Abstract
E3 ubiquitin ligases are attractive drug targets due to their specificity to the ubiquitin machinery. However, the development of E3 ligase inhibitors has proven challenging for the fact that they must disrupt protein–protein interactions (PPIs). The E3 ligase involved in interactome provide new hope for the discovery of the E3 ligase inhibitors. These currently known natural binding partners of the E3 ligase can benefit the discovery of other unknown substrates and also the E3 ligase inhibitors. Herein, we present a novel strategy that using multiple substrates to elucidate the molecular recognition mechanism of E3 ubiquitin ligase. Molecular dynamics simulation, molecular mechanics-generalized born surface area (MM-GBSA) binding energy calculation and energy decomposition scheme were incorporated to evaluate the quantitative contributions of sub-pocket and per-residue to binding. In this case, Kelch-like ECH-associated protein-1 (Keap1), a substrate adaptor component of the Cullin–RING ubiquitin ligases complex, is applied for the investigation of how it recognize its substrates, especially Nrf2, a master regulator of the antioxidant response. By analyzing multiple substrates binding determinants, we found that both the polar sub-pockets (P1 and P2) and the nonpolar sub-pockets (P4 and P5) of Keap1 can make remarkable contributions to intermolecular interactions. This finding stresses the requirement for substrates to interact with the polar and nonpolar sub-pockets simultaneously. The results discussed in this paper not only show the binding determinants of the Keap1 substrates but also provide valuable implications for both Keap1 substrate discovery and PPI inhibitor design.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ubiquitination is one of the most important post-translation modifications in eukaryotes [1]. With the exception of the most known function as regulating protein turnover inside the cell [2, 3], ubiquitination can also induce conformational changes that alter the biological function. Furthermore, different kinds of ubiquitination with different types, lengths, connectivities, and anchoring sites play distinct roles in the cell and regulate diverse areas of biology [4]. Like phosphoration, ubiquitination is reversible and can be occurred in only one or several amino acid residues on the same protein. Thus the ubiquitin system could be the prime candidate for these new targets following the kinase superfamily [5].
Ubiquitination of a protein target is tightly regulated through a cascade of enzyme activities (E1 → E2 → E3), which can link the C-terminal Gly residue of ubiquitin (Ub) to the Lys side chain of the target protein through an isopeptide bond [6, 7]. The transfer of ubiquitin from the E2 enzyme to substrates mediated by the ubiquitin E3 ligase is the key step for the appropriate ubiquitination of target protein which confers specificity to ubiquitination. In addition, the E3 ubiquitin ligases are specified by over 600 human genes, surpassing the 518 protein kinase genes [8]. Thereby, E3 ubiquitin ligases are more attractive therapeutic targets than general proteasome inhibitors for modulating the ubiquitin system [5, 8, 9]. However, the development of E3 ligase inhibitors has proven challenging, in part due to the fact that they must disrupt protein–protein interactions (PPI) [10]. Finding active small molecular compounds to disrupt the interface of two proteins can be intrinsically more difficult than searching for small molecules that block catalytic activity [11]. So by now, only a few additional E3 ligase inhibitors have been reported that target inhibitors of P53–MDM2 [12], apoptosis proteins (IAPs) [13], Skp1–Cullin–F-box (SCF)Cdc40 [14], SCFMet30 [15], and von Hippel–Lindau (VHL) [16, 17]. Therefore, this field remains unmined. Researchers have still not developed a general approach for identifying inhibitors of many E3 ubiquitin ligases, while the E3 ligase involved in interactome provides the new hope. Each ligase degrades multiple proteins and many other proteins have been found to inhibit ubiquitination through disrupting the PPI of E3 ligase and its substrate.
These natural binding partners of the E3 ligase not only enrich the regulatory network of the E3 ligase, but also can benefit the discovery of the E3 ligase inhibitors. Herein, we present a novel strategy that using multiple substrates to elucidate the sub-pocket-activity and per-residue-activity relationship of the E3 ligase. Molecular dynamics (MD) simulation, molecular mechanics-generalized born surface area (MM-GBSA) binding energy calculation and energy decomposition scheme were incorporated to give the quantitative contributions of sub-pocket and per-residue to binding. Using different substrates of the E3 ligase as the input structures, the detailed information of sub-pocket-activity and per-residue-activity relationship can be obtained. These quantified binding elucidations of the E3 ubiquitin ligase and substrate can provide direct information about the binding determinants of E3 ligase. These information can guide the discovery of novel substrate and benefit the discovery of the E3 ligase inhibitor. For example, the key residue of the substrate identified from the quantified analysis can give the detailed structure fragment to design the inhibitor.
