Abstract
We report an investigation designed to explore alternative approaches for ranking of docking poses in the search for antagonists of the adenosine A2A receptor, an attractive target for structure-based virtual screening. Calculation of 3D similarity of docking poses to crystallographic ligand(s) as well as similarity of receptor–ligand interaction patterns was consistently superior to conventional scoring functions for prioritizing antagonists over decoys. Moreover, the use of crystallographic antagonists and agonists, a core fragment of an antagonist, and a model of an agonist placed into the binding site of an antagonist-bound form of the receptor resulted in a significant early enrichment of antagonists in compound rankings. Taken together, these findings showed that the use of binding modes of agonists and/or antagonists, even if they were only approximate, for similarity assessment of docking poses or comparison of interaction patterns increased the odds of identifying new active compounds over conventional scoring.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Virtual screening (VS) of compound databases is a widely applied approach in pharmaceutical research to complement biological screening and aid in the identification of active compounds [1, 2]. Typically, VS is used to prioritize database compounds for in vitro assays. VS approaches can roughly be divided into structure—and ligand-based methods. Structure-based (SB) methods [3] such as docking [4] make use of the target structure-usually a protein—as a screening template, whereas ligand-based (LB) methods [5] utilize structural information from known active compounds in the search for new chemical entities. SBVS and LBVS approaches are not mutually exclusive and can be combined in a variety of ways [6]. For example, we have recently introduced a hybrid SB/LBVS methodology consisting of docking and ligand 3D similarity analysis [7]. This approach involved docking and the generation of ligand poses followed by assessing the 3D similarity of modeled poses and crystallographic binding modes of known active compounds. 3D similarity analysis was facilitated by calculating atomic property density functions of test compounds and quantifying their overlap [8, 9]. The underlying idea is to replace conventional force field-based scoring functions, yielding energy scores, with a 3D similarity measure for compound ranking. In benchmark calculations, 3D similarity-based compound rankings were found to produce higher recall of active compounds than scoring functions for a variety of targets [7]. By design, this hybrid docking/similarity approach is knowledge based, as it explicitly takes known crystallographic binding modes into account. Accordingly, it is not applicable in cases where no X-ray structures of ligand–target complexes are available as references.
In the present work, we have applied and further extended this modeling and ranking protocol in a case study on one of the structurally best characterized G protein coupled receptor (GPCR) [10], i.e. the adenosine A2A receptor [11]. In recent years, GPCR structural biology has provided a much improved basis for SBVS [12, 13] applications on this eminent family of therapeutic targets for the treatment of a variety of central nervous system disorders, cancers, or inflammatory diseases [11]. X-ray structures have provided unprecedented insights into the conformational plasticity of ligand binding sites in GPCRs and the often delicate balance of and subtle differences between agonistic and antagonistic ligand effects. Such structural insights have opened the door for SBVS applications [14]. However, major challenges for SBVS include the variety of conformational effects that have been observed upon ligand binding as well as the wide spectrum of conformational states available to these integral membrane proteins [10]. For the A2A receptor, several complex X-ray structures have been determined with either agonists or antagonists present bound in the orthosteric ligand binding site, hence providing a sound basis for exploring and exploiting binding modes of functionally distinct ligands.
As an extension of 3D similarity analysis, we have also used a protein–ligand interaction fingerprint (PLIF) [15] to compare modeled and crystallographic ligand poses in the search for A2A receptor antagonists. The calculation of PLIFs also critically depends on ligand poses but does not capture molecular similarity such as atomic property density functions. Rather, PLIFs account for interactions within the target binding site that result from given binding poses. Accordingly, the use of PLIFs is more related to pharmacophore methods than whole-molecule similarity assessment. Hence, comparison of PLIFs computed for models and experimental reference ligands evaluates to what extent crystallographic interactions are mimicked by predicted poses, in accord with the pharmacophore concept.
