Abstract
Purpose
Worldwide publications follow the gold standard method—the polymerase chain reaction (PCR)—for detecting Y-chromosome microdeletions; however, markers are frequently variable between the studies. Can we detect the deletions by another molecular method with more genomic coverage? The Y chromosome harbors several different genes responsible for testicular development and spermatogenesis, and its repetitive conformation predisposes it to complex rearrangements that have clinical impact. Our aim was to evaluate a molecular diagnostic method, the Multiplex Ligand Probe-dependent Amplification (MLPA), which is also a valuable ancillary method for the identification of deletions, duplications, and rearrangements in a single and faster reaction, leading to a better comprehension of patients’ phenotypes, and should be considered a useful tool for detection of Y chromosome deletions.
Methods
This is a study of diagnostic accuracy (transversal prospective study) conducted to investigate Y-chromosome deletions in 84 individuals through PCR and MLPA methods. Forty-three infertile men (azoospermic and oligozoospermic) and 41 controls (40 fertile men and 1 normal karyotyped woman) were analyzed by PCR and MLPA techniques.
Results
We diagnosed seven (7) deletions (16.2%) by PCR and 9 with MLPA (21%). In addition, we found five (5) duplications and a suggestive mosaic.
Conclusion
Our results demonstrate that MLPA technique is valuable in the investigation of microdeletions and microduplications. Besides deletions, duplications can cause instability of chromosome genes, possibly leading to infertility. Both studied techniques provide an advantageous diagnostic strategy, thus enabling a better genetic counseling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infertility affects about 15% of couples attempting pregnancy, and, in approximately 50% of these cases, male factors are responsible. The second most frequent genetic cause of male infertility [1], which is clinically characterized by azoospermia and oligozoospermia depending on the amount of lost genetic material and the size of the affected region, is Y-chromosome microdeletions [2]. The majority of genes located in the Y chromosome are involved in male-related functions, such as gonadal differentiation and spermatogenesis [3,4,5].
The human Y chromosome is a small structure around 60 Mb comprising 63 genes, and it is basically composed of pseudoautosomal regions (PAR), euchromatin, and heterochromatin. The euchromatic region of the Y chromosome includes many pseudogenes or amplified genes [4, 6, 7].
The spermatogenesis locus was mapped in the euchromatic portion of Yq and was named azoospermia factor (AZF), because the first six men observed with terminal deletions in Yq were azoospermic [8].
The AZF region architecture contains repetitive homologous sequences that predispose it to chromosomal rearrangements. These have long been known to significantly impact fertility, causing pathogenic alterations such as deletions or duplications [4]. Microdeletions in the AZF sub-regions a, b, or c, lead to different clinical phenotypes, namely Sertoli cell-only syndrome (SCOS), spermatogenesis arrest, and hypospermatogenesis.
These deletions are usually de novo events, since fathers of affected patients usually do not present any microdeletions [9]. Given the Y chromosome’s vertical transmission to male offspring, all male descendants will inherit the microdeletions [10, 11]. This emphasizes the importance of genetic counseling for these patients.
The European Academy of Andrology (EAA) and the European Molecular Genetics Quality Network (EMQN) [1, 12] recommend the STS-PCR (sequence tagged sites–polymerase chain reaction) assays for detecting Y-chromosome microdeletions. However, STS-PCR detects deletions only in a specific portion of the Y chromosome, thus limiting the detection of other pathogenic copy-number variations (CNVs). In the literature, several reports associated the duplicated CNVs of the Y chromosome with spermatogenic failure [13,14,15,16]. Hence, new tests are needed to better evaluate these genomic variations.
Meeting such a need is the Multiplex Ligand Probe-dependent Amplification (MLPA) assay, which comprises up to 43 probes (mostly exons of a target gene) capable of detecting deletions and duplications in a single reaction. Each probe is specific for a different known DNA sequence for the purpose of evaluating the CNVs of the targets [17].
In this study, we analyzed azoospermic and oligozoospermic patients by both methods, aiming to determine if MLPA was more effective than the gold standard method (STS-PCR) in diagnosing Y-chromosome microdeletions.
Materials and methods
This is a transversal prospective study. Our study comprised two groups. The patients group had as inclusion criteria oligozoospermic and azoospermic men, who had their semen samples analyzed by the WHO criteria [18, 19], and the control group, whose inclusion criterium was fertile men who had undergone vasectomy. All the participants were attended in Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil, in the period of September, 2014 to January, 2018, forming consecutive series.
All individuals that agreed to participate had signed the informed consent and had their blood samples collected in EDTA tubes. Blood was used for genomic DNA extraction with the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA concentration and purity were evaluated by spectrophotometry (Nanodrop ND-2000, Thermo Fisher Scientific Inc., USA), and then DNA samples were analyzed by STS-PCR and MLPA techniques. Exclusion criteria were poor DNA concentration and purity.
