Abstract
The corrosion inhibition behaviour of four selected amino acid compounds, namely l-cysteine, l-histidine, l-tryptophan and l-serine on mild steel surface in deaerated 1 M HCl solution were studied electrochemically by Tafel polarization and electrochemical impedance spectroscopy methods and computationally by the quantum chemical calculation and molecular dynamics simulation. Electrochemical results show that these amino acid compounds inhibit the corrosion of mild steel in 1 M HCl solution significantly. The order of inhibition efficiency of these inhibitors follows the sequence: l-tryptophan > l-histidine > l-cysteine > l-serine. The quantum chemical calculations were performed to characterize the electronic parameters which are associated with inhibition efficiency. The molecular dynamics simulations were applied to find the equilibrium adsorption configurations and calculate the interaction energy between inhibitors and iron surface. Results obtained from Tafel and impedance methods are in good agreement. The electrochemical experimental results are supported by the theoretical data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The use of corrosion inhibitors is one of the most effective and economic methods to protect metal surfaces against corrosion in aggressive media [1–3]. Most of the efficient pickling inhibitors are organic compounds that contain mainly nitrogen, oxygen, sulphur phosphorus, and multiple bonds or aromatic rings in their structures. The number of lone pairs of electrons and loosely bound π-electrons in these functional groups are the key structural features that determine the inhibition efficiency [4, 5]. However, the toxicity of these compounds is also documented [6]. Environmental restrictions imposed on heavy-metal and phosphonate-based corrosion inhibitors, guided scientific researches towards studying non-toxic and non-phosphor environmental-friendly corrosion inhibitors [7–9].
Amino acids are non-toxic, relatively cheap, and easy to produce with purities greater than 99%. They are considered to be more promising green inhibitors [10]. Amino acid compounds were reported to show corrosion resistant behaviour on copper, mild steel and aluminium alloy [11–17]. In the previous literature, most attention was paid to the mechanism of adsorption and also to the relationship between the structure of inhibitors, their adsorption on the metal surface, and their inhibition efficiencies on the macroscopic level. With the in-depth study of organic corrosion inhibitors, it has been found that the inhibition efficiency depends on some physicochemical properties of inhibitors, such as functional groups, steric effects, electronic density at the donor atom, π orbital character and corrosive environment [18–21]. Therefore, theoretical prediction of the efficiency of corrosion inhibitors has become very popular [22–25]. Quantum chemical calculation and molecular dynamic simulation have become an effective way to study the correlation between molecular structure and inhibition properties on the microcosmic level. Quantum chemical calculations have proved to be a very powerful tool for studying corrosion inhibition mechanism and helped to design the novel high efficiency inhibitors by the quantitative structure–activity relationship (QSAR) method [26–28]. Molecular dynamic simulation is used as a beneficial supplement of quantum chemical method, which is applied to study the interaction between inhibitors and metal surface. This provides a convenient way to study the complex system, which includes one inhibitor molecule, hundreds of solvent molecules and the metal surface. It also solves the problem that quantum chemical method is usually used for small systems containing between 10 and 100 atoms [29–32]. Due to expensive and time-consuming of experimental methods, it can be inferred that these two theoretical methods will play more and more important role in studying corrosion inhibitors with the continuous development of computer hardware and software technology.
The purpose of this paper is to evaluate l-cysteine (l-Cys), l-histidine (l-His), l-tryptophan (l-Try) and l-serine (l-Ser) as corrosion inhibitor for mild steel in 1 M deaerated HCl solution by potentiodynamic polarization and electrochemical impedance spectroscopy (EIS) techniques. Another objective in this work is to discuss the relationship between quantum chemical parameters, such as the highest occupied molecular orbital (E HOMO), lowest unoccupied molecular orbital (E LUMO), the energy gap (ΔE, ΔE = E LUMO − E HOMO), dipole moment (μ) and the experimental inhibition efficiencies. Furthermore, the adsorption characteristics of these amino acid compounds on the iron surface using molecular dynamics simulations were also performed.
Experimental
Materials
l-Cys, l-His, l-Try and l-Ser were purchased from Acros Organics. The chemical structures, names and abbreviations of amino acid compounds were given in Table 1. The 1 M HCl solution was made with AR grade HCl and double distilled water. The composition of mild steel used in the experiments was shown in Table 2. The working electrode (WE) was in the form of a rod machined into a cylindrical form embedded in epoxy resin leaving an open surface area of 1 cm2. The exposed surface was abraded with emery paper (from grade #600 to #2000), rinsed by double distilled water, degreased with acetone, dried at room temperature before electrochemical use.
Electrochemical measurements
Electrochemical measurements were performed in a conventional three-electrode cell of capacity 500 mL. The WE, a reference saturated calomel electrode (SCE) and platinum counter electrode were placed inside a three-electrode cell. The HCl solution was degreased with ultra-pure nitrogen bubbling for 30 min to avoid any reactions with dissolved oxygen. Measurements were performed at room temperature, 25 °C with PARSTAT 2273 (Princeton Applied Research Company). The WE was immersed in 1 M HCl solution for 30 min until a steady-state open circuit potential (OCP) was obtained. Tafel polarization scans were carried out by changing the electrode potential automatically from −250 to +250 mV versus OCP with a scan rate of 0.166 mV/s. The data recordings and the calculation of the electrochemical parameters were performed using PowerSuite software. EIS were carried out at OCP over a frequency range of 100 kHz–10 MHz. The sinusoidal potential perturbation was 5 mV in amplitude. The data were collected using PowerSuite software and interpreted with Zsimpwin software. Each test was run in triplicate to verify the reproducibility and the average values reported. All potentials were referred to the SCE.
Quantum chemistry calculations
The amino acid compounds have been fully optimized at the using B3LYP Density Functional Theory (DFT) with 6-311G(d,p) basis set with Gaussian 03W program. This basis set provided accurate geometry and electronic properties for a wide range of organic compounds. The following quantum chemical parameters, which indicate structural characteristics of these inhibitors, were considered: E HOMO, E LUMO, ΔE and μ.
DFT has also been found to be successful in providing insights into the chemical reactivity and selectivity, in terms of global parameters such as electronegativity (χ), hardness (η) and local ones such as the Fukui function \( \left( {f(\overrightarrow {r} )} \right) \). The local reactivity of the molecules was analyzed through an evaluation of the Fukui indices. These are a measurement of the chemical reactivity, as well as an indicative of the reactive regions and the nucleophilic and electrophilic behaviour of the molecule. The Fukui function \( f(\overrightarrow {r} ) \) is defined as the derivative of the electronic density \( \rho (\overrightarrow {r} ) \) with respect to the number of electrons N at a constant external potential \( v(\overrightarrow {r} ) \) [33]:
If the effects of relaxation associated with the addition or removal of electronic charges are not considered, then
where \( \rho_{\text{LUMO}} (\overrightarrow {r} ) \) is the density of the first unoccupied molecular orbital and \( \rho_{\text{HOMO}} (\overrightarrow {r} ) \) is the density of the highest occupied molecular orbital.
The condensed Fukui function calculations are based on the finite difference approximations and partitioning of the electron density \( \rho (\overrightarrow {r} ) \) between atoms in a molecular system.
where q k is the gross charge of atom k in the molecule and N is the number of electrons. The condensed Fukui function is local reactivity descriptor and can be used only for comparing reactive atomic centres within the same molecule [34].
The Fukui function calculations were performed with DMol3 version 4.3 available in Material Studio software (Accelrys, San Diego, CA), using a PBE (Perdew, Burke and Enzerhof) functional and a double-numeric quality basis set with polarization functions (DND). Dmol3 use a Mulliken population analysis.
Molecular dynamics simulations
The molecular dynamics simulations were performed using the software, Material Studio 4.3. As to the three kinds of Fe surface (1 1 0, 1 0 0, 1 1 1), Fe (1 1 1) and Fe (1 0 0) surfaces have relatively open structures whereas Fe (1 1 0) is a densely packed surface and has the most stabilization. Therefore, the surface of Fe (1 1 0) was chosen to simulate the adsorption process. The molecular dynamics simulation of the interaction between the inhibitor molecule dissolved in H2O and the Fe (1 1 0) surface was carried out in a simulation box (32.3 Å × 32.3 Å × 30.9 Å) with periodic boundary conditions in order to simulate a representative part of an interface devoid of any arbitrary boundary effects. The inhibitor molecule was energy optimized, Fe (1 1 0) surface and water layers was constructed using the amorphous cell module. The molecular dynamics simulation was performed under 25 °C, NVT ensemble, with a time step of 0.1 fs and simulation time of 50 ps. Details of simulation process can be referred to some previous literature [35, 36].