In the present work, Kelch-like ECH-associated protein-1 (Keap1), a substrate adaptor component of the Cullin–RING ubiquitin ligases (CRL) complex, was considered for investigation. In the complex, Cul3 (Cullin 3) and Rbx1 (Ring box 1) form the catalytic component of the enzyme complex and interact with an E2 ubiquitin ligase to transfer ubiquitin to the substrate. Keap1 is the adaptor component of the complex which is in charge of recognizing ubiquitination substrate. The well-known substrate for the Keap1–Cul3–RING ubiquitin E3 ligase complex is nuclear factor erythroid-2-related factor 2 (Nrf2) [18], which is an attractive target for the prevention and treatment of oxidative stress-related diseases and conditions including cancer, neurodegenerative, cardiovascular, metabolic and inflammatory diseases [19]. Besides, Keap1 also mediates the ubiquitination of several other important proteins including IKKβ [20, 21] (IκB kinase β) and Bcl-2 [22]. Moreover, many other proteins have been found to inhibit the Keap1 mediated ubiquitination through disrupting the PPI of Keap1 and its substrate. For example, the selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through disrupting the PPI of Keap1–Nrf2 [23, 24]. The pathological process associated with p62 accumulation can result in hyperactivation of Nrf2, delineating unexpected roles of selective autophagy in controlling the transcription of cellular defence enzyme gene [24]. Thus, understanding of the intermolecular recognition mechanism between Keap1 and substrates is important for the study of Keap1-related interactome.
Previously, we have investigated the Keap1–Nrf2 ETGE motif interaction using the MD simulations [25]. Key electrostatic interactions between arginines of Keap1 and glutamates of Nrf2 play an important role in Keap1–Nrf2 ETGE motif PPI. However, using one substrate of Keap1 could not give the whole understanding of the substrate recognition mechanism. In order to deeply understand the recognition mode and the crucial interactions between Keap1 and substrate, four different substrates in complex with Keap1 DC domain were included to perform MD simulations combining with MM-GBSA free energy calculations. Total energy was decomposed on per residue contribution, and hydrogen bond occupancies were monitored throughout the simulations. The results provide detailed information of sub-pocket-activity relationship of Keap1 and per-residue-activity relationship of substrates, which can directly guide the discovery of Keap1 substrates. What’s more, these critical insights into the binding determinants of the Keap1 DC domain structure can facilitate the design of the peptide inhibitors [26] and further the small molecular inhibitory compounds of Keap1, which has been proven successful [25, 27].
Materials and methods
General procedure and system preparations
The currently known structures of Keap1–substrate complex were obtained from protein data bank (PDB). The detailed information of crystal structures was listed in Table 1. All structures obtained from PDB were corrected using clean protein tool in Discovery Studio (DS) 3.0 package (Accelrys Inc., San Diego, CA, USA). All calculations were conducted using Dawning TC2600 cluster. Except for otherwise mentioned, other parameters were set as default.
Preparation of MD starting structures
Four known substrates with reported crystal structures were downloaded from PDB for further analysis. These peptides were LDEETGEFL (the Nrf2 ETGE motif, PDB ID 1X2R), WRQDLD (the Nrf2 DLG motif, PDB ID 2DYH), QNEENGEQE (a nuclear oncoprotein, prothymosin α, PDB ID 2Z32) and VDPSTGEL (the selective autophagy substrate p62, PDB ID 3ADE). To avoid terminal charges on the peptides, the N-terminal residues were capped with acetyl (ACE). All crystallographic water molecules were removed from the coordinate set.
Molecular dynamics simulations
MD simulation of Keap1 DC domain bound to four different peptide substrates were performed using PMEMD module of AMBER 12 with ff99SB modifications [31, 32] of the Cornell et al. [33] force field. TIP3P water molecules were applied to solvate the complex, extending at least 12 Å from the protein. The counterions were added to the solvent to keep the system neutral (twelve Na+ for 1X2R, nine Na+ for 2DYH, twelve Na+ for 2Z32 and ten Na+ for 3ADE). The geometry of the system was minimized in two steps before MD simulation. First, the water molecules were refined through 2,500 steps of steepest descent followed by 2,500 steps of conjugate gradient, keeping the protein fixed with a constraint of 2.0 kcal mol−1 Å2. Second, the complexes were relaxed by 10,000 cycles of minimization procedure (5,000 cycles of steepest descent and 5,000 cycles of conjugate gradient minimization). During the simulation, the particle mesh Ewald method [34] was employed to calculate the long-range electrostatic interactions, while the SHAKE method [35] was applied to constrain all covalent bonds involving hydrogen atoms to allow the time step of 2 fs. A 10 Å cutoff value was used for the nonbonded interactions. The whole system was heated from 0 to 300 K running 50 ps molecular dynamics with position restraints at constant volume. Subsequent isothermal isobaric ensemble (NPT)-MD was performed for 500 ps to adjust the solvent density with a time constant of 1.0 ps for pressure relaxation. Harmonic restraints with force constants of 2 kcal/(mol Å2) were applied to all receptor and peptide atoms in this step. An additional 500 ps of unconstrained NPT-MD at 300 K with a time constant of 2.0 ps for pressure relaxation was performed to relax the system without constraints. The production dynamics at constant pressure achieved lengths of 40 ns of which snapshots saved at 20 ps intervals were used for further analysis.
Analysis of MD trajectories and binding energy calculation
To explore the stability of complexes in the simulations and to ensure the rationality of the sampling method, the ‘ptraj’ tool in Amber12 was used to analyze the time-dependence of the RMSD of the backbone atoms. Furthermore, the hydrogen bonds were defined by a distance cutoff of 3.2 Å and an angle cutoff of 120°. Hydrogen bonds were only considered if their occupancies attained >20 % (the percentage of simulation time in which the hydrogen bond is formed).
The MM-GBSA method [36, 37], implemented in the AMBER program, has been used for free energy calculation and to investigate the energetic contributions of each residue to binding. The basic theory of the MM-GBSA approach is that the free energy of binding can be obtained through calculating only the end points of the thermodynamical cycle of ligand binding. The binding free energy, Δ Gbind, can be calculated as:
Each term can be estimated as follows:
where ΔGMM is the molecular mechanics free energy, ΔGsol is the solvation free energy, and TΔS represents the entropy term. The molecular mechanics energy was calculated by the electrostatic and van der Waals interactions, while the solvation free energy was composed of the polar and the nonpolar contributions:
The contribution of polar solvation energy, ΔGele,sol, is calculated with the generalized born (GB) implicit solvent model, whereas the nonpolar part of the solvation energy, ΔGnonpol,sol, is dependent on the solvent accessible surface area (SA). TΔS are the contributions arising from changes in the degrees of freedom of the solute molecules, which were not considered here. Thus, values reported for the MM-GBSA calculations should be considered as “effective energies” (ΔGEff) rather than the free energies [38].
For MM-GBSA analysis, snapshots at 20 ps intervals were extracted from the last 20 ns of the MD trajectory, and the binding free energies were averaged over the ensemble of conformers produced. In an attempt to detect the hot spot residues, the effective binding energies were decomposed into contributions of individual residues using the MM-GBSA energy decomposition scheme introduced by Gohlke et al. [39].
Results
In order to fully understand the binding mode and key interactions of Keap1–peptide substrates, MD simulations of Keap1 bound to four known peptide substrates were carried out in combination with free energy calculations. These peptides were LDEETGEFL (the Nrf2 ETGE motif, PDB ID 1X2R), WRQDLD (the Nrf2 DLG motif, PDB ID 2DYH), QNEENGEQE (a nuclear oncoprotein, prothymosin α, PDB ID 2Z32) and VDPSTGEL (the selective autophagy substrate p62, PDB ID 3ADE). The Nrf2 ETGE motif and the Nrf2 DLG motif together function as a “hinge and latch model” that senses the oxidative/electrophilic stress [40–42]. While both prothymosin α and p62 have been found to participate in dissociating Nrf2 from the Keap1–Nrf2 complex through competitive binding to the Keap1 DC domain. Thus, deeply understanding of the binding mode and key interactions between Keap1 and known substrates can contribute to the rational design of PPI inhibitor of Keap1.
Static structure analysis of binding mode
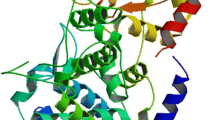
The binding interface in the X-ray crystal structure of the Keap1–peptide has been widely reported in the literature. In brief, the binding cavity of Keap1 contain five sub pockets as P1, P2, P3, P4 and P5 (shown in Fig. 1). The polar sub pockets, P1 and P2, have been proven of great importance in binding substrate. The P1, formed by residues Ser508, Phe478, Ile461, Arg483, Arg415 and Gly462, is highly positively charged and the electrostatic interactions with the Arg483 and Arg415 are significant for binding. In the four peptide substrates, the residues occupying this sub pocket are Glu, Gln, Glu and Asp, separately. The Gln in Nrf2 DLG motif which lacks the key carboxyl group makes the binding affinity getting an order of magnitude reduction. The P2 sub pocket, consisting of Ser363, Arg380, Asn382 and Asn414, is also positively charged. Moreover, the residues which occupy this sub pocket are all glutamates, indicating its conservation and important role in substrate recognition. Noteworthily, the Arg415 can take part in the P1 and P2 simultaneously. The P3 sub pocket, formed by Gly509 Ala556, Ser555, Ser602, Gly603 and Gly571, is occupied by the peptide backbone. These small-sized residues make this pocket sensitive to the steric hindrance. For example, the Ala mutation of the Gly in the Nrf2 ETGE motif nearly abolishes binding to Keap1. Unlike the P1, P2 and P3, the hydrophobic site P4 and P5 have been poorly investigated. In the case of Nrf2 ETGE motif, the side chains of Leu76, Glu78 and Phe83 cover both two sites, while for Nrf2 DLG motif, the two sites are nearly vacant. It may be one of the reasons why the ETGE motif and DLG motif show distinct binding affinity for Keap1. Although both P62 and Prothymosin α occupy the two hydrophobic sub-pockets, the binding modes show some differences. The hydrophobic residues of Prothymosin α, including Val348, Pro350 and Leu355, occupy the P4 and P5. On the contrary, P62 uses polar residues, including Gln40, Glu42 and Gln47, to occupy the two hydrophobic sub-pockets.
System stability examination
The convergence and stability of the simulations were monitored through the examination of the root-mean-square deviation (RMSD) of backbone atoms with respect to the structures obtained at the end of the equilibration procedure. The detailed results can be found in Fig. 2. As can be seen in the plots, the β-propeller structure of Keap1 shows good structure stability in all cases. In order to ensure the system stable and well equilibrated, only the MD trajectories from the last 20 ns of simulation of all systems were taken for MM-GBSA analysis.
Hydrogen bond interactions
With the aim of highlighting key binding determinants of the Keap1–substrate, we investigated the hydrogen bond interactions formed between Keap1 and the simulated peptide substrates along the MD trajectories. The hydrogen bonds of which occupancies were more than 20 % were considered and shown in the Fig. 3. The hydrogen bonds are colored as red (occupancy >80 %), purple (occupancy of 60–80 %), blue (occupancy of 40–60 %) and green (occupancy of 20–40 %). The detailed information can be found in Supporting Information Table S4.
Hydrogen bonds formed between Keap1 and peptide substrates along the MD trajectories. The hydrogen bonds of which occupancies were more than 20 % were considered and shown in the Figure. The hydrogen bonds are colored as red (occupancy >80 %), purple (occupancy of 60–80 %), blue (occupancy of 40–60 %) and green (occupancy of 20–40 %)
Hydrogen bonds formed via the peptide backbone fix the conformation of the peptide
Two strong canonical hydrogen bonds are formed in the P3 sub-pocket which is occupied by the peptide backbone. Both of the two strong hydrogen bonds are formed between the hydroxyl hydrogen of the serine, namely Ser555, Ser602, and the carbonyl oxygen of the peptide backbone. Strikingly, the two hydrogen bonds are located between the two polar cavities, P1 and P2. These conservative hydrogen bonds can stabilize the binding conformation of the peptide. While concerning Nrf2 DLG motif, one of the canonical hydrogen bonds, formed by Ser602 is absent, partly owing to the lack of the acidic amino acid in the P1 cavity. The residues connected after the Ser602 interacting carbonyl group are glycine and glutamate in all of the three cases indicating the structure conservation.
Another strong canonical hydrogen bond is formed between Gln530 (side chain NH2) and the carbonyl oxygen of the peptide backbone, of which occupancies were over 80 % in all cases (for P62, the occupancies are the sum of two amide hydrogens in the side chain). The Gln530, in the P4 sub-pocket, is located on the edge of the binding cavity, which can restrain the N-terminal of the peptide. The three conserved canonical hydrogen bonds together determinate the appropriate conformation of the peptide and drive the acidic residues of the peptide into the polar cavity. Besides, in the case of Nrf2 ETGE motif, the C-terminal carbonyl oxygen of the peptide backbone can also interact with the Asn382 (side chain NH2), which can make the binding conformation more stable.
Hydrogen bonds formed via the side chain of the peptide mainly interact with the polar sub pockets
The polar sub-pockets, P1 and P2, both form strong polar interactions with the peptide side chain. In the P1 sub-pocket, the hydrogen bond involved in the Arg415 can be found in the three complexes except the Nrf2 DLG motif which lacks the acidic res idues in this site. The Nrf2 ETGE motif could interact with all of the three polar residues, namely Arg415, Arg483 and Ser508, in the P1, indicating the strong binding in this site. Nonetheless, P62 only can form stable hydrogen bond with Arg415, suggesting the weak binding of Keap1–P62 in the P1. In the P2, the hydrogen bonds are more conservative than those in the P1. The hydrogen bonds formed between the acidic residues in the peptide and the Arg380, Asn382 and Ser508 in the Keap1 can be found in all cases except the Nrf2 DLG motif, which does not interact with Ser508. The occupancies of the Nrf2 DLG motif are also the smallest in the four complexes. Thus, the binding strength between the Nrf2 DLG motif and Keap1 in P2 cavity may be the weakest among the four cases. Noteworthily, the Prothymosin α can form stable hydrogen bond with the Tyr525 (side chain OH) using the side chain carboxyl group and this strong hydrogen bond cannot be found in other cases.
Binding energy calculation
Previous study [43] showed that MM-PBSA performed better in calculating the absolute binding free energies but that MM-GBSA performed better in calculating relative free energies. Herein, we focus on the insight into the binding energy origination of peptide ligands and the main driving force for their binding through comparison of four different peptide substrates, thus the MM-GBSA method was chosen to calculate binding energy. Binding energy calculated by MM-PBSA was also available in Table S1 for comparison.
Overall, the calculated effective binding energies of the four complexes all show negative values, indicating the favorable PPI in all cases. The Nrf2 ETGE and DLG motif gave big differences in the effective binding energies, consistent with the reported results that the binding constants of the two motifs show two orders of magnitude differences. However, the calculated values overestimate the binding free energy which can be partly explained due to two missing contributions: the lack of entropic contributions, which can be expected to be unfavorable in the case of the flexible peptides; and the lack of energetic contributions due to conformational changes, which were not considered here because of the use of the single trajectory approach [38].
As shown in Table 2, the decomposition of the binding free energy showed that the nonpolar contribution was the major part of the binding energy, but it was little changed among the four ligands. In contrast to the nonpolar contribution, the electrostatic contribution varied widely within the four substrates and it is the primary origin of the binding energy differences. The electrostatic contribution in the gas phase was favorable in all cases, indicating the strong polar interaction in Keap1–substrate binding. However, the desolvation penalties associated with the binding event was also huge, especially for the Nrf2 DLG motif. Only for the Keap1–Nrf2 ETGE motif complex, the total electrostatic contribution of binding was favorable.
Per-residue contributions
Next, we examined the energetic contributions of individual residue to seek the key one that dominates the Keap1–substrate binding.
Per-residue contributions of the peptides
The free energy decomposition of peptide ligand can give the energetic contributions of individual residue in the ligand. These information can quantitatively evaluate the importance of individual residue and sequentially guide the identification of small molecular inhibitor that mimic the binding determinants of protein–protein complexes. The per-residue contributions to the binding effective energy of the peptide ligand were listed in the Fig. 4. The most favorable interactions are formed by the Glu79 in the Nrf2 ETGE motif, whose contribution to ΔGeff is as low as −11.7 kcal/mol.
This agrees with an experimental mutation study on the residues in the Nrf2 ETGE motif [29]. The large contribution of Glu79 to the binding free energy is attributed to both the total nonpolar contributions and the total electrostatic contribution. The favorable electrostatic interaction as calculated by the molecular mechanics force field (ΔGELE) can offset the unfavorable electrostatic contribution due to desolvation (ΔGele,sol). The total electrostatic contribution is more than half of the effective binding energy (Supporting Information Table S2). Besides, the van der Waals contribution and the nonpolar part of solvation free energy are both favorable for binding. The huge contribution of the Glu79 in the Nrf2 ETGE motif indicates that the proper occupation of the P1 sub-pocket by the aliphatic carboxylic acid group should be addressed in the rational design of the Keap1 PPI inhibitors. The Glu43 of Prothymosin α also gave the similar result. The Asp349 of P62, which only forms strong canonical hydrogen bond with Arg380 in the P1 cavity, shows much lower contribution to the effective binding energy. The residue occupying the P2 cavity shows moderate effective binding energy and the binding free energy was attributed to the van der Waals contribution and the nonpolar desolvation term. It is noteworthy that some nonpolar residues show great influence on binding energy, such as Phe83 in Nrf2 ETGE motif, Ile28 in Nrf2 DLG motif and Leu355 in P62. The contributions of these nonpolar residues are mainly from the favorable van der Waals contribution. It partly explains why the nonpolar contribution dominated the effective binding energies of Keap1-Ligand.
Sub-pocket contribution of Keap1 binding cavity
In order to find the hot-spot residues in the Keap1 substrate binding cavity, we also evaluated the per-residue contribution of the residues on the binding surface. Each sub-pocket contributions were calculated and shown in Fig. 5 as the sum of component residues (detailed information of per-residue effective energy can be found in the Supporting Information). The most favorable interactions are formed in the P1 sub pocket in the Keap1–Nrf2 ETGE motif complex. This sub pocket’s contribution to binding is as low as −17.3 kcal/mol. It is consistent with that the most favorable residue, Glu79 in Nrf2 ETGE motif, occupied the P1 sub pocket. The mutation of key Arg415 and Arg483 of Keap1, which are responsible for the electrostatic interactions with the substrate, has also been found in human cancer cells [44, 45]. As shown in the Fig. 5, the Nrf2 ETGE motif and the Prothymosin α make use of the whole four sub pockets which are responsible for the most of the effective binding energies and the two peptides are also more potent than the others.
Per-residue contribution to the binding effective energy of the Keap1 bound to different peptide substrates. The per-residue contributions were calculated by applying the MM-GBSA decomposition approach to MD trajectories of Keap1 in complex with a Nrf2 ETGE motif, b Nrf2 DLG motif, c Prothymosin α peptide, and d P62 peptide. The per-residue contributions are mapped onto the starting structures of the simulation using a color code with a linear scale. Residues whose contributions to the effective free energy ΔGEff ≤ −1 kcal/mol (Table S3 in the Supporting Information) are labeled. e Per-pocket contribution to the effective binding energy. Keap1–Nrf2 ETGE motif was colored as blue, Keap1–Nrf2 DLG motif was colored as red, Keap1–Prothymosin α was colored as green and the Keap1–P62 was colored as purple
The polar sub pockets, P1 and P2, are the main source of effective binding energies. While the P3 pocket, which is occupied by the peptide backbone, is important for binding, but it has little contribution to the effective binding energy. Noteworthily, the hydrophobic sub pockets are also important for the binding energy, especially for the P4 sub pocket which is used by all of the four peptide ligands.