Sample size calculation
According to the prevalence of Y-chromosome microdeletions in the infertile population, the sample size required for the study was a minimum of 38 individuals in each group, given a statistical power of 90% at a 5% significance level.
Molecular analysis
STS-PCR analysis
PCR was performed for the following specific STS markers of Y chromosome: SY84, SY86 (AZFa region); SY127, SY134 (AZFb region); SY254, SY255 (AZFc region); and SRY and ZFX/Y (short arm of Y chromosome) for controls [12]. This technique is, nowadays, the gold standard. Data were presented as absence or presence of Y-chromosome microdeletions.
MLPA analysis
The MLPA technique was performed using the SALSA MLPA probe-mix P360 version B1 (MRC Holland, Amsterdam, The Netherlands) kit following the manufacturer’s instructions. The kit contained 55 probes, of which 12 were located in autosomal chromosomes (for internal control reaction), and 43 were located in Y-chromosome AZF regions (16 AZFa, 15 AZFb, and 12 AZFc regions). Moreover, 9 control fragments were generated (with amplification products smaller than 120 nucleotides) to ensure the quality of the denaturation reaction and of DNA samples [17].
Separation of the amplification products via electrophoresis was performed using an ABI 3500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and the data were analyzed using GeneMarker software, version 1.6 (www.softgenetics.com-Softgenetics, State College, Pennsylvania, USA).
The peak area of each fragment was compared with that of a control sample, and the results were considered abnormal when the relative peak-height ratio was less than 0.75 (deletion) or greater than 1.25 (duplication). (www.mlpa.com).
Considering the genomic map calculated by the distance between the Y-chromosome telomere of the short arm and MLPA probes or STSs, it should be kept in mind that the sub-regions analyzed by both techniques are not the same, but side by side (Table 1).
Cytogenetics and fish analysis
We evaluated a single patient with abnormal results by MLPA, using two different methodologies. We analyzed 20 metaphases cells using G-Band. Subsequently, we use fluorescent in vitro hybridization (FISH) with the Probe LPE0XYc—Chromosome X Alpha and Y Alpha Satellite Probes (Cytocell, Cambridge, UK) in order to improve the results.
Statistical analysis
Statistical analysis was performed with SPSS (SPSS for the Social Sciences, version 14.0) software.
Ethics approval
The Research Ethics Committee of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo (HC-FMUSP) approved this study, and written informed consent for publication was obtained from the patients (CAPPesq # 535.321).
Results
The intended number of patients to participate was 86 people, independent of racial or demographic status, and after eligibility criteria, the final number was 84 people, summarized in Fig. 1.
The present study included 84 individuals as follows: 43 infertile men (azoospermic and oligozoospermic) and 41 controls (40 fertile men, and one (1) healthy woman). All the DNA samples were capable to be analyzed by both techniques, and there were no excluded patients.
The study population is admixed, including Caucasian, Black, Yellow, and Pardo ethnicities.
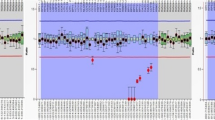
We detected seven (7) deletions by the PCR method (16.3%) and nine (9) by MLPA (21%). Furthermore, MLPA detected five (5) duplications (being one an extra X chromosome, a control probe) and one (1) case suggestive of mosaic (Table 2, Fig. 2).
None of the availed patients revealed partial AZFc deletions, such as described by Rozen et al. [20]. In MLPA technique, the presence of partial AZFc deletions is given by the exact copy number of some probes, as described by manufacturers.
The PCR revealed a sensitivity of 77% with 95% of accuracy for these patients when compared to the MLPA.
PCR results from azoospermic patients revealed one (1) patient with AZFa deletion (7.1%), three (3) patients with AZFbc deletion (21.4%), and one (1) patient with AZFc (7.1%). Results obtained from oligozoospermic patients revealed only two (2) AZFc deletions (6.9%).
The use of MLPA enabled the detection of an extra X chromosome, corroborating the diagnosis of Klinefelter Syndrome (47, XXY) for patient M37, obtained also by G-banding karyotype. Three (3) of our patients (M7, M14, and M28) presented the same duplication in the AZFc region, which involved probes located at the BPY2, DAZ, and CDY1B genes. One patient (M29) had duplications in the SRY probe (short arm of Y chromosome) and in all of the AZFa probes, along with deletion in AZFb and nearly complete deletion in AZFc. Another patient with normal PCR results (M43) had only one copy of all of the 43 Y-chromosome probes detected. These data suggested the presence of a possible mosaic, confirmed later by G-banding karyotype (Fig. 3) and FISH results revealing ishX(DXZ1x1,DYZ3x0)[283]/X(DXZ1x1),Y(DYZ3x1)[212]/X(DXZ1x2),Y(DYZ3x0) [2]/X(DXZ1x2),Y(DYZ3x1) [2] (Fig. 4).