The interaction energy E Fe-inhibitor of iron surface with the inhibitor was calculated according to the following equation:
where E complex is the total energy of the Fe crystal together with the adsorbed inhibitor molecule, E Fe and E Inhibitor is the total energy of the iron crystal and free inhibitor molecule, respectively. And the binding energy is the negative value of the interaction energy, E binding = −E Fe-inhibitor.
Results and discussions
Tafel polarization studies
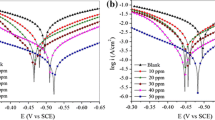
Figure 1 show the cathodic and anodic polarization curves of the mild steel immersed in 1 M HCl at 25 °C in absence and presence of various concentrations of l-Cys, l-His, l-Try and l-Ser, respectively. Electrochemical parameters such as corrosion potentials (E corr), anodic and cathodic Tafel slopes (β a and β c), corrosion current densities (j corr), percentage inhibition efficiencies (IE p, %), and degrees of surface coverage (θ), for various concentrations of these amino acid compounds were given in Table 3. The percentage inhibition efficiency and the degree of surface coverage were calculated using the following formulas:
where j 0 and j 1 are the corrosion current densities for without and with inhibitors, respectively.
From the results in Table 3, it can be easily seen that these four amino acid compounds under investigation conditions are efficient inhibitors. It is also found that j corr values decreased considerably in presence of inhibitors and decreased with increasing inhibitor concentration except for l-Ser with the concentration of 1.0 × 10−4 mol L−1. In this case, the stimulating effect on corrosion process was stronger than the suppression effect [37]. By comparing the j corr in Table 3, the inhibition efficiency of these four amino acid compounds decreases in the order: l-Try > l-His > l-Cys > l-Ser under the same conditions. l-Try shows the maximum inhibition efficiency among all the other investigated compounds at the concentration of 1.0 × 10−2 mol L−1.
As seen from Fig. 1, the anodic and cathodic polarization curves were inhibited after the addition of four amino acid compounds. This means that the addition of the investigated inhibitors not only reduce anodic dissolution but also retard the hydrogen evolution reaction. The unchanged anodic and cathodic Tafel slopes indicated that the four compounds act as adsorptive inhibitors, suppressing both anodic and cathodic reaction by adsorbing on mild steel surface and blocking the active sites [21]. Furthermore, E corr shifted slightly (<30 mV) in all cases. Based on these experimental results, the four amino acid compounds can be regarded as mixed-type inhibitors [38]. However, the changing trends of E corr for four inhibitors are not the same. No definite trend was observed in the shift of E corr values, in the presence of various concentrations of l-Cys and l-Ser. But for l-His and l-Try, the E corr shifted slightly in the positive direction with increase in l-His concentration, while E corr shifted slightly in the negative direction with increase in l-Try concentration. Therefore, it can be inferred that l-Try acts more as a cathodic than anodic inhibitor, and in the contrary, l-His acts more as an anodic inhibitor than cathodic inhibitor.
Electrochemical impedance spectroscopy studies
Nyquist plots for mild steel in 1 M HCl in the absence and presence of the four amino acid compounds in the concentration range of 1.0 × 10−4–1.0 × 10−2 mol L−1 at 25 °C were shown in Fig. 2. It is apparent from this figure that all Nyquist plots are similar in shape and consist of a single capacitive loop, which mean only one charge transfer reaction during metal dissolution process [39]. The simple equivalent circuit model for this system is shown in Fig. 3. The circuit comprises a solution resistance R s, in series with the parallel combination of the charge transfer resistance R ct and a double layer capacitance (C dl). C dl values were calculated from the frequency at which the imaginary component of impedance was maximum using the following equation:
where f max is the frequency at which the imaginary component of impedance is maximum.
