine correlation peaks. The two cross-peaks corresponding to two expected Cβ-alanine signals are labeled. The spectrum shows that a significant amount of 13C from exogeneous 13C-alanine has been incorporated in other amino acids. The scrambling level reaches 25% in methyls of Leucine, Valine and Isoleucine (Ile-γ2 only) and 5–15% for other methyl, methylene and methines sites. The locations of two selected traces, (a) and (b) presented below the HSQC spectra, are indicated by a dashed line on the 2D spectrum. On the 1D traces the first contour level plotted in the 2D spectrum is represented by a dashed line. The intensities of trace (b) relative to trace (a) has been multiply by a factor 12. B Schematic illustration of the major metabolic pathways involved in amino acid biosynthesis in E. coli. The level of the scrambling of 13C from 3-[13C] l-alanine is represented by the thickness of the arrows. Three transaminases are responsible for the conversion of 13C-alanine into pyruvate. The methyl group of pyruvate is directly utilised for the synthesis of Leucine, Valine and Isoleucine (Ile-γ2 only), and gives rise to an important (~25%) 13C scrambling in these methyl groups. Pyruvate is also incorporated in the TCA cycle and glycolytic pathways. These reactions are responsible of the background 13C labeling of most of the amino-acids sites (5–15%)
Reduction of isotopic scrambling
The scrambling of 13C observed in Fig. 2A arises from the metabolic interconversion of alanine and pyruvate (Fig. 2B). The simplest way to reduce the scrambling would be to add unlabeled pyruvate to the cell culture at the same time as the exogenous 3-[13C] l-alanine. Indeed, addition of 3 g/l of pyruvate with 3 g/l of acetate reduced scrambling to a level lower than 5% for all methine, methylene and methyl sites (Figure S1). However, due to the efficiency of the three transaminases, pyruvate is still converted into alanine. The addition of exogenous unlabeled pyruvate also reduces the level of incorporation of 3-[13C] l-alanine to approximately 60% (Figure S1). Therefore, to maintain a high level of incorporation of labeled alanine, the reduction of isotopic scrambling should be achieved without the addition of exogenous pyruvate.
The methyl groups of leucine, valine and isoleucine (Ile-γ2 only) are directly transferred from pyruvate (Fig. 2B) and therefore these sites are more severely affected by isotopic scrambling. Addition of α-ketoisovalerate, at a concentration higher than 100 mg/l of culture, has been shown to give near complete labeling of leucine and valine methyl groups (Goto et al. 1999). To attempt to reduce isotope scrambling in leucine and valine methyl groups, the medium was supplemented with unlabeled α-ketoisovalerate in addition to 13C-alanine. α-ketobutyrate has also been used for specific labeling of the isoleucine-δ1 methyl group (Gardner and Kay 1997). However, as the Ile-γ2 methyl group is derived directly from the methyl group of pyruvate, the use of deuterated α-ketobutyrate will not reduce isotope scrambling on Ile-γ2 methyl sites. To overcome this issue, isoleucine can be added directly to the culture to further reduce isotope scrambling. As illustrated in Figure S2, the addition of unlabeled isoleucine at 60 mg/l reduces the undesired secondary labeling of isoleucine methyl groups to a level lower than 3%.
13C labeled atoms of pyruvate are also incorporated in other amino acids (Fig. 2B) via the glycolytic pathway (H, S, G, C, W, F, Y) and the tricarboxylic acid (TCA) cycle (E, N, Q, P, R, D, K, M, T, I). Efficient deuteration of proteins can be realized using perdeuterated glucose, glycerol, acetate (Venters et al. 1995) or succinate (Vanatalu et al. 1993) as the source of carbon. Glucose and glycerol are incorporated in to the glycolytic pathway (Fig. 2B), while acetate (converted in to acyl-CoA) and succinate are directly incorporated in TCA cycle. Perdeuterated forms of the three last compounds are available at very competitive price, compared to glucose-d7. These three metabolites are thus good candidates for carbon sources to complement glucose in order to reduce background isotope scrambling in methine, methylene and methyl groups of isoleucine (Ile-δ1), threonine and methionine.
Figure 3A shows a region of an 1H-13C-HSQC spectrum of ubiquitin produced in E. coli in M9/H2O medium using a combination of 1 g/l of glucose, 2.5 g/l glycerol as sources of carbon. One hour prior to induction, 800 mg/l of 3-[13C] l-alanine together with 2.5 g/l of succinate, 60 mg/l of isoleucine and 200 mg/l of α-ketoisovalerate were added to the culture. NMR spectra and extracted 1D traces show near complete incorporation of labeled alanine is achieved with a concomitant reduction in 13C scrambling of all other methine, methylene and methyl sites to a level lower than 5%.
2D 1H-13C HSQC spectra of ubiquitin with methyl-specific alanine labeling. The spectra were recorded at 25°C in D2O on a NMR spectrometer operating at a proton frequency of 600 MHz. Ubiquitin has been overexpressed in E. coli in presence of labeled alanine at 800 mg/l, 2.5 g/l of succinate, 60 mg/l of isoleucine and 200 mg/l of α-ketoisovalerate. In A U-[1H], U-[15N], U-[12C], U-[13C1H3]-Ala-β ubiquitin overexpressed in M9/H2O medium using protonated glucose (1 g/l) glycerol (5 g/l), and protonated precursors. B U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β ubiquitin overexpresed in M9/D2O medium using glucose-d7, glycerol-d5 with the addition one hour prior to induction of 2-[2H], 3-[13C] l-alanine, together with succinate-d4, isoleucine-d10 and α-ketoisovalerate-d7. In both spectra, the first contour level corresponds to a signal equal to 5% of the maximal intensities of Cβ-alanine correlation peaks. The two cross-peaks corresponding to Cβ-alanine signals are labeled. Two selected traces (a) and (b) located by a dashed line on the 2D spectra, are presented below the HSQC spectra. On each 1D trace the first contour level plotted in the 2D spectrum is represented by a dashed line. The intensities of trace (b) relative to trace (a) has been multiply by a factor 12
Production of U-[2H], U-[15N], U-[12C], U-[13C1H3]-β-Ala proteins in M9/D2O culture medium
Commercially available 3-[13C] l-alanine was used to report isotope scrambling in order to develop an efficient protocol to achieve a high level of incorporation of methyl-labeled alanine with reduced recycling into other amino-acids. To ensure that perdeuterated proteins are specifically protonated on the alanine methyl group, the culture should be prepared in 100% D2O using perdeuterated carbon sources and alanine deuterated at the Hα position. For this purpose the use of fully protonated 3-[13C] l-alanine in M9/D2O growth medium would be inadequate and would result in the production of proteins with a high level of protonation at the alanine Hα position. All the precursors and carbon sources in the optimized protocol described above have been selected not only for their efficiency to reduce isotope scrambling but also for their commercial availability in perdeuterated form at a relatively low cost (see section “Materials and Methods” for details). As 2-[2H], 3-[13C] l-alanine is not available commercially, a simple and cost effective protocol was established to specifically exchange the alpha proton of the 3-[13C] l-alanine by deuterium, with retention of configuration, using tryptophan synthase (Nibeilliu and Malthouse 2004; Isaacson et al. 2007; see section “Materials and Methods” for further details).
Specific methyl-protonated alanine was used to produce U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β ubiquitin. Figure 3B shows an extract of a 1H-13C-HSQC spectrum of ubiquitin produced in M9/D2O with 2-[2H], 3-[13C] l-alanine and perdeuterated forms of all supplemented precursors and carbon sources (a detailed protocol is given in the Materials and Methods section). As it can be seen, complete incorporation of 2-[2H], 3-[13C] l-alanine is achieved with negligible labeling at other sites. Quantitative analysis of NMR spectra demonstrated that the level of alanine methyl group labeling is more than 95% without any detectable level 13CDH2 or 13CD2H isotopomers. Isotopic scrambling has been considerably reduced and none of the other methine, methylene or methyl sites give rise to peaks intensities higher than 0.5%. The reduction of background labeling (compare Fig. 3A, B) is primarily due to the fact that in deuterated M9, the only source of protons is the alanine methyl group. In order to be incorporated in other residues, alanine methyl protons would have to be transferred to pyruvate. In vivo the protons of pyruvate spontaneously exchange with deuterium from the D2O used to prepare the culture medium. Moreover, protonated pyruvate is recycled through the glycolysis pathway or TCA cycle for use in the synthesis of other amino acids. These reactions involve enzyme-catalyzed exchange of protons with deuterium from the D2O solvent. As a result the few 13C-methyl groups derived from exogenous 2-[2H], 3-[13C] l-alanine (<5% of all amino acid carbon sites) lose their protons during the process of being incorporated into other amino acids.