Discussion
The integrity of Y chromosome is critical for spermatogenesis and sexual differentiation and determination. Due to advances in molecular techniques, abnormalities of Y chromosome have been more accurately identified, therefore reducing underdiagnosed pathogenic changes and improving the genotype-phenotype relation [4, 21].
Several authors report that the increased number of regions investigated in Y chromosome lead to an improvement of diagnosis. Thus, our findings corroborate with literature data and showed that MLPA is also a useful molecular tool for detecting Y chromosome microdeletions in AZF regions. In addition, using the MLPA technique, it is possible to identify other types of abnormalities, such as duplication, mosaicism, and complex rearrangements [22, 23].
This study corroborates the findings of pathogenic CNVs in AZF regions by the few reports in the literature discussing such data [15, 16].
Success in finding mature sperm cells in azoospermic patients is dependent on the deleted region. A few years ago, men who showed complete or partial AZFb or AZFbc deletions had no hope of finding sperm cells in testicular sperm extraction (TESE) [24, 25]. However, there are studies reporting the detection of these cells in the patients, even in the penile ejaculate [26,27,28,29], pointing to the need for a careful reevaluation of these cases. These findings are possibly due to advances in detection techniques, which have more genomic coverage.
The MLPA technique has two main limitations: a mutation or polymorphism in the sequence detected by a probe may cause a reduction in relative height peak, even if the mutation is not located at the binding site. In addition, probe signal intensity may vary according to DNA purity, and this variation may be associated with the extraction method, elution solution, degradation degree, and presence of contaminants, such as residual reagents, RNA, or others [17, 30].
Duplication findings are controversial. Some authors suggest that duplications may affect male fertility [31], or are secondary to partial AZFc deletions could restore the concentration of motile spermatozoa to the normal value [32], while others suggest that duplications in the AZFc region do not affect spermatogenesis [33]. We must also consider the presence of several polymorphic deletions in fertile men [34]. The aforementioned results may justify the presence of duplications in two of our fertile control men.
Moreover, Lu et al. (2014) evaluated the degree of spermatogenic involvement of the multiple copies in AZFc genes by gene dosage in this region of eight families, and they found that only the CNVs of the DAZ and BPY2 genes were associated with spermatogenic failure. This finding may explain the infertility of our three patients who presented duplication in these same gene probes.
Notwithstanding all of the genomic data generated by the most recent cytogenomic techniques, it is still a challenge to correlate these data to new phenotypic profiles, clearly showing the need for more studies for a fuller understanding of such effects, allowing patients to receive increasingly individualized and more effective therapy.
Conclusion
This study demonstrated that MLPA analysis of Y chromosome is a valuable ancillary method for the identification of micro alterations associated with infertility in Brazilian patients.
References
Krausz C, Hoefsloot L, Simoni M, Tüttelmann F. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: State-of-the-art 2013. Andrology. 2014;2:5–19.
Esteves SC, Agarwal A. Novel concepts in male infertility. Int Braz J Urol. 2011;37:5–15.
Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–72.
Krausz C, Casamonti E. Spermatogenic failure and the Y chromosome. Hum. Genet. 2017;136:637–55.
Quintana-Murci L, Krausz C, McElreavey K. The human Y chromosome: function, evolution and disease. Forensic Sci Int. 2001;118:169–81.
Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–6.
Vogt PH. Molecular genetics of human male infertility: from genes to new therapeutic perspectives. Curr Pharm Des. 2004;10:471–500.
Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum. Genet. 1976;34:119–24.
Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenes Y chromosome microdeletions and alterations of spermatogenesisis. Endocr Rev. 2001;22:226–39.
Kühnert B, Gromoll J, Kostova E, Tschanter P, Luetjens CM, Simoni M, et al. Case report: natural transmission of an AZFc Y-chromosomal microdeletion from father to his sons. Hum Reprod. 2004;19:886–8.
Cram DS, Ma K, Bhasin S, Ariasc J, Pandjaitanc M, Chu B, et al. Y chromosome analysis of infertile men and their sons conceived through intracytoplasmic sperm injection: vertical transmission of deletions and rarity of de novo deletions. Fertil Steril. 2000;74:909–15.
Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int. J. Androl. 2004;2:240–9.
Vaszkó T, Papp J, Krausz C, Casamonti E, Géczi L, Olah E. Discrimination of deletion and duplication subtypes of the deleