Abstract
This paper demonstrated a new method for preparing fluorescent molecularly imprinted polymer (MIP) for specific recognition of a target protein. The MIP-based fluorescent receptor was developed by coating MIP layer on the surface of l-cysteine modified Mn2+-doped ZnS quantum dots (QDs) using the surface molecular imprinting process. These MIP-QDs composites demonstrated fast adsorption kinetics, high stability, and good dispersibility in aqueous media. Since the fluorescence quenching of MIP-QDs composites is proportional to the concentration of the lysozyme, the MIP-based fluorescent receptor was successfully applied to the direct fluorescence quantification of lysozyme without further pretreatment. The MIP-based fluorescent receptor exhibited good selectivity and sensitivity for lysozyme detection. The optimum fluorescence intensity of the MIP-QDs was found to be at pH 6.0, and the linear range of lysozyme was from 0.1 to 2.0 μM with the detection limit of 25.2 nM. Moreover, the proposed fluorescent receptor was satisfactorily applied to the determination of lysozyme in real samples. This study provides a new approach for recognizing and detecting of specific proteins in biological samples.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lysozyme (lyz), which widely exists in body tissues and secretions, plays an important physiological role in the innate immune system [1]. It is a small single chain protein consisting of 129 amino acids that can break down bacteria by hydrolyzing the major component of bacterial cell walls [2]. The abnormal concentration of lysozyme in body fluid and tissues is the potential indicator for many diseases, such as leukemia, renal diseases, and meningitis [3]. To date, extensive of strategies for the lysozyme detection have been reported, including enzyme-linked immunosorbent assays (ELISAs) [4, 5], resonance rayleigh scattering method (RRS) [6, 7], quartz crystal microbalance (QCM) [8], surface plasmon resonance (SPR) [9], colorimetric [10], chemiluminescence (CL) [11, 12], mass spectrometric detection (MASS) [13], high performance liquid chromatography (HPLC) [14], and electrochemistry [15, 16]. However, many of these methods have its own limitations such as complex sample pre-treatment, higher cost, unsatisfactory detection limit, and lower selectivity [17–21]. Therefore, a low-cost, reliable, and selective strategy for lysozyme detection is in high demand.
Molecular imprinting technique (MIT) is a process to synthesize artificial receptors with specific binding sites that are complementary to target molecule [22, 23]. Due to their significant advantages, such as highly physical/mechanical stability, high selectivity, and low cost for preparation, molecularly imprinted polymers (MIPs) are successfully applied in analysis of various analytes [24, 25]. The imprinting of small molecules is well achieved and straightforward. However, imprinting of protein is still a challenge to people. The difficulty of this process is mainly caused by the large molecular size, the structural complexity, and diversity of functional groups of the protein, which leads to more non-specific absorption, mass transfer blocking and increases the difficulty in removing template protein from interior binding sites [26–28]. Surface imprinting on nanoparticles has high surface area to volume ratio and high binding capacity, which facilitates easy removal of the template protein, fast rebinding kinetics and good accessibility of the target protein to the recognition sites [29]. The design of chemical sensors based on surface imprinting on the nanostructured materials can provide an effective approach to overcome many of these difficulties in imprinting of proteins.
In most cases, synthesis of MIP-based sensor mainly combined imprinted polymers and a transducer [30]. As a kind of nanomaterial, quantum dots (QDs) have the potential as a fluorescent transducer for molecular imprinting sensing of proteins due to their unique optical properties, such as high luminescence efficiency, good photo stability, narrow and symmetric emission spectra, and size-dependent emission wavelengths [31–33]. The unique size of QDs allows them both as a fluorescence signal source and a multifunctional support for the binding with biomolecules or other analyte [34]. Surface imprinting proteins on quantum dots are providing new forms of transducing the biological recognition process into an optical analytical signal [35, 36]. The coupling of MIPs and QDs into one system produces a new fluorescent sensor that not only makes protein imprinting possible or easier but also improves the specific recognition and provides more sensitive detection [37].