The impedance diagram does not give a perfect semicircle, which has been attributed to frequency dispersion of interfacial impedance. The percentage Inhibition efficiency (IE E, %) can be calculated as following equation:
where R ct and R 0ct are the polarization resistance of mild steel with and without inhibitor molecules. Inhibition efficiencies and the impedance parameters derived from Nyquist plots are given in Table 4. The charge transfer resistance R ct values increased and the capacitance values C dl decreased with increasing concentration of four amino acid compounds. The increase in R ct is attributed to the formation of protective film on the metal/solution interface. The decrease in C dl can be interpreted as a decrease in local dielectric constant and/or an increase in the thickness of electronic double layer, which result from the adsorption of the amino acid molecules on the metal surface [40]. It can be seen that the IE E values obtained from the EIS are in complete agreement with those obtained from Tafel polarization measurements.
Quantum chemistry calculation
To investigate the relationship between the molecular electronic structure of inhibitors and their inhibition efficiencies, quantum chemical calculations were performed. The optimized structures of the four amino acid compounds were shown in Fig. 4. Quantum chemical parameters such as E HOMO, E LUMO, ΔE and μ were given in Table 5. Additionally, the orbital density distributions of E HOMO and E LUMO for amino acid compounds were shown in Fig. 5.
According to the frontier orbital theory, E HOMO is often associated with the electron donating ability of a molecule. High values of E HOMO indicate a tendency of the molecule to donate electrons to appropriate acceptor molecules with low-energy, empty electron orbital. Similarly, E LUMO represents the ability of the molecule to accept electrons. The lower the values of E LUMO, the more probable it is that the molecule would accept electrons [41]. ΔE, the difference of E LUMO and E HOMO, is another important factor that should be considered. It has been reported that low values of ΔE will provide good inhibition efficiency, because the energy for removing an electron from the last occupied orbital will be low [42].
In Table 5, the values of E HOMO of four amino acid compounds increased in the following order: l-Try > l-His > l-Cys > l-Ser. The values of ΔE decreased in the following order: l-Try < l-His < l-Cys < l-Ser. The sequences of E HOMO and ΔE support the results obtained from the electrochemical measurements. The inhibition efficiency increased with increase in the E HOMO and decrease in ΔE. l-Try has the highest E HOMO and the lowest ΔE among these four amino acid compounds. Therefore, l-Try has the strongest interaction with iron surface and the best inhibition effect. However, it can be seen in Table 5, there are no obvious linear correlation between E LUMO, μ and inhibition efficiency. Similar observations have been reported in some previous literature [43, 44].
The number of electrons transferred (ΔN) from the inhibitor molecule to the metallic atom was also calculated using the following equation:
where χFe and χinh represent the absolute electronegativity of iron and the inhibitor molecule, respectively; ηCu and ηinh represent the absolute hardness of iron and the inhibitor molecule. These quantities are associated with electron affinity (A) and ionization potential (I) which are useful in their ability to help chemical behaviour [45].
I and A are related in turn to EHOMO and ELUMO as following equations:
Values of χ and η were calculated by using the values of I and A obtained from quantum chemical calculations. In order to calculate ΔN, a theoretical value for the electronegativity of bulk iron was used χ Fe ≈ 7 eV, and a global hardness of η Fe ≈ 0, by assuming that for a metallic bulk I = A, because they are softer than the neutral metallic atoms. The values of ΔN for four amino acid compounds were also listed in Table 5.
According to Lukovits, if ΔN < 3.6, the inhibition efficiency increased with increasing electron donating ability at the metal surface [46]. It can be inferred from the calculation results that inhibitors investigated in this study were donors of electrons, and the iron surface was the acceptor. Order of ΔN is as follow: l-Try > l-His > l-Cys > l-Ser, which is in accordance with the change trends of E HOMO and inhibition efficiency. The highest inhibition efficiency of l-Try can be attributed to the strongest coordinate bonds formed between the lone electron pairs of heterocyclic atom/π electrons of indole ring and the vacant d-orbitals of the metal surface.
The local reactivity was analyzed by means of the Fukui Indices. Fukui functions are a qualitative way of measuring and displaying the reactivity of a molecule [47]. The molecule sites with large values of f +k are the sites where the molecule will receive charge, when attacked by a nucleophilic reagents, and the molecule sites with large values of f −k are the preferred sites where the molecule will donate charge when attacked by an electrophilic reagent [48]. The values of Fukui functions for a nucleophilic and electrophilic attack were shown in Table 6 for the four amino acid compounds, in which only the largest values were presented. For nucleophilic attack, the most reactive site of l-Cys is on the C4, O5 and O7 atom, l-His is on the C8, O9 and O10 atom, l-Ser is on O1, C2 and O3 atom and l-Try is on C9, C12 and C15 atom. For electrophilic attack, the most reactive site of l-Cys is on the S1 atom, l-His is on N11 atom, l-Ser is on O1, O2 and N7 atoms and l-Try is on C6, C9 and C12 atom of indole ring. The distributions of HOMO and LUMO on each inhibitor are basically consistent with the atoms that exhibit greatest values of f −k and f +k , which demonstrate that these active atoms will play a significant role in the interaction with the iron surface.
Molecular dynamics simulation
The further information on the interaction between the inhibitor and the iron surface can be provided by the molecular dynamics simulations. Figure 6 shows the different adsorption equilibrium configurations of four amino acid compounds. Through the simulation process, each configuration changed greatly. By comparing the equilibrium configurations of four amino acid compounds adsorbed on Fe (1 1 0) surface, it can be conclude that l-Cys and l-Ser can be adsorbed on the iron surface through the heterocyclic atoms in their molecules, while l-His and l-Try can be adsorbed on the iron surface through not only the heterocyclic atoms but also the imidazole ring and the indole ring, respectively. Both interactions can make it possible for amino acid compounds to form coordinated bonds with iron surface, and form the compact film preventing the corrosive media to move to the metal surface.
The values of the interaction energy and the binding energy of the four amino acid compounds on iron surface were listed in Table 7. In all cases, the binding energy has a positive value. Generally, the binding energy increases, the more stable the inhibitor adsorbs on the metal surface, and the high inhibition efficiency. Based on the calculations, l-Ser presented the lowest binding energy value of 256 kJ mol−1, followed by l-Cys with 275 kJ mol−1 and l-His with 289 kJ mol−1. Finally, l-Try displayed the highest value of 402 kJ mol−1. l-His has the highest binding energy during the simulation process. A number of lone pair of electrons on the O and N atoms as well as the π-electron clouds on the indole ring can provide electrons to the unfilled 3d-orbitals of iron surface to form protective layer. High values of binding energy obtained with l-Try molecule explain its highest inhibition efficiency from the electrochemical measurements.
Summary and Conclusions
-
1.
Tafel polarization and EIS technique were used to characterize the corrosion inhibition of mild steel in 1MHCl by the four amino acid compounds. The inhibition efficiency increased as follows, l-Ser < l-Cys < l-His < l-Try under the same experimental condition.
-
2.
Calculations of the E HOMO, ΔE and ΔN illuminate that the order of the inhibition efficiency of these inhibitors follows the sequence: l-Try > l-His > l-Cys > l-Ser, and this result consists well with the experimental results.
-
3.
The molecular dynamics simulation results show that these four amino acid compounds can absorb on the iron surface through the heteroatoms and heterocyclic ring in their structures.
References
Fouda AS, Mostafa HA, El-Abbasy HM (2010) J Appl Electrochem 40:163
Amin MA, Abd El Rehim SS, El-Naggar MM, Abdel-Fatah HTM (2010) J Mater Sci 44:6258. doi:10.1007/s10853-009-3856-2
Abelev E, Sellberg J, Ramanarayanan TA, Bernasek SL (2010) J Mater Sci 44:6167. doi:10.1007/s10853-009-3854-4
Shukla SK, Singh AK, Ahamad I, Quraishi MA (2009) Mater Lett 63:819
Shukla SK, Quraishi MA (2009) J Appl Electrochem 39:1517
Sinko J (2001) Prog Org Coat 42:267
Bothi Raja P, Sethuraman MG (2008) Mater Lett 62:1602
Ketsetzi A, Stathoulopoulou A, Demadis KD (2008) Desalination 223:487
Umoren SA, Obot IB, Obi-Egbedi NO (2009) J Mater Sci 44:274. doi:10.1007/s10853-008-3045-8
Lyon S (2004) Nature 427:406