Discussions and conclusions
E3 ubiquitin ligases have been expected as attractive candidates of drug targets for a long time [5]. However, the successful cases are limited. The biggest obstacle may be that modulation of E3 ligase activities requires the targeting of PPI, which is highly challenging drug targets. Different from other kinds of PPI, the E3 ligase involved interactome has some promising characters. Each ligase degrades multiple proteins and many other proteins have been found to inhibit ubiquitination through disrupting the PPI of E3 ligase and its substrate. These natural binding partners of the E3 ligase can not only guide the discovery of novel substrates, but also benefit the discovery of the E3 ligase inhibitors. Herein, we present a novel strategy that using multiple substrates to elucidate the sub-pocket-activity and per-residue-activity relationship of the E3 ligase and substrate. MD simulation, MM-GBSA binding energy calculation and energy decomposition scheme were incorporated to give the quantitative contributions of sub-pocket and per-residue to binding. With the help of the results of multiple substrates, the hot spot of the E3 ligase can be obtained in the quantitative descriptions and the key residue of the substrate can be identified to directly design the inhibitors. The method was used for the elucidation of the intermolecular recognition between the E3 ligase Keap1 and its substrates, which can directly guide the discovery of novel Keap1 substrates. Several determinants were firstly identified for the modulator design, and the strong binding motif of Keap1 was firstly proposed for the substrate discovery.
Nonpolar interactions are the main source of the binding energy
Previous research has identified that the electrostatic interactions between the two acidic glutamates of the ETGE motif and conserved arginines (Arg380, Arg415, and Arg483) at the entrance of the central cavity on the bottom side of the β-propeller structure of Keap1-DC are critical for binding [41]. However, in this paper, we showed that the effective binding energy was mainly from the nonpolar contribution, especially the van der Waals interactions. Not only the hydrophobic amino acids, but also these polar residues with the carbon chains do contribute to the total nonpolar part of the binding energy. Both of the hydrophobic sub pockets, P4 and P5, can gave the positive and statistically significant impact on the binding. The residues located in the P4 and P5 sub pockets, such as Leu and Phe, indicate that these hydrophobic sites favor the large hydrophobic group.
Electrostatic interactions are the driving force for Keap1 binding
Although the electrostatic contribution of the effective binding energy was not in a dominant position, it varied widely within the four substrates and it played an important role in the distinction of the binding affinity. The total electrostatic contribution varied from −13.7 to 28.9 kcal/mol, while the total nonpolar only varied from −56.7 to −49.7 kcal/mol. Noteworthily, the polar P1 sub pocket shows extremely huge difference in the binding energy between different substrates, from −17.3 kcal/mol in Keap1–Nrf2 ETGE motif to −4.2 kcal/mol in P62, indicating that only the effective occupation of the P1 sub pocket could cause the favorable binding energy changes. While in the case of P62, though the Asp349 could interact with Arg415 in Keap1, it was not large enough to compensate the desolvation penalties associated with the binding event. Thus, in order to obtain the energy advantages in the polar sub pockets, the carboxy group in the polar sub pockets should be well positioned and properly linked by the carbon chains. Recently, the further study [46] of P62–Keap1 showed that Ser351 phosphorylation of the autophagy-adaptor protein P62 markedly increases P62’s binding affinity for Keap1. Phosphorylated Ser351 formed extra hydrogen bonds with Arg483 and Ser508 of Keap1 and showed similar binding mode compared to Glu79 in ETGE of Nrf2. ITC experiment results also showed that phosphorylated P62 for Keap1-DC is approximately 30-fold higher than that of nonphosphorylated P62 but fivefold lower than that of ETGE. Our research combining with these experimental results further confirmed that the electrostatic interactions involved in key arginines, Arg380, Arg415, Arg483, are the driving force for Keap1 binding.
Key Glu residue and its mimic are the core residues of the Keap1 binding motif
Previous studies have identified that the two Glu residues of the ETGE motif are important for binding depending on the experimental Alanine mutation results. Herein, we explained why the proper positioned glutamate was so important for Keap1 binding in the dynamic process. The carboxyl group of the glutamic acids can form multiple stable electrostatic interactions with the key arginines along the MD simulation. In addition, the hydrophobic carbon chain are also important for binding. The carbon chain not only contributes to the binding energy through the favorable vdW interaction, but also can keep the carboxyl group in the appropriate place. On contrast, in the case of the Nrf2 DLG peptide, the Asp residue did not make as much contribution as the Glu residue in the Nrf2 ETGE peptide to binding, which further indicated the core role of the Glu residue. The research of phosphorylated and nonphosphorylated P62 gave an alternative way to investigate the Keap1 substrate. Phosphorylated serine can mimic the Glu residue and form multiple hydrogen bonds with the polar residues of Keap1. Phosphorylated P62 has similar binding affinity compared to Keap1. Our results also showed that the Glu residue’s contributions to binding mainly originated from the electrostatic interactions formed by the carboxyl group and the nonpolar interactions involved in the carbon chain. It is reasonable that the phosphorylated Ser residue can replace the Glu in the ETGE motif. The well-known binding motif of Keap1 is the (D/N)XETGE, which has been validated by several substrates, such as IKK beta, PGAM5 and PALB2. However, inspired by these research results, all of these proteins that contain the motif (D/N)X(E/S)TG(E/S) could be the Keap1’s substrate. It can be used to find the potential Keap1 substrates and fulfill the Keap1 related interactome.