In this study, a facile approach to imprint protein on the surface of l-cysteine-modified Mn2+-doped ZnS QDs was developed. l-cysteine was used to modify the surface of ZnS QDs by the ligand competition. This process increased the water dispersion ability, reduced cytotoxicity, and provided carboxylic acid group that can bind with amino group of template protein [38]. The fluorescent receptor was fabricated by surface graft imprinting in aqueous solutions using l-cysteine modified Mn2+-doped ZnS QDs as supports, acrylamide as functional monomers, N, N′-methylenediacrylamide as cross-linker and lysozyme as template. These MIP-based QDs composites combined the advantages of molecular imprinting with the optical properties of the QDs, which could be a potential as fluorescence sensor for recognizing and detecting of proteins [39, 40]. The feasibility of using the MIP-based fluorescent sensor for lysozyme detection is mainly based on the fact that the l-cysteine-capped Mn-doped ZnS QDs acts as receptor for recognizing signal amplification and optical readout, while the MIP shell on the QDs provides analyte selectivity and prevents interfering molecules from binding with the fluorescent receptor. The MIP-QDs composites were more strongly quenched by the template molecule (lysozyme) and the fluorescence quenching was proportional to the concentration of lysozyme. Based on the efficient quenching of the MIP-QDs composites by target protein, this MIP-based fluorescent receptor provides a simple strategy for selective, sensitive, and direct fluorescence sensing of target protein in biological samples.
Experimental
Reagents and materials
Zinc sulfate heptahydrate, sodium sulfide nonahydrate, manganese (II) chloride tetrahydrate, and acetic acid were obtained from Beijing Chemical Works. l-cysteine was obtained from J&K Scientific Ltd. Acrylamide (AAM), sodium dodecyl sulfate (SDS), N, N′- methylenediacrylamide (MBAAM), ammoniumpersulfate (APS), and N,N,N′,N′,-tetramethylethylethylenediamine (TEMED) were purchased from Tianjin Fine Chemical Research Institute. Lysozyme (Lyz, MW 14.4 kDa, pI 11.2), bovine serum albumin (BSA, MW 68 kDa, pI 4.9), bovine hemoglobin (BHb, MW 66 kDa, pI 6.7), and ovalbumin (OVA, MW 45 kDa, pI 4.7) were purchased from Sigma-Aldrich stock. All chemicals were analytical grade, and all the solutions were prepared with double distilled deionized water.
Characterization
All the fluorescence measurements were performed on a RF-5301 spectrofluorometer (Shimadzu, Japan) equipped with a quartz cell (1 × 1 cm). The UV spectra were recorded on a UV-1800 spectrophotometer (Shimadzu, Japan). The morphology of the l-cysteine-capped ZnS QDs and MIP-coated QDs were characterized by high-resolution transmission electron microscopy (HRTEM) on a H9000 (Hitachi, Japan), operating at a 300 kV accelerating voltage.
Synthesis of l-cysteine-capped ZnS QDs
l-cysteine-capped ZnS QDs were prepared based on previous publication procedures with some modifications [41, 42]. In brief, an aqueous solution of ZnSO4·7H2O (0.25 M), MnCl2·4H2O (0.02 M), l-cysteine (0.15 M) was added to a three-necked flask. The mixture was stirred under dry nitrogen at room temperature for 90 min. Then 5 mL of the Na2S·9H2O aqueous solution (0.25 M) was dropwise added. The above solutions were kept stirring for another 12 h under dark conditions. The obtained l-cysteine-capped ZnS QDs were precipitated with ethanol, separated by centrifuging, washed with water and ethanol for three times to remove the unreacted substance, and dried under vacuum for further use.