Different PLIF designs have been reported and applied in docking and SBVS [16]. For example, Marcou and Rognan used PLIFs to prioritize inhibitors of cyclin-dependent kinase 2 over molecular decoys, demonstrating superior performance compared to force field scoring [17]. Furthermore, Desaphy et al. [18] devised a PLIF-based protocol to post-process docking poses. Moreover, Da and Kireev reported the design of so-called structural protein–ligand interaction fingerprints (SPLIF) [19], which were then applied by Da et al. [20] to discover a series of Mer kinase inhibitors.
In addition to evaluating alternative measures of binding mode resemblance for ranking of docking poses, we have analyzed whether or not the structure of an inactive form of the adenosine A2A receptor provided an advantage over an active conformation when searching for antagonists. Furthermore, both antagonists and agonists as well as molecular models derived from them were evaluated as reference compounds for binding mode assessment, yielding some surprising findings.
Materials and methods
Receptor structure selection and preparation
Fourteen structures [21–28] of the A2A receptor are currently available in the Protein DataBank (PDB) (Table 1). These X-ray structures include complexes with five unique antagonists and four agonists. For our analysis, we selected an active (agonist-bound) [21] and an inactive (antagonist-bound) [26] conformation of the receptor, with PDB codes 2YDV and 4EIY, respectively, on the basis of highest resolution an lowest crystallographic R-factors. The selected structures were prepared for docking using Molecular Operating Environment (MOE) 2014.09 [15]. Bound ions, organic solvent, and water molecules were removed from the receptor ligand binding domains used as a template for docking. Potential benefits of including water molecules in docking studies on the adenosine A2A receptor have been stated previously [29]. Since we used chemically diverse sets of ligands for docking, structural water molecules interacting with specific agonists or antagonists were not considered to avoid potential bias of benchmark calculations. Other preparation steps included the addition of hydrogen atoms, computation of protonation states and tautomers, assignment of partial charges, and mild energy minimization (structural relaxation) using the Amber10 force field until a root mean square (RMS) gradient of 0.1 kcal/mol/Å2 was reached.
Ligand preparation
A benchmark set for the A2A receptor was extracted from the DEKOIS 2.0 [30]. This set included 40 active compounds and 1200 decoys. Activity annotations were confirmed on the basis of corresponding BindingDB [31] records. Our analysis was focused on antagonists that provide a much larger knowledge base for docking studies than agonists. In addition, the only three known agonists (BDB50085666, BDB50085668, and BDB50309479) present in the benchmark set were retained together with 100 decoys selected from the ZINC database [32] by the developers of the DEKOIS database that matched physicochemical properties of these agonists. Hence, a final set of 37 antagonists, three agonists, and 1200 decoys was obtained. An initial low-energy 3D conformation of each ligand was generated with MOE and protonation states and partial charges were assigned using the AM1-BCC implementation in MOE following a previously reported protocol [33], which was also was applied to prepare crystallographic ligands for docking.
Docking and scoring
All docking trials were carried out using the Dock module of MOE [15]. Coordinates of crystallographic ligands were used to define the docking sites. A triangle matcher function was used to generate and 1000 docking poses for each ligand. The top-30 best solutions according to the London dG scoring function were pre-selected and further refined and subjected to re-scoring using the GBVI/WSA dG scoring function. On the basis of these two-step posing protocol, the best pose of each ligand was used for ranking. Additional force field-based rankings were generated by scoring the selected poses using the London dG and Alpha HB scoring functions [34, 35]. Force field-based rankings were then compared to similarity—and PLIF-based rankings, as specified in the following.
Similarity calculations
Similarity to experimental binding modes was quantified using the property density function-based 3D similarity measure [6] and the PLIF implementation of MOE [15]. On the basis of this implementation, receptor–ligand contacts were classified into six types of interactions including side chain-mediated hydrogen bonds (donor and acceptor), backbone-mediated hydrogen bonds (donor and acceptor), ionic interactions, and surface interactions. PLIFs were calculated with default settings and compared using the Tanimoto coefficient. Normalized overlap of property density functions (ranging from 0 to 1) was calculated as a measure of 3D similarity. For each similarity measure, compound rankings were calculated.
As crystallographic reference compounds, the bound antagonist ZM24138521 (PDB code 4EIY) and agonist NECA [26] (2YDV) were used as well as other [22–25, 27, 28] structurally characterized agonists and antagonists (Table 1). Ligand overlays were generated by superposing alpha carbon atoms of corresponding A2A receptor structures.
Performance evaluation
Receiver operator characteristic (ROC) plots were generated to evaluate compound rankings. ROC curves monitor the percentage of known active compounds retrieved at each position of the ranking. The area under the ROC curve (AUC) was computed as a measure of the enrichment of active compounds in a ranking applying the composite trapezoidal rule. AUC values of 0.5 correspond to a random compound distribution, while further increasing values indicate the enrichment of active compounds at high rank positions. An AUC value of 1 would be produced by a ranking in which all active compounds would be ranked higher than the best scoring decoys. In addition, to assess early enrichment of active compounds, the enrichment factor for 10 % of the ranked database (Ef10 %) was computed [36].
Results and discussion
Docking reproduces experimental binding modes
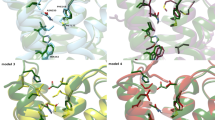
The A2A receptor has canonical GPCR topology with seven transmembrane helices connected by three intracellular and three extracellular loops [10]. The antagonist ZM241385 [21] and agonist NECA [26] bind to the orthosteric site of the receptor (Fig. 1a). The triazolotriazine core of antagonist ZM241385 approximately aligns with the adenine moiety of agonist NECA, which form comparable hydrogen bonds with Glu1695.30 and Asn2536.55 (superscripts refer to conventional Ballesteros–Weinstein numbering system [37]). Otherwise, the antagonist and agonist form distinct interactions. For example, the ribose moiety of NECA forms additional hydrogen bonds with Ser2777.42 and His2787.43 (Fig. 1b). These interactions are thought to stabilize the active conformation of the receptor [26].
Adenosine A2A receptor in complex with an agonist and antagonist. a The superposition of the agonist—(orange) and antagonist-bound (green) forms of the receptor. The structure and molecular surface of co-crystallized ligands in the orthosteric binding site are displayed. b Receptor–ligand interaction details for both complexes. In c, d the overlay of docked (magenta) and crystallographic binding modes of NECA (orange) and ZM241385 (green) is shown, respectively
Re-docking and cross-docking of ZM241385 and NECA were carried out using the antagonist-(4EIY) and agonist-bound (2YDV) form of the receptor. The best poses from re-docking reproduced the experimental binding modes with RMS deviations of only of 0.4 Å (ZM241385) 0.3 Å (NECA), as reported in Table 2, and key interactions were modeled correctly (Fig. 1c, d); an encouraging finding. Cross-docking calculations were performed as a control to assess the influence of different conformations of several binding site residues observed in these structures on posing. These conformational differences are illustrated in Fig. 1b. As anticipated, cross-docking resulted in poses that were inconsistent with X-ray binding modes, yielding RMDS deviations larger than 2 Å in both cases (Table 2). Hence, conformational differences of binding site residues impaired accurate modeling of the antagonist into the binding site of the agonist-bound form of the receptor and vice versa, as expected on the basis of the X-ray structures and confirmed by cross-docking.
The set of 37 known antagonists, three agonists, and 1200 decoys was then docked into the antagonist—and agonist-bound form of the A2A receptor from which ligands had been removed and alternative rankings of ligand poses were generated, as detailed in the “Materials and methods” section.
Ranking on the basis of standard scoring functions
Modeled binding modes were first evaluated using three different scoring functions (London dG, GBVI/WSA dG, and Alpha HB). Only low to moderate enrichment of known antagonists was detected in each ranking. AUC values ranged from 0.57 to 0.64 and Ef10 % enrichment factors from 1.62 to 2.16 (Table 3). The best performance was achieved by scoring with London dG resulting in an AUC values of 0.64 (4EIY) and 0.62 (2YDV) (Fig. 2). Thus, recall of known antagonists was only slightly higher for the antagonist-compared to the agonist-bound form of the receptor.
ROC plots for force field and 3D similarity rankings. Blue curves represent the results for the best-performing scoring function London dG (Docking), green curves for 3D similarity to ZM241385, green dashed curves for 3D similarity to a fragment of ZM241385 (consisting of the triazolotriazine core and the furan ring), cyan curves for highest 3D similarity to any of the five available crystallographic antagonists, orange curves for 3D similarity to NECA, and yellow curves for highest 3D similarity to any of the four available crystallographic agonists. The red lines provide a reference for random compound selection. In addition to X-ray binding modes, transferred ZM241385 and NECA (as described in the text) were also used as reference compounds
Ranking on the basis of 3D similarity
Next 3D similarity calculations comparing docked and crystallographic binding modes were carried out. Compounds docked into 2YDV were ranked by calculating 3D similarity to NECA resulting in an AUC value of 0.71 and Ef10 % factor of 3.78 (Table 3), thus yielding a notable improvement over force field-based rankings, although the binding mode of an agonist was used as a reference. In this context, we note that Rodríguez et al. [38] reported a successful prospective SBVS campaign using agonist-bound structures of the A2A receptor. However, all newly identified active compounds were found to be antagonists [38]; a rather surprising finding. However, in the case of 3D similarity comparison of binding models, a rationale for the ability of the agonist NECA to select antagonists was provided by considering that the adenine core of NECA resembled the triazolotriazine core of ZM241385, as illustrated in Fig. 1b. Hence, binding modes of these core fragments corresponded to each other, which led to the successful identification of antagonists on the basis of both references. Surprisingly, however, when compounds were docked into 4EIY and ranked by calculating 3D similarity to the antagonist ZM241385, lower search performance was observed than in the case of 2YDV/NECA, with an AUC of only 0.57 (Table 3). Searching for a possible structural rationale for these findings, we observed that the phenol ring and ethylamine group of ZM241385 were located in a region proximal to the extracellular loops of the receptor where no clear interactions were formed and several other known A2A antagonists lacked corresponding groups [24, 25], suggesting that these moieties were conformationally unrestraint and not essential for binding. Hence, for whole-molecule similarity calculations, the presence of the phenol ring and ethylamine group was considered a potential caveat. Therefore, as a control, a fragment of ZM241385 was constructed only containing the triazolotriazine core and furan ring that were involved in well-defined interactions with the receptor and had corresponding moieties in other antagonists [24, 25]. There is evidence in the literature that this fragment itself might be active because the phenol ring and ethylamine group are not essential for binding [39]. However, we consider such fragments generated for similarity calculations as “artificial” ligands. When the ZM241385 fragment was used as a reference for 3D similarity calculations, a clear improvement in search performance was observed, with an AUC value of 0.75 and an Ef10 % factor of 3.24 (Table 3; Fig. 2). These findings supported the view that overlapping core structures of NECA and ZM241385 were responsible for the observed enrichment of antagonists in 3D similarity calculations. Furthermore, the results showed that an artificial A2A receptor ligand representing an important core fragment of active compounds could be effectively used as a reference for similarity scoring, which we considered an important result in the context of our investigation. It is expected that core structure fragment constructs can also be used for similarity-based ranking in the case of other targets, even if these artificial ligands might not be active.