Keap1 inhibitor design based on the substrate recognition mechanism
Enhancing Nrf2 activity through modulating the Keap1 mediated Nrf2 inhibition has been identified as an important approach for prevention of cancer and other chronic diseases in which oxidative and inflammatory stress contribute [47]. However, most known Nrf2 activators are electrophilic species or metabolically activated to become electrophilic [48] and the molecular mechanism of these activators is the covalent binding to the thiol of the cysteine, which does not possess selectivity and specificity for the ubiquitous cysteines in cells. Thus, the targets of currently known Nrf2 activators could be promiscuous, which may lead to intolerable side effects. Intriguingly, PPI of Keap1–Nrf2 has evolved as a direct molecular target of ARE activation and opens up a new direction for the design of reversible ARE inducers with high specificity and potency [19]. These results reported in this paper can contribute to the rational design of PPI inhibitors of Keap1–Nrf2, which has been used in the Keap1–Nrf2 PPI inhibitor discovery [25]. Several tips could be suggested for the future design of PPI inhibitors of Keap1.
-
1.
The main driving force for the substrate binding is hydrophobic interactions. The total nonpolar part, especially the van der Walls contributions, is the main source of the binding energy. Thus fully occupying the five sub pockets is important to the design of potent PPI inhibitors.
-
2.
Effective occupying the pol ar sub pockets could effectively improve the binding affinity. Stable strong polar interactions with polar pockets do not mean the favorable binding energy changes. Only the carboxyl group in the P1 or P2 which is well positioned and appropriately linked to the scaffold could contribute to the total binding energy. The Glu79 of the Nrf2 ETGE motif in the P1 pocket is an excellent example.
-
3.
The scaffold of the inhibitor should occupy the P3 sub pocket to stabilize the binding conformation. Although the P3 sub pocket does not contribute to the total binding energy, it is important to fix the conformation of the ligand. The scaffold of the PPI inhibitors should occupy this cavity to enable the side chain with the appropriate conformation.
-
4.
The hydrophobic fragments located in the P4 and P5 sub pockets could give extra binding energy contributions. The total contributions of P4 and P5 are more than one-third of the total binding energy in all cases. The potent PPI inhibitors should make full use of the two pockets. The Leu, Phe and Ile used by the peptide ligands indicate that the hydrophobic sites favor the large hydrophobic group.
Abbreviations
- Keap1:
-
Kelch-like ECH-associated protein-1
- Nrf2:
-
Nuclear factor erythroid 2-related factor 2
- MD:
-
Molecular dynamics
- RMSD:
-
Root-mean square deviation
- MM-GBSA:
-
Molecular mechanics generalized born surface area
- PPI:
-
Protein–protein interaction
- CRL:
-
Cullin–RING ubiquitin ligases
- Cul3:
-
Cullin 3
- Rbx1:
-
Ring box 1
References
Spasser L, Brik A (2012) Chemistry and biology of the ubiquitin signal. Angew Chem Int Ed Engl 51(28):6840–6862
Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243(4898):1576–1583
Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19(1):94–102
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229
Cohen P, Tcherpakov M (2010) Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143(5):686–693
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2(3):169–178
Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78(1):399–434. doi:10.1146/annurev.biochem.78.101807.093809
Lydeard JR, Harper JW (2010) Inhibitors for E3 ubiquitin ligases. Nat Biotechnol 28(7):682–684
Garber K (2005) Missing the target: ubiquitin ligase drugs stall. J Natl Cancer Inst 97(3):166–167
Wells JA, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450(7172):1001–1009
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303(5659):844–848
Sun H, Lu J, Liu L, Yi H, Qiu S, Yang C-Y, Deschamps JR, Wang S (2010) Nonpeptidic and potent small-molecule inhibitors of cIAP-1/2 and XIAP proteins. J Med Chem 53(17):6361–6367
Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M (2010) An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol 28(7):733–737
Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J (2010) Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol 28(7):738–742
Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, Noblin DJ, Jorgensen WL, Ciulli A, Crews CM (2012) Targeting the von Hippel–Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J Am Chem Soc 134(10):4465–4468
Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, Gareiss PC, Jorgensen WL, Ciulli A, Crews CM (2012) Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew Chem Int Ed Engl 51(46):11463–11467
Motohashi H, Katsuoka F, Engel JD, Yamamoto M (2004) Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1–Nrf2 regulatory pathway. PNAS 101(17):6379–6384
Magesh S, Chen Y, Hu L (2012) Small molecule modulators of Keap1–Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev 32(4):687–726
Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, Li CW, Ding Q, Liao TL, Lai CC, Lin AC, Chang YH, Tsai SF, Li LY, Hung MC (2009) KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 36(1):131–140
Jiang Z-Y, Chu H-X, Xi M-Y, Yang T-T, Jia J-M, Huang J-J, Guo X-K, Zhang X-J, You Q-D, Sun H-P (2013) Insight into the intermolecular recognition mechanism between Keap1 and IKKβ combining homology modelling, protein–protein docking, molecular dynamics simulations and virtual alanine mutation. PLoS ONE 8(9):e75076
Niture SK, Jaiswal AK (2011) INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ 18(3):439–451
Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD (2013) Arsenic inhibits autophagic flux, activating the Nrf2–Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33(12):2436–2446
Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12(3):213–223
Jiang Z-Y, Lu M-C, Xu LL, Yang T-T, Xi M-Y, Xu X-L, Guo X-K, Zhang X-J, You Q-D, Sun H-P (2014) Discovery of potent Keap1–Nrf2 protein–protein interaction inhibitor based on molecular binding determinants analysis. J Med Chem 57(6):2736–2745
Hancock R, Bertrand HC, Tsujita T, Naz S, El-Bakry A, Laoruchupong J, Hayes JD, Wells G (2011) Peptide inhibitors of the Keap1–Nrf2 protein–protein interaction. Free Radic Biol Med 52(2):444–451
Sun H-P, Jiang Z-Y, Zhang M-Y, Lu M-C, Yang T-T, Pan Y, Huang H-Z, Zhang X-J, You Q-d (2014) Novel protein–protein interaction inhibitor of Nrf2–Keap1 discovered by structure-based virtual screening. MedChemComm 5(1):93–98
Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang M-I, Kobayashi A, Yokoyama S, Yamamoto M (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21(5):689–700
Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M (2007) Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27(21):7511–7521
Padmanabhan B, Nakamura Y, Yokoyama S (2008) Structural analysis of the complex of Keap1 with a prothymosin alpha peptide. Acta Crystallogr, Sect F: Struct Biol Cryst Commun 64(4):233–238
Simmerling C, Strockbine B, Roitberg AE (2002) All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc 124(38):11258–11259
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65(3):712–725
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A Second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117(19):5179–5197
Cheatham TE III, Miller JL, Fox T, Darden TA, Kollman PA (1995) Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc 117(14):4193–4194
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE III (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897
Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein–protein complex Ras–Raf. J Comput Chem 25(2):238–250
Fulle S, Withers-Martinez C, Blackman MJ, Morris GM, Finn PW (2013) Molecular determinants of binding to the plasmodium subtilisin-like protease 1. J Chem Inf Model 53(3):573–583
Gohlke H, Kiel C, Case DA (2003) Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. J Mol Biol 330(4):891–913
Tong KI, Kobayashi A, Katsuoka F, Yamamoto M (2006) Two-site substrate recognition model for the Keap1–Nrf2 system: a hinge and latch mechanism. Biol Chem 387(10–11):1311–1320
Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M (2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 26(8):2887–2900
Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M (2010) Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci USA 107(7):2842–2847
Hou T, Wang J, Li Y, Wang W (2010) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51(1):69–82
Takahashi T, Sonobe M, Menju T, Nakayama E, Mino N, Iwakiri S, Nagai S, Sato K, Miyahara R, Okubo K, Hirata T, Date H, Wada H (2010) Mutations in Keap1 are a potential prognostic factor in resected non-small cell lung cancer. J Surg Oncol 101(6):500–506
Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S (2008) Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res 68(5):1303–1309
Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M (2013) Phosphorylation of p62 activates the Keap1–Nrf2 pathway during selective autophagy. Mol Cell 51(5):618–631
Sporn MB, Liby KT (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12(8):564–571
Hur W, Gray NS (2011) Small molecule modulators of antioxidant response pathway. Curr Opin Chem Biol 15(1):162–173
Acknowledgments
This work is supported by the Project 81230078 (key program), 81202463 (youth foundation), 81173087 and 91129732 of National Natural Science Foundation of China, 2014ZX09507002-005-015, 2013ZX09402102-001-005 and 2010ZX09401-401 of the National Major Science and Technology Project of China (Innovation and Development of New Drugs)
Conflict of interest
The authors declare no other conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, ZY., Xu, LL., Lu, MC. et al. Investigation of the intermolecular recognition mechanism between the E3 ubiquitin ligase Keap1 and substrate based on multiple substrates analysis. J Comput Aided Mol Des 28, 1233–1245 (2014). https://doi.org/10.1007/s10822-014-9799-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9799-y