Preparation of MIP-coated QDs composites
The MIP-coated l-cysteine-capped ZnS QDs composites (MIP-QDs) were prepared by a surface molecularly imprinting process. To a 50-mL flask, acrylamide monomer (200 mg) and lysozyme (30 mg) were first dissolved in 25 mL phosphate buffer (0.02 mol/L, pH 6.0) and stirred for 20 min. Then, 280 mg l-cysteine-capped Mn-doped QDs, N, N′- methylenediacrylamide (200 mg) was added. After continuously stirring for another 30 min, 15-mg ammonium persulphate and 10 μL N, N, N′, N′-tetramethylethylenediamine (TEMED) were added into the mixture in sequence. Subsequently, the mixture solution was deoxygenated by purging with nitrogen, and the reaction proceeded at room temperature for 20 h. As a control group, non-imprinted polymer-coated l-cysteine-capped ZnS QDs composites (NIP-QDs) were prepared through the same procedure without using lysozyme. The MIP- and NIP-coated QDs were centrifuging, washed by water several times to remove the unreacted substance. The resultant MIP- and NIP-QDs were washed repeatedly with a mixture of sodium dodecyl sulfate (10 %, w:v) and acetic acid (10 %, v:v) solution to remove the lysozyme until the fluorescence intensity of MIP-QDs similar to that of their non-imprinted counterparts. Finally, the products were washed by methanol, and dried under vacuum.
Response time of the MIP-capped ZnS QDs
The incubation time influence on the fluorescence quenching efficiency of MIP-QDs composites was first examined. To carry out the response time test, the as-prepared MIP- or NIP-coated QDs were placed into lysozyme solutions (1 μM), and then were shaken at 25 °C for different periods of time. At last, the incubated composites were taken out, and the fluorescence intensity was recorded at different times.
Effect of pH on the fluorescence emission of MIP-capped ZnS QDs
The effect of pH on the fluorescence intensity change of MIP-QDs was investigated. For a 10-mL calibrated test tube, 1 mL of 0.6 g L−1 MIP-coated QDs was diluted to volume with different buffer solutions (pH 4.0–11.0). After 25-min incubation at 25 °C, the change in fluorescence intensity of MIP-coated QDs in the presence and absence of lysozyme in different buffer solutions was recorded by a spectrofluorometer.
Measurement procedure
To carry out the rebinding experiments, 1 mL of 0.6 g L−1 MIP- or NIP-coated QDs, a lysozyme standard solution with a given concentration was sequentially added in a 10-mL calibrated test tube. The mixture was diluted to 10 mL with phosphate buffer (pH 6.0), and the concentration of lysozyme ranging from 0.1 to 2.0 μM. After being mixed thoroughly, the mixture incubated for 25 min at 25 °C, the change in the fluorescence spectra of the MIP- and NIP QDs were measured by using a spectrofluorometer.
Rebinding specificity
The selectivity experiments were carried out by using BSA, BHB, and OVA as references. To a 10-mL calibrated test tube, MIP- or NIP-coated QDs, a lysozyme with a given concentration or reference proteins (BHB, OVA, and BSA) were sequentially added. Various samples with the concentration ranging from 0.1 to 2.0 μM were prepared with phosphate buffer (0.02 mol L−1, pH 6.0). After being shaken for 25 min at room temperature, the fluorescent intensity of the solution was recorded at excitation wavelength of 302 nm.
Real samples
In this study, two different chicken egg whites were used as real samples for analysis. For each sample, the chicken egg white was separated from a fresh egg, and homogenized in an ice bath for 30 min. Then 3 mL of the homogenized egg was diluted to 600 mL with phosphate buffer (0.02 mol L−1, pH 6.0). After the diluted solution was immersed in an ice bath, and centrifuged at 6000 rpm for 30 min, the supernatant solution was used as a lysozyme source. All of the samples were filtered through a 0.45 μm filter. The MIP-QDs were added into the samples, and incubated for 25 min at room temperature. The changes in fluorescence intensity of the MIP-QDs were recorded, and the concentrations of lysozyme in egg white samples were detected by the proposed method. To evaluate the accuracy of the developed method, a recovery test was also carried out by the samples spiked with lysozyme standard solution.