Fu JJ, Li SN, Cao LH, Wang Y, Yan LH, Lu LD (2010) J Mater Sci 45:979. doi:10.1007/s10853-009-4028-0
Saifi H, Bernard MC, Joiret S, Rahmouni K, Takenouti H, Talhi B (2010) Mater Chem Phys 120:661
Zhang DQ, Gao LX, Zhou GD (2005) J Appl Electrochem 35:1081
Zhang DQ, Cai QR, Gao LX, Lee KY (2008) Corros Sci 12:3615
Zerfaoui M, Oudda H, Hammouti B, Kertit S, Benkaddour M (2004) Prog Org Coat 51:134
Ashassi-Sorkhabi H, Ghasemi Z, Seifzadeh D (2005) Appl Surf Sci 249:408
Diab ASM, Abdel-Azzem M, Mandour H (2005) Bull Electrochem 21:97
Issa RM, Awad MK, Atlam FM (2008) Appl Surf Sci 255:2433
Avci G (2008) Colloids Surf A 317:730
Granero MFL, Matai PHLS, Aoki IV, Guedes IC (2009) J Appl Electrochem 39:1199
Behpour M, Ghoreishi SM, Gandomi-Niasar A, Soltani N, Salavati-Niasari M (2009) J Mater Sci 44:2444. doi:10.1007/s10853-009-3309-y
Gece G (2008) Corros Sci 50:2981
Jamalizadeh E, Jafari AH, Hosseini SMA (2008) J Mol Struct 870:23
Lukovits I, Shaban A, Kálmán E (2005) Electrochim Acta 50:4128
Duda Y, Govea-Rueda R, Galicia M, Beltraén HI, Zamudio-Rivera LS (2005) J Phys Chem B 109:22674
Gómez B, Likhanova NV, Aguilar MAD, Olivares O, Hallen JM, Martínez-Magadán JM (2005) J Phys Chem A 109:8950
Zhang SG, Lei W, Xia MZ, Wang FY (2005) J Mol Struct 732:173
Tang YM, Chen Y, Yang WZ, Liu Y, Yin XS, Wang JT (2008) J Appl Electrochem 38:1553
Feng Y, Chen S, Guo W, Liu G, Ma H, Wu L (2007) Appl Surf Sci 253:8734
Khaled KF (2009) J Solid State Electrochem 13:1743
Khaled KF, Amin MA (2009) Corros Sci 51:1964
Khaled KF, Amin MA (2009) J Appl Electrochem 39:2553
Rodríguez-Valdez LM, Villamisar W, Casales M, González-Rodriguez JG, Martínez-Villafañe A, Martinez L, Glossman-Mitnik D (2006) Corros Sci 48:4053
Wang H, Wang X, Wang H, Wang L, Liu A (2007) J Mol Model 13:147
Xia S, Qiu M, Yu L, Liu F, Zhao H (2008) Corros Sci 50:2021
Khaled KF (2008) Electrochim Acta 53:3484
Oguzie EE, Li Y, Wang FH (2007) Electrochim Acta 53:909
Oguzie EE, Wang SG, Li Y, Wang FH (2008) J Solid State Electrochem 12:721
Gopi D, Govindaraju KM, Kavitha L (2010) J Appl Electrochem. doi:10.1007/s10800-010-0092-z
Prabhu RA, Venkatesha TV, Shanbhag AV, Praveen BM, Kulkarni GM, Kalkhambkar RG (2008) Mater Chem Phys 108:283
Şahin M, Gece G, KarcI F, Bilgiç S (2008) J Appl Electrochem 38:809
Gece G, Bilgiç S (2009) Corros Sci 51:1876
Özcan M, Dehri I, Erbil M (2004) Appl Surf Sci 236:155
Li W, He Q, Pei C, Hou B (2007) Electrochim Acta 52:6386
Obot IB, Obi-Egbedi NO (2010) Corros Sci 52:198
Larabi L, Benali O, Mekelleche SM, Harek Y (2006) Appl Surf Sci 253:1371
Cruz J, Martínez-Aguilera LMR, Salcedo R, Castro M (2001) Int J Quantum Chem 85:546
Khaled KF, Amin MA (2009) Corros Sci 51:2098
Acknowledgement
The authors are pleased to acknowledge the financial support provided by Specialized Research Fund for the Doctoral Program of Higher Education, China (No. 20093219120014) and NUST Research Funding, China (No. 2010ZYTS016).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fu, Jj., Li, Sn., Wang, Y. et al. Computational and electrochemical studies of some amino acid compounds as corrosion inhibitors for mild steel in hydrochloric acid solution. J Mater Sci 45, 6255–6265 (2010). https://doi.org/10.1007/s10853-010-4720-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10853-010-4720-0