In addition, we also used all five structurally characterized antagonists of the A2A receptor (Table 1) superposed in the binding site of 4EIY as reference compounds and selected the highest similarity value as the final score for ranking (akin to a 1-nearest neighbor approach). Corresponding calculations were carried out using the four structurally characterized agonists (Table 1) superposed in the binding site of 2YDV. In the former case, search performance was increased compared to the complete structure of ZM241385, with an AUC of 0.71, but remained lower than for the ZM241385 core fragment. In the latter case, the use of multiple agonists as references did not yield a further improvement over NECA (Table 3; Fig. 2).
NECA was also positioned in the binding site of 4EIY following superposition of the agonist—and antagonist-bound form of the receptor, as shown in Fig. 1a, b. This reference structure was termed “transferred NECA”. Analogously, ZM241385 was transferred into 2YDV based on the same superposition (transferred ZM241385). Transferred NECA, transferred ZM241385, and its corresponding core fragment were then used as reference compounds. These calculations produced in part surprising results. ZM241385 placed into the binding site of agonist-bound form of the receptor de-selected known antagonists with an AUC value of 0.45 (i.e. lower than for random selection). The ability to enrich known antagonists was again recovered, albeit to a lesser extent than before, when the core fragment of transferred ZM241385 was used as reference, yielding an AUC value of 0.64. By contrast, when transferred NECA was used as a reference compound, a significant prioritization of antagonists over decoys was detected, with an AUC value of 0.74 and an Ef10 % factor of 3.78 (Table 3; Fig. 2). Hence, a model of an agonist in the antagonist-bound form of the receptor was capable of prioritizing antagonists in compound rankings when used as a reference for 3D similarity calculations. A plausible rationale for these findings was provided by the partial binding mode resemblance of NECA and ZM241385 in combination with a key structural difference. As shown in Fig. 1b, the binding modes of the antagonist and agonist overlapped in their core regions, but NECA lacked the terminal unconstrained terminal moieties of ZM241385 that compromised whole-molecule similarity calculations, as discussed above.
Ranking on the basis of interaction fingerprints
Modeled and experimental reference binding modes were also compared using PLIFs, which provided a pharmacophore-centric assessment of interaction similarity and thus an additional-and conceptually different-approach to evaluate 3D similarity. PLIF-based Tanimoto similarity to crystallographic reference(s) was calculated to rank docking poses. ROC plots for all PLIF-based rankings are represented in Fig. 3. The use of ZM241385 and transferred ZM241385 as references for PLIF calculations yielded AUC values comparable to those obtained by using the ZM241385 fragment in 3D similarity-based rankings (Table 3; Fig. 3). Hence, the ability of PLIFs to capture relevant interactions with the receptor enabled this approach to circumvent the caveat associated with the terminal moieties of ZM241385.
ROC plots for PLIF-based rankings. Reported are the results for PLIF-based compound rankings compared to 3D similarity rankings on the basis of the ZM241385 fragment and NECA according to Fig. 2
Furthermore, the use of NECA as a reference in both structures also resulted in AUC values comparable to those obtained in the 3D similarity-based rankings. In the antagonist-bound form of the receptor, further increased Ef10 % factors of 4.59 (ZM241385) and 4.05 (NECA) were observed (Table 3). On the other hand, in the agonist-bound form, an Ef10 % factor of 2.43 was determined for ZM241385 while a lower Ef10 % factor of 1.89 was obtained for NECA. Furthermore, AUC values and enrichment factors comparable to single reference compound present in the antagonist-bound form of the receptor were obtained when multiple ligands were used. Taken together, these results indicated that a single antagonist-or even agonist-used as a reference was already sufficient to capture interactions using PLIFs that prioritized antagonists over decoys when ranking docking poses. On the basis of these findings, 3D interaction analysis provided an attractive complement to 3D similarity calculations for ranking of poses.