Results and discussion
Preparation of MIP-QDs fluorescence biosensors
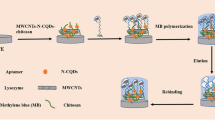
The proposed fluorescence biosensor fabrication process is mainly based on coating molecularly imprinted polymer on the surface of QDs, as illustrated in Fig. 1. First, l-cysteine was used as a non-toxic material to coat the surface of Mn2+-doped ZnS QDs to form l-cysteine-capped ZnS QDs by covalent bonds. This process indicates that carboxylic acid group to QDs not only improves the water dispersion ability for chemical sensing, but also makes it available to bind with amino group of lysozyme. Besides, l-cysteine-modified ZnS QDs can act as assistant monomers to create effective recognition sites. Secondly, acrylamide was chosen as a functional monomer as acrylamide had multiple hydrogen binding sites with the lysozyme. N, N′- methylenediacrylamide and ammonium persulphate were used as a cross-linker and catalyst, respectively. The molecularly imprinted polymer layer was immobilized on l-cysteine-capped ZnS QDs via a facile surface imprinting process. After the removal of the template molecular, the binding sites are exposed on the surface of polymers, which are complementary to the template in size, shape, and position of the functional groups, and are able to selectively rebind with the template molecule.
Template removal
To investigate the effect of template removal, the emission spectra of MIP- and NIP-QDs were recorded before and after removing the lysozyme. In this work, direct fluorescence detection to examine the template removal was more convenient and sensitive than the indirect UV detection. As shown in Fig. 2, prior to the removing of the lysozyme, the fluorescence emission intensity of the MIP-QDs was approximately 40.6 % of their non-imprinted ones. It is mainly because of the excitation electron transfers from QDs to lysozyme, which results in weak emission. After template removal, the fluorescence intensity of the MIP-QDs was restored dramatically to 92.8 % compared to their non-imprinted ones. Based on this fluorescence quenching model, the selective and sensitive MIP-QDs biosensors for the direct fluorescence detection of lysozyme were developed.
Characterization
Figure 3 depicts the excitation spectra and emission spectra of the as-prepared l-cysteine-capped Mn-doped ZnS QDs and MIP-QDs nanosphere. As shown in Fig. 3, the maximum emission of both the l-cysteine-capped ZnS QDs and MIP-QDs is located at 586 nm. The excitation wavelength of l-cysteine-capped ZnS QDs is centered at 307 nm; however, the maximum excitation wavelength of MIP-QDs blue shifts to 302 nm. The MIP-QDs exhibit novel optical properties, including wide excitation, as well as narrow and symmetric emission spectra. Figure 4 shows the ultraviolet absorption spectrum of MIP-QDs and lysozyme. The maximum UV absorption of lysozyme is located at 282 nm and the absorption of the MIP-QDs is centered at 305 nm. There were no overlap bands between the absorbance of lysozyme and the excitation of the QDs; the excitation energy is unlikely to transfer from QDs to lysozyme. The fluorescence quenching mechanism may be caused by the transfer of the electron of the conductive bands of the QDs to the UV band of lysozyme molecules, which hinders the electronic transition of the d electrons of the Mn2+ to the excited state, resulting in the orange fluorescence emission quenching of the Mn2+-ZnS QDs [43].
The shape and size distribution of the obtained l-cysteine-capped Mn-doped ZnS QDs and MIP-QDs were characterized by high-resolution transmission electron microscopy. As shown in Fig. 5a, the l-cysteine-capped ZnS QDs are close to spherical morphology, aggregated on a rough surface with diameters ranging from 3 to 4 nm. Figure 5b displays that the MIP-QDs particles sizes are larger and the average diameter is about 6 nm, indicating that the MIP successfully coats on the l-cysteine-capped Mn-doped ZnS QDs through surface molecular imprinting process. The results of TEM image show that the fluorescent sensing receptor is successfully prepared by embedded the QDs into the molecularly imprinted polymer matrix.
Response time
In order to use MIP-QDs for fluorescence detection of lysozyme, the time response of the biosensor was first investigated. Figure 6 shows the kinetics of MIP-QDs composites. As shown in Fig. 6, both the MIP- and NIP-QDs had a rapidly absorb kinetics for lysozyme. They reached the maximum adsorption capacity in a short time. There is a significant initial decrease in fluorescence intensity after the addition of lysozyme to the MIP-QDs composites. After incubation for 21 min, the MIP-QDs obtained stable fluorescence intensity, which indicated that the MIP-QDs reached the adsorption equilibrium for adsorption and desorption. As a control, the adsorption equilibrium of NIP-QDs emerged at eight minutes, and the change in fluorescence intensity of NIP-QDs was lower than that of MIP ones. It indicates that no binding sites were formed in the NIP-QDs; thus the fluorescence quenching of NIP processed slightly. However, in MIP-QDs, more lysozyme molecule specific binding with the recognition sites makes obvious fluorescence quenching.