Comparison with previously proposed Alanine labeling protocol
Isaacson et al. (2007) proposed a method for specifically protonating the methyl group of alanine in which proteins are overexpressed in perdeutered rich culture medium supplemented with methyl-protonated alanine (50 mg/l). The presence of a mix of perdeuterated amino-acids in the culture medium ensures that the recycling of 13CH3 groups to other residues is considerably reduced. However, rich medium also contains a source of fully deuterated alanine, which would also be incorporated into the target protein. Therefore, the overall result is a protein containing a mixture of alanine labeling with incorporation levels of the desired 3-[13C,1H]-labeled alanine diluted by the background levels of 3-[12C,2H]-labeled alanine.
In order to compare the protocol presented here with the method proposed by Isaacson et al. (2007), the same protein (i.e. ubiquitin) was overexpressed using both protocols and analyzed by NMR spectroscopy. Production of perdeuterated ubiqutin using rich perdeuterated culture medium as previously described (Isaacson et al. 2007) substantially enhanced protein expression. Ubiquitin prepared by either method yielded high quality NMR spectra, with narrow lines for the two alanine methyl groups and without noticeable isotope scrambling in other residues. Nevertheless, at identical protein concentration the intensity of the alanine signals in the protein prepared in rich perdeuterated medium is substantially weaker (Fig. 4). This difference of intensity suggests that the incorporation level of methyl-specific labeled alanine when [13C]-labeled alanine is used in combination with deuterated Spectra 9 based rich medium is only ~30% (Isaacson et al. 2007). This result is consistent with the low level of incorporation observed in our systematic study where various amounts of 13C-alanine were added in M9/H2O culture medium (Fig. 1). While isotope incorporation levels are dependent on the different growth/expression conditions used, the primary reason for the lower of isotope incorporation is the presence of perdeuterated alanine in the Spectra 9 rich deuterated medium (a 2H-Celtone/D2O derived culture medium). According to the manufacturer’s analysis Spectra 9 medium contains 60 ± 10 mg of deuterated alanine (for additional information see the manufacturer’s website: www.spectrastableisotopes.com/Catalog/Celtone_Powder_Information.aspx). Furthermore, we have established that when 2-keto, 3-[2H2], 4-[13C]-butyrate precursor (50 mg/l) is added to Spectra 9 perdeuterated rich medium the level of incorporation of isoleucine is reduced to 70% compared to the level achieved using α-keto-acid in M9 medium (Gardner and Kay 1997; Sounier et al. 2007).
Comparison of 2D 1H-13C HSQC spectra of U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β ubiquitin overexpressed in M9/D2O medium (A) or Spectra-9/D2O rich medium (B). Spectra have been recorded at 25°C in D2O on a NMR spectrometer operating at a proton frequency of 600 MHz. Ubiquitin used for spectra A has been produced in E. coli in M9/D2O medium using glucose-d7 (1 g/l) and glycerol-d5 (2.5 g/l) with an addition one hour prior to the induction of 2-[2H], 3-[13C] l-alanine at 800 mg/l, together with 2.5 g/l of succinate-d4, 60 mg/l of isoleucine-d10 and 200 mg/l of α-ketoisovalerate-d7. Ubiquitin used for spectra B has been produced in Spectra-9/D2O rich medium with an addition 1 h prior to induction of 2-2H, 3-13C l-alanine at 50 mg/l as described by Isaacson et al. (2007). Spectra B has been scaled to take into account the difference in sample concentration used for acquisition of spectra A. The first contour level (of both spectra A and B) corresponds to a signal equal at 5% of the maximal intensities of Cβ-alanine correlation peaks detected in spectra A. The two cross-peaks corresponding to Cβ-alanine signals are labeled. Selected traces located by a dashed line on the 2D spectra, are presented below the HSQC spectra. On each 1D trace the first contour level plotted in the 2D spectrum is represented by a dashed line. The intensities of trace (b) relative to trace (a) has been multiply by a factor 12
A reduction in the level of isotope incorporation will directly affect the sensitivity of NMR experiments. The active alanine concentration (i.e. the amount of sites with the desired isotope incorporation) in proteins prepared using the method of Issacson et al. is essentially 30% of the total protein concentration. Protein samples with methyl-specific protonation have been shown to be useful for extracting precise long-range inter-methyl group distances (Sounier et al. 2007). The signal of an NOE cross-peak between two protonated alanines methyl groups decreases with the square of the level of incorporation. Therefore the preparation of proteins with methyl-specific protonated alanines prepared using the M9-based method presented here will enhance the sensitivity of NOESY experiments by one order of magnitude compared to the scheme previously proposed (Isaacson et al. 2007).
Application to malate synthase G
One of the major applications of methyl-specific labeling strategy is the study of large proteins and protein complexes. MSG, a monomeric protein of 82 kDa, was produced using the new protocol presented here. Pure MSG specifically 1H and 13C labeled on the methyl group of alanine was produced at a yield of 80 mg/l. A 13C-1H-HMQC spectrum of MSG recorded at 800 MHz is presented in Fig. 5A. The signals of the 73 alanine residues of MSG can be detected with high signal to noise ratio and resolution. The method described in this manuscript is equally applicable for the introduction of 2-3-[13C2] l-alanine or U-[13C] l-alanine in perdeuterated proteins. One should note that acquisition of [13C,1H]-HMQC spectra acquired of samples produced using 2-3-[13C2] l-alanine or [13C3] l-alanine requires CT carbon frequency editing or homonuclear carbon-carbon decoupling to maintain the high resolution of the spectra. Cα and Cβ resonance assignments available from standard triple resonance experiments acquired on a U-[13C,15N, 2H]-labeled protein sample can be used a starting point for alanine methyl resonance assignment. These data in combination with, for example, HCC spectra recorded of a perdeuterated protein specifically labeled with 2-[2H], 2-3-[13C2] L-alanine would permit residue-specific assignment of alanine methyl [13C,1H] resonances. Introduction of U-[13C] l-alanine would also be possible using the protocol outlined here but at significantly higher cost. To facilitate the assignment of alanine methyl resonances of MSG, 2-[2H], 2-3-[13C2] l-alanine was incorporated in to perdeuterated MSG. An HCC experiment was used to correlate 1Hβ and 13Cβ frequencies to the corresponding 13Cα resonance. 2D extracts of this experiment are presented in Fig. 5B. Using this experiment together with Cα and Cβ assignments of alanine, previously reported by Tugarinov et al. (2002), it was possible to assign all the alanine methyl resonances observed in HMQC spectra of MSG (Fig. 5A).
A 2D 1H-13C HMQC spectrum of alanine methyl group in Malate Synthase G (MSG—82 kDa) recorded on a 0.8 mM sample of U-[2H], U-[15N], U-[12C], U-[13C1H3]-Ala-β MSG. An expansion of the central part of the spectrum is displayed in the bottom right corner. B 2D-(F1,F3) extracts of 3D HCC experiment correlating Cα (F1), Cβ (F2) and Hβ (F3) frequencies of MSG alanine residues recorded on a 1 mM sample of U-[2H], U-[15N], U-[12C], U-[13C2H]-Ala-α, U-[13C1H3]-Ala-β MSG. Both spectra have been recorded at 37°C in D2O on a NMR spectrometer operating at a proton frequency of 800 MHz. All the cross-peaks in spectra A and B have been labeled with the corresponding residue number. Asterisk corresponds to minor form of MSG as previously described by Tugarinov et al. (2002)
Conclusion
A cost effective method for the preparation of methyl-specific 13C,1H-labeled alanine in M9/D2O media was developed. Simultaneous introduction in the culture medium of specifically labeled alanine with perdeuterated precursors allowed isotope scrambling to be reduced to <0.5%. The protocol presented here will allow incorporation of specifically methyl-protonated alanine at a level higher than 95% in an otherwise perdeuterated protein. Combined with recently developed methyl TROSY approach, this specific methyl-labeling protocol allows the acquisition of high quality correlation spectra of alanine methyl groups in high molecular weight proteins. This new labeling protocol is expected to be particularly useful as the incorporation of protonated alanine methyl groups offers a unique opportunity to probe structural and dynamical properties of protein backbone in large biomolecular assemblies.
References
Arora A, Abildgaard F, Bushweller JH, Tamm LK (2001) Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol 8:334–338
Brutscher B, Boisbouvier J, Pardi A, Marion D, Simorre JP (1998) Improved sensitivity and resolution in 1H-13C NMR experiments of RNA. J Am Chem Soc 120:11845–11851
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Fernandez C, Adeishvili K, Wüthrich K (2001) Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX in dihexanoyl phosphatidylcholine micelles. Proc Natl Acad Sci USA 98:2358–2363
Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2002) NMR analysis of a 900 kDa GroEL GroES complex. Nature 418:207–211
Gardner KH, Kay LE (1997) Production and incorporation of 15N, 13C, 2H (1H–δ1 Methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc 119:7599–7600
Gardner KH, Kay LE (1998) The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Ann Rev Biophys Biomol Struct 27:357–406
Goss RJM, Newill PLA (2006) A convenient enzymatic synthesis of l-halotryptophans. Chem Commun 4924–4925
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13:369–374
Hamel DJ, Dahlquist FW (2005) The contact interface of a 120 kD CheA-CheW complex by methyl TROSY interaction spectroscopy. J Am Chem Soc 127:9676–9677
Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S (2007) A new labelling method for methyl transverse relaxation-optimized spectroscopy NMR spectra of Alanine residues. J Am Chem Soc 129:15428–15429
McCaldon P, Argos P (1988) Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide-sequences. Proteins Struct Funct Genet 4:99–122
Miclet E, Williams JDC, Clore GM, Bryce DL, Boisbouvier J, Bax A (2004) Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J Am Chem Soc 126:10560–10570
Miclet E, Boisbouvier J, Bax A (2005) Measurement of eight scalar and dipolar couplings for methine-methylene pairs in proteins and nucleic acids. J Biomol NMR 31:201–216
Muchmore DC, McIntosh LP, Russel CB, Anderson DE, Dahlquist FW (1989) Expression and 15N labelling of proteins for proton and 15N nuclear magnetic resonance. Meth Enzymol 177:44–73
Nibeilliu ME, Malthouse PG (2004) The stereospecificity and catalytic efficiency of the tryptophan synthase-catalysed exchange of the α-protons of amino acids. Biochem J 381:847–852
Pervushin K, Riek R, Wider G, Wuthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole- dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Pervushin K, Riek R, Wider G, Wuthrich K (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J Am Chem Soc 120:6394–6400
Riek R, Wider G, Pervushin K, Wüthrich K (1999) Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA 96:4918–4923
Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE (1996) Selective methyl group protonation of perdeuterated proteins. J Mol Biol 263:627–636
Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K (2000) NMR assignment and secondary structure determination of an octameric 110 kDa protein using TROSY in triple resonance experiments. J Am Chem Soc 122:7543–7548
Sounier R, Blanchard L, Wu Z, Boisbouvier J (2007) High-accuracy distance measurement between remote methyls in specifically protonated proteins. J Am Chem Soc 129:472–473
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445:618–622
Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE (2005) Quantitative NMR spectroscopy of supramolecular complexes: Dynamics side pores in ClpP are important for product release. Proc Natl Acad Sci USA 102:16678–16683
Sprangers R, Velyvis A, Kay LE (2007) Solution NMR of supramolecular complexes: providing new insights into function. Nat Meth 4:697–703
Tugarinov V, Kay LE (2004) An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR 28:165–172
Tugarinov V, Muhandiram DR, Ayed A, Kay LE (2002) Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase G. J Am Chem Soc 124:10025–10035
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Tugarinov V, Choy WY, Orekhov VY, Kay LE (2005) Solution NMR-derived global folded of a monomeric 82-kDA enzyme. Proc Natl Acad Sci USA 102:622–627
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754
Vanatalu K, Paalme T, Vilu R, Burkhardt N, Junemann R, May R, Ruhl M, Wadzack J, Nierhaus KH (1993) Large-scale preparation of fully deuterated cell components—ribosomes from Escherichia coli with high biological activity. Eur J Biochem 216:315–321
Venters RA, Huang C-C, Farmer BTII, Trolard R, Spicer LD, Fierke CA (1995) High-level 2H/13C/15N labeling of proteins for NMR studies. J Biomol NMR 5:339–344
Acknowledgements
This work was supported by the CEA, the CNRS, and the UJF. J.B acknowledges funding from HFSP (CDA #0029/2004) and CNRS (Interdisciplinary program interface between physics, biology and chemistry). The authors thank Drs. Kay and Remington for providing clones of Malate Synthase G, Drs. Amero and Marion for stimulating discussion, and Dr. Plevin for suggestions and careful reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ayala, I., Sounier, R., Usé, N. et al. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR 43, 111–119 (2009). https://doi.org/10.1007/s10858-008-9294-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-008-9294-7