Multiple receptor conformations
The use of multiple receptor conformations in docking (i.e. ensemble docking) might further improve the enrichment of ligands by indirectly taking structural flexibility of the target into account [40–44]. GPCRs, which transition through a spectrum of active or inactive conformations, are a paramount case of target flexibility. Hence, we have also investigated whether ensemble docking using both 2YDV and 4EIY structures improves the enrichment of known antagonists following our approach. To assess this possibility, the overall best score obtained for each ligand and decoy was used to generate new rankings. Interestingly, the resulting ranks of antagonists based on scoring functions were lower than those obtained using 2YDV or 4EIY alone (Table 4). By contrast, 3D similarity—or PLIF-based ranking on the basis of ensemble docking essentially averaged the enrichments obtained for the individual structures. Thus, in this case, the use of two receptor conformations did not results in a notable improvement of rank positions of known active compounds. For target structure selection, different strategies were introduced previously [41, 42, 44]. In case of A2A receptor, several structures have been determined at a relatively low resolution (Table 1), making them less suitable templates for docking and hence limiting the potential of ensemble docking. Structural genomics efforts focusing on GPCRs [45] are likely to provide further candidate structures for ensemble docking.
Ranking of agonists
As an additional control, rankings were also generated by including the three known agonists and corresponding decoys in the docking calculations. Compounds were ranked using scoring functions, 3D similarity, and PLIF similarity. The rank positions of the docked agonists are reported in Table 5. In general, agonists tended to rank higher when docked into the active conformation, as also observed in our cross-docking calculations. However, some exceptions are evident in Table 5 for highly ranked agonists docked into 4EIY. This provides further evidence that reasonable binding modes might be obtained even when the receptor conformation does not correspond to the activity state of the docked ligand.
Conclusions
Herein we have reported a case study on the adenosine A2A receptor to further explore alternative ranking schemes for docking poses of putative ligands. Structural information for GPCRs is still limited but the A2A receptor is well characterized-including a number of milestone structures for the GPCR field-such that systematic SBVS trials could be carried out using alternative structural templates and information provided by multiple crystallographic agonists and antagonists. The docking and ranking calculations have focused on the search for antagonists, which are available in sufficient numbers to enable a meaningful statistical assessment of benchmark calculations.
In our case study, ranking schemes for docking poses using an antagonist-bound as well as agonist-bound form of the A2A receptor and different reference compounds were compared. It was found that ranking of docking poses on the basis of calculated 3D similarity to crystallographic reference compounds yielded consistently higher enrichment of antagonists than conventional force field-based scoring. Because these calculations quantified whole-molecule similarity, we also investigated interaction fingerprints for binding mode comparisons. This approach represented an indirect assessment of “local” similarities that were also pose-dependent, by positioning of functional groups, resulting in specific receptor–ligand interactions. PLIF calculations resulted in compound rankings that were consistently better than those obtained by force field-based scoring and at least comparable and in part superior to explicit 3D similarity assessment. Specifically, PLIF-based rankings mostly resulted in further improved early enrichment of antagonists compared to 3D similarity rankings. Taken together, both “global” and “local” similarities between crystallographic reference compounds and docking poses were effectively exploited to improve the quality of compound rankings over force field-based scoring.
Moreover, several important lessons were learned concerning the choice of reference ligands for pose comparisons. First, corresponding core fragments of an antagonist as well as an agonist were sufficient to prioritize A2A antagonists over decoys. In fact, the presence of a flexible moiety in the antagonist ZM241385, which was not present in several other antagonists, illustrated a potential caveat associated with whole-molecule similarity calculations that was counter-balanced by focusing on the core fragment of the antagonist or by using interaction fingerprints as an alternative. Since the non-conserved flexible moiety in ZM241385 was not involved in significant receptor–ligand interactions, it did not affect PLIF calculations.