Effect of pH
The effect of pH on the fluorescence intensity of MIP-and NIP-QDs were investigated in the presence and absence of lysozyme. As shown in Fig. 7, the fluorescence intensity of both MIP- and NIP-QDs was obviously decreased in the range of pH 6.0–11.0 before the addition of lysozyme. After adding lysozyme, both the MIP- and NIP-QDs were obviously quenched, although the change in fluorescence intensity for the MIP-QDs composites is larger than that of the NIP-QDs ones. As is known, lysozyme is a amphoteric protein, the pI of lysozyme is 11.2. Thus, when the pH < 11.2, the lysozyme molecules were positively charged and l-cysteine-modified QDs were negatively charged. Therefore, the lysozyme molecule was more easily bonded with MIP-QDs composites in the weak acid environment. The experiment results showed that the maximum change of the fluorescence intensity of MIP-QDs composites occurred at pH 6.0 when lysozyme were added (Table 1, ΔFMIP). The best recognition ability of the MIP-QDs composites was also achieved at pH 6.0 (IF1), indicating that the best lysozyme binding generated at pH 6.0. The results above suggest that pH 6.0 was the optimal pH value for further experiments.
Fluorescence sensing of lysozyme by MIP-QDs composites
Orange fluorescence is generated by the photoexcitation of the host nanocrystal (ZnS) recombined by the lower-lying states of Mn2+ ion (4T1–6A1). When the target protein are trapped by the molecularly imprinted polymer shell of MIP-QDs composites, the electron of QDs transfers to the lysozyme resulting in the fluorescence quenching of the MIP-QDs composites. Figure 8 depicts the results of fluorescence sensing of lysozyme by using MIP-QDs composites. As shown in Fig. 8, both the MIP- and NIP-coated QDs composites were quenched by adding lysozyme. However, the MIP-QDs composites showed more than two times of fluorescence quenching efficiency on lysozyme (F 0/F − 1) than that of NIP ones. The noticeable quenching efficiency of the MIP-QDs composites was mainly achieved by the specific binding of imprinted cavities with lysozyme in the aqueous media. These analysis results showed that the fluorescence quenching depends on the adsorptive of lysozyme. In the case of the MIP-coated QDs, the fluorescence quenching efficiency is proportional to the concentration of lysozyme. Based on the fluorescence quenching system, a facile and direct fluorescent quantification method for detecting lysozyme has been successfully developed. The fluorescence quenching in this system follows the Stern–Volmer equation:
where F 0 is the initial fluorescence intensity in the absence of the lysozyme, F is the fluorescence intensity in the presence of the quencher, K sv is the quenching constant of the quencher, and Q is the concentration of the quencher.
Under the optimal conditions, the calibration plot of F 0/F − 1 versus Q showed a good linear relationship in the range from 0.1 to 2.0 μM with a correlation of 0.9863. The linear regression equation is F 0/F − 1 = 0.6251 Q + 0.0709. The 3σ limit of detection (IUPAC criteria, 3σ/S) was 25.2 nM, where σ is the standard deviation of the blank signal, and S is the slope of the calibration plot. The precision for five replicate detections of 1.0 μM of lysozyme was 0.94 % (relative standard deviation). These results suggest that the MIP-QDs composites were successfully applied to selective, sensitive, and direct fluorescence detection of lysozyme without any preconcentration and expensive instruments.