It is noted that docking and ranking calculations can probably not be expected to distinguish between fine interaction details and binding mode discrepancies that differentiate between GPCR antagonists and agonists. However, for practical SBVS applications on GPCRs, the finding that a single crystallographic binding mode of an antagonist or agonist was capable to prioritize antagonists over decoys has important implications. Even a transferred antagonist or agonist used as a reference for global and/or local similarity assessment might be sufficient to improve compound rankings over conventional scoring. Strongest support for this conjecture was provided by the results of calculations using the agonist transferred NECA whose core partly resembled ZM241385. This model compound was capable of significantly enriching antagonists in rankings.
References
Lavecchia A, Di Giovanni C (2013) Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 20:2839–2860
Heikamp K, Bajorath J (2013) The future of virtual compound screening. Chem Biol Drug Des 81:33–40
Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH (2012) Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J 14:133–141
Irwin JJ, Shoichet BK (2016) Docking screens for novel ligands conferring new biology. J Med Chem. doi:10.1021/acs.jmedchem.5b02008
Ripphausen P, Nisius B, Bajorath J (2011) State-of-the-art in ligand-based virtual screening. Drug Discov Today 16:372–376
Drwal MN, Griffith R (2013) Combination of ligand—and structure-based methods in virtual screening. Drug Discov Today Technol 10:e395–e401
Anighoro A, Bajorath J (2016) Three-dimensional similarity in molecular docking: prioritizing ligand poses on the basis of experimental binding modes. J Chem Inf Model 56:580–587
Hu Y, Furtmann N, Gütschow M, Bajorath J (2012) Systematic identification and classification of three-dimensional activity cliffs. J Chem Inf Model 52:1490–1498
Peltason L, Bajorath J (2007) Molecular similarity analysis uncovers heterogeneous structure-activity relationships and variable activity landscapes. Chem Biol 14:489–497
Kobilka BK (2007) G protein coupled receptor structure and activation. Biochim Biophys Acta Biomembr 1768:794–807
de Lera Ruiz M, Lim Y-H, Zheng J (2014) Adenosine A2A receptor as a drug discovery target. J Med Chem 57:3623–3650
Shoichet BK, Kobilka BK (2012) Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci 33:268–272
Tautermann CS (2014) GPCR structures in drug design, emerging opportunities with new structures. Bioorg Med Chem Lett 24:4073–4079
Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA (2010) Structure-based discovery of A2A adenosine receptor ligands. J Med Chem 53:3748–3755
Chemical Computing Group, Inc. Molecular operating environment, version 2014.09
Mpamhanga CP, Chen B, McLay IM, Willett P (2006) Knowledge-based interaction fingerprint scoring: a simple method for improving the effectiveness of fast scoring functions. J Chem Inf Model 46:686–698
Marcou G, Rognan D (2007) Optimizing fragment and scaffold docking by use of molecular interaction fingerprints. J Chem Inf Model 47:195–207
Desaphy J, Raimbaud E, Ducrot P, Rognan D (2013) Encoding protein–ligand interaction patterns in fingerprints and graphs. J Chem Inf Model 53:623–637
Da C, Kireev D (2014) Structural protein–ligand interaction fingerprints (SPLIF) for structure-based virtual screening: method and benchmark study. J Chem Inf Model 54:2555–2561
Da C, Stashko M, Jayakody C, Wang X, Janzen W, Frye S, Kireev D (2015) Discovery of mer kinase inhibitors by virtual screening using structural protein–ligand interaction fingerprints. Bioorg Med Chem 23:1096–1101
Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, Ijzerman AP, Cherezov V, Stevens RC (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–236
Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC (2008) The 2.6 Ångstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322:1211–1217
Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N, Cameron AD, Kobayashi T, Hamakubo T, Iwata S, Murata T (2012) G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 482:237–240
Congreve M, Andrews SP, Doré AS, Hollenstein K, Hurrell E, Langmead CJ, Mason JS, Ng IW, Tehan B, Zhukov A, Weir M, Marshall FH (2012) Discovery of 1,2,4-triazine derivatives as adenosine A2A antagonists using structure based drug design. J Med Chem 55:1898–1903
Doré AS, Robertson N, Errey JC, Ng I, Hollenstein K, Tehan B, Hurrell E, Bennett K, Congreve M, Magnani F, Tate CG, Weir M, Marshall FH (2011) Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure 19:1283–1293
Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AGW, Tate CG (2011) Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474:521–525
Lebon G, Edwards PC, Leslie AGW, Tate CG (2015) Molecular determinants of CGS21680 binding to the human adenosine A2A receptor. Mol Pharmacol 87:907–915
Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao Z-G, Cherezov V, Stevens RC (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332:322–327
Lenselink EB, Beuming T, Sherman W, van Vlijmen HWT, Ijzerman AP (2014) Selecting an optimal number of binding site waters to improve virtual screening enrichments against the adenosine A2A receptor. J Chem Inf Model 54:1737–1746
Bauer MR, Ibrahim TM, Vogel SM, Boeckler FM (2013) Evaluation and optimization of virtual screening workflows with DEKOIS 2.0—a public library of challenging docking benchmark sets. J Chem Inf Model 53:1447–1462
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 35:D198–D201
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Anighoro A, Rastelli G (2013) Enrichment factor analyses on G-protein coupled receptors with known crystal structure. J Chem Inf Model 53:739–743
Corbeil CR, Williams CI, Labute P (2012) Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des 26:775–786
Planesas JM, Claramunt RM, Teixidó J, Borrell JI, Pérez-Nueno VI (2011) Improving VEGFR-2 docking-based screening by pharmacophore postfiltering and similarity search postprocessing. J Chem Inf Model 51:777–787
Bender A, Glen RC (2005) A discussion of measures of enrichment in virtual screening: comparing the information content of descriptors with increasing levels of sophistication. J Chem Inf Model 45:1369–1375
Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in g protein coupled receptors. Neurosci Methods 25:366–428
Rodríguez D, Gao Z-G, Moss SM, Jacobson KA, Carlsson J (2015) Molecular docking screening using agonist-bound GPCR structures: probing the A2A adenosine receptor. J Chem Inf Model 55:550–563
Federico S, Paoletta S, Cheong SL, Pastorin G, Cacciari B, Stragliotto S, Klotz KN, Siegel J, Gao Z-G, Jacobson KA, Moro S, Spalluto G (2011) Synthesis and biological evaluation of a new series of 1,2,4-triazolo[1,5-a]-1,3,5-triazines as human A2A adenosine receptor antagonists with improved water solubility. J Med Chem 54:877–889
Bottegoni G, Kufareva I, Totrov M, Abagyan R (2009) Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J Med Chem 52:397–406
Rueda M, Bottegoni G, Abagyan R (2010) Recipes for the selection of experimental protein conformations for virtual screening. J Chem Inf Model 50:186–193
Bottegoni G, Rocchia W, Rueda M, Abagyan R, Cavalli A (2011) Systematic exploitation of multiple receptor conformations for virtual ligand screening. PLoS One 6:e18845
Sgobba M, Caporuscio F, Anighoro A, Portioli C, Rastelli G (2012) Application of a post-docking procedure based on MM–PBSA and MM–GBSA on single and multiple protein conformations. Eur J Med Chem 58:431–440
Hou X, Li K, Yu X, Sun J-P, Fang H (2015) Protein flexibility in docking-based virtual screening: discovery of novel lymphoid-specific tyrosine phosphatase inhibitors using multiple crystal structures. J Chem Inf Model 55:1973–1983
Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, Rosen H, Wüthrich K (2013) The GPCR network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov 12:25–34
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anighoro, A., Bajorath, J. Binding mode similarity measures for ranking of docking poses: a case study on the adenosine A2A receptor. J Comput Aided Mol Des 30, 447–456 (2016). https://doi.org/10.1007/s10822-016-9918-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9918-z