Specificity
During the imprinting process, the MIP shell of QDs formed a stable recognition sites between the monomers and the template protein through strong molecular interactions. This interaction makes good recognition selectivity of the MIP-QDs composites, which are supported by the data shown in Fig. 9. As shown in Fig. 9, three kinds of proteins, BHB, OVA, and BSA, were chosen as references to evaluate the selectivity of the MIP-QDs composites. Compared to the reference protein, lysozyme exhibits a significant quenching effect on the fluorescence of the MIP-QDs. The target analyte has more chance to access the MIP-QDs composites because of the formed imprinting cavity by the removal of the template, so the MIP-QDs show larger K SV than the NIP-QDs. The K sv,MIP of lysozyme is much larger than the reference proteins due to the selectively rebinding of MIP-QDs. The imprinting factor (IF) was used to evaluate the selectivity of the materials:
The imprinting factor for lysozyme was 4.65, which was higher than that of BSA, OVA, and BHB (1.42, 1.29, and 1.28, respectively). These results show that the MIP-QDs composites display selectivity to the template protein (lysozyme), and have the ability to discriminate the interference proteins and target protein on the basis of different molecular shape, size, and structure.
Real sample analysis
Application in real samples is an important facet of the fluorescence sensor. As shown in Table 2, the concentrations of lysozyme in the diluted chicken egg whites samples were 1.15 and 1.09 μM, respectively. A recovery test was also carried out by standard addition method in order to evaluate the accuracy of the developed method. The recovery of lysozyme spiked samples ranged from 100.15 to 103.21 %, and the relative standard deviations were between 1.13 and 1.85 % (n = 3).
A recycling test was also implemented to evaluate the recyclability of the prepared MIP-QDs composites. As shown in Fig. 10, in the first three rebind/elution cycles, the fluorescence intensity of MIP-QDs composites was slightly changed. After six regeneration cycles, the fluorescence intensity of the MIP-QDs composites decreased 15.5 %. It might be a partial destruction of their structure by excessive washing. Additionally, the MIP-QDs composites could be stored under dry and dark conditions for a long period. The fluorescence intensity of MIP-capped QDs did not change significantly change after 30 days. These results suggested that the MIP-QDs composites were successfully fabricated, and could detect lysozyme with good regeneration and stability.
The analytical characteristics of the proposed methods were compared with previously reported techniques for lysozyme detection (Table 3). Each method has its advantages and limitations, the most of reported techniques need the preconcentration and purification pretreatment. The MIP-QDs-based fluorescence sensor shows rapid respond time, low cost, better or comparable limit of detection, and good selectivity compared to the other reported techniques. The results above suggest that the MIP-QDs has the potential as a lysozyme sensor since it could be selective, accurate and does not need further pretreatment for detecting of trace lysozyme in biologic samples.
Conclusion
In this paper, a new type of MIP-coated l-cysteine-capped Mn-doped ZnS QDs composites was successfully prepared as a fluorescent sensor for the specific recognition and determination of lysozyme. The prepared fluorescent receptor can selectively capture the target protein (lysozyme), and then the electron transfer between the QDs and the target protein is able to result in the fluorescence quenching of the QDs. Based on the fluorescence quenching mechanism, the fluorescent receptors can be successfully used to recognize the target protein specifically. Furthermore, a series of rebinding experiments show that the MIP-QDs-based fluorescence sensor has good selectivity and sensitivity for lysozyme over references proteins. The MIP-QDs composites also display good practicability, stability, and reusability. By combining the selectivity of MIPs with the excellent optics properties of QDs, the MIP-QDs composites can successfully detect of trace lysozyme in biological fluids without any expensive instruments or time-consuming analysis processes involved.
References
Harrison JF, Lunt GS, Scott P, Blainey JD (1968) Urinary lysozyme, ribonuclease, and low-molecular-weight protein in renal disease. Lancet 1:371–375
Cheng AKH, Ge B, Yu HZ (2007) Aptamer-based biosensors for label-free voltammetric detection of lysozyme. Anal Chem 79:5158–5164
Levinson SS, Elin RJ, Yam L (2002) Light chain proteinuria and lysozymuria in a patient with acute monocytic leukemia. Clin Chem 48:1131–1132
Carstens C, Deckwart M, Webber-Witt M, Schäfer V, Eichhorn L, Brockow K, Fischer M, Christmann M, Paschke-Kratzin A (2014) Evaluation of the efficiency of enological procedures on lysozyme depletion in wine by an indirect ELISA method. J Agric Food Chem 62:6247–6253
Viadl ML, Gautron J, Nys Y (2005) Development of an ELISA for quantifying lysozyme in hen egg white. J Agric Food Chem 53:2379–2385
Cai Z, Chen G, Huang X, Ma M (2011) Determination of lysozyme at the nanogram level in chicken egg white using Resonance Rayleigh-scattering method with Cd-doped ZnSe quantum dots as probe. Sensor Actuat B Chem 157:368–373
Lu X, Luo Z, Liu C, Zhao S (2008) Resonance Rayleigh scattering for detection of proteins in HPLC. J Sep Sci 31:2988–2993
Sener G, Ozgur E, Yilmaz E, Uzun L, Say R, Denizli A (2010) Quartz crystal microbalance based nanosensor for lysozyme detection with lysozyme imprinted nanoparticles. Biosens Bioelectron 26:815–821
Mihai I, Vezeanu A, Polonschii C, Albu C, Radu G-L, Vasilescu A (2015) Label-free detection of lysozyme in wines using an aptamer based biosensor and SPR detection. Sensors Actuat B Chem 206:198–204
Chen YM, Yu CJ, Cheng TL, Tseng WL (2008) Colorimetric detection of lysozyme based on electrostatic interaction with human serum albumin-modified gold nanoparticles. Langmuir 24:3654–3660
Liu S, Na W, Pang S, Shi F, Su X (2014) A label-free fluorescence detection strategy for lysozyme assay using CuInS(2) quantum dots. Analyst 139:3048–3054
Li S, Gao Z, Shao N (2014) Non-covalent conjugation of CdTe QDs with lysozyme binding DNA for fluorescent sensing of lysozyme in complex biological sample. Talanta 129:86–92
Kondekova M, Maier V, Ginterova P, Marak J, Sevcik J (2014) Analysis of lysozyme in cheese samples by on-line combination of capillary zone electrophoresis and mass spectrometry. Food Chem 153:398–440
Liao YH, Brown MB, Martin GP (2001) Turbidimetric and HPLC assays for the determination of formulated lysozyme activity. J Pharmacy and Pharmacology 53:549–554
Rodriguez MC, Rivas GA (2009) Label-free electrochemical aptasensor for the detection of lysozyme. Talanta 78:212–216
Rohrbach F, Karadeniz H, Erdem A, Famulok M, Mayer G (2012) Label-free impedimetric aptasensor for lysozyme detection based on carbon nanotube-modified screen-printed electrodes. Anal Biochem 421:454–459
Vasilescu A, Gaspar S, Mihai I, Tache A, Litescu SC (2013) Development of a label-free aptasensor for monitoring the self-association of lysozyme. Analyst 138:3530–3537
Wang J, Liu B (2009) Fluorescence resonance energy transfer between an anionic conjugated polymer and a dye-labeled lysozyme aptamer for specific lysozyme detection. Chem Commun 17:2284–2286. doi:10.1039/B820001G
Pellegrino L, Tirelli A (2000) A sensitive HPLC method to detect hen’s egg white lysozyme in milk and dairy products. Int Dairy J 10:435–442
Kondeková M, Maierb V, Ginterováb P, Marák J (2014) Analysis of lysozyme in cheese samples by on-line combination of capillary zone electrophoresis and mass spectrometry. Food Chem 153:398–404
Erdema A, Eksina E, Mutia M (2014) Chitosan–graphene oxide based aptasensor for the impedimetric detection of lysozyme. Colloid Surfaces B 115:205–211
Haupt K (2003) Peer reviewed: molecularly imprinted polymers: the next generation. Anal Chem 75:376A–383A
Gn Wulff (2002) Enzyme-like catalysis by molecularly imprinted polymers. Chem Rev 102:1–28
Haupt K, Mosbac K (2000) Molecularly imprinted polymers and their use in biomimetic sensors. Chem Rev 100:2495–2504
Lin CI, Joseph AK, Chang CK, Lee YD (2004) Molecularly imprinted polymeric film on semiconductor nanoparticles: analyte detection by quantum dot photoluminescence. J Chromatogr A 1027:259–262
Bossi A, Bonini F, Turner AP, Piletsky SA (2007) Molecularly imprinted polymers for the recognition of proteins: the state of the art. Biosens Bioelectron 22:1131–1137
Janiak DS, Kofinas P (2007) Molecular imprinting of peptides and proteins in aqueous media. Anal Bioanal Chem 389:399–404
Ge Y, Turner AP (2008) Too large to fit? Recent developments in macromolecular imprinting, Trends Biotechnol 26:218–224
Díaz-Díaz G, Antuña-Jiménez D, Carmen Blanco-López M, Jesús Lobo-Castañón M, Jand Miranda-Ordieres A, Tuñón-Blanco P (2012) New materials for analytical biomimetic assays based on affinity and catalytic receptors prepared by molecular imprinting. Trend Anal Chem 33:68–80
Ding Z, Annie Bligh SW, Tao L et al (2015) Molecularly imprinted polymer based on MWCNT-QDs as fluorescent biomimetic sensor for specific recognition of target protein. Mat Sci Eng C 48:469–479
Alivisatos AP (1996) Semiconductor clusters, nanocrystals, and quantum dots. Science 271:933
Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T (2008) Quantum dots versus organic dyes as fluorescent labels. Nat Methods 5:763–775
Algar WR, Massey M, Krull UJ (2009) The application of quantum dots, gold nanoparticles and molecular switches to optical nucleic-acid diagnostics. Trends Anal Chem 28:292–306
Basabe-Desmonts L, Reinhoudt DN, Crego-Calama M (2007) Design of fluorescent materials for chemical sensing. Chem Soc Rev 36:993–1017
Wei X, Zhou Z, Hao T, Li H, Yan Y (2015) Molecularly imprinted polymer nanospheres based on Mn-doped ZnS QDs via precipitation polymerization for room-temperature phosphorescence probing of 2,6-dichlorophenol. RSC Adv 5:19799–19806
Zhang W, He XW, Li WY, Zhang YK (2012) Thermo-sensitive imprinted polymer coating CdTe quantum dots for target protein specific recognition. Chem Commun 48:1757–1759
You CC, Miranda OR, Gider B, Ghosh PS, Kim I, Erdogan B, Krovi SA, Bunz UHF, Rotello VM (2007) Detection and identification of proteins using nanoparticle-fluorescent polymer ‘chemical nose’ sensors. Nat Nanotechnol 2:318–323
Koneswaran M, Narayanaswamy R (2009) l-cysteine-capped ZnS quantum dots based fluorescence sensor for Cu2+ ion. Sensor Actuat B Chem 139:104–109
Bian W, Wang F, Wei Y, Wang L, Liu Q, Dong W, Shuang S, Choi MMF (2015) Doped zinc sulfide quantum dots based phosphorescence turn-off/on probe for detecting histidine in biological fluid. Anal Chim Acta 856:82–89
Tan L, Huang C, Peng R, Tang Y, Li W (2014) Development of hybrid organic-inorganic surface imprinted Mn-doped ZnS QDs and their application as a sensing material for target proteins. Biosens Bioelectron 61:506–511
Wang HF, He Y, Ji TR, Yan XP (2009) Surface molecular imprinting on Mn-doped ZnS quantum dots for room-temperature phosphorescence optosensing of pentachlorophenol in water. Anal Chem 81:1615–1621
Gattás-Asfura KM, Leblanc RM (2003) Peptide-coated CdS quantum dots for the optical detection of copper(ii) and silver(i). Chem Commun 21:2684–2685
Tan L, Kang C, Xu S, Tang Y (2013) Selective room temperature phosphorescence sensing of target protein using Mn-doped ZnS QDs-embedded molecularly imprinted polymer. Biosens Bioelectron 48:216–223
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, X., Yang, S., Sun, L. et al. Surface-imprinted polymer coating l-cysteine-capped ZnS quantum dots for target protein specific recognition. J Mater Sci 51, 6075–6085 (2016). https://doi.org/10.1007/s10853-016-9914-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10853-016-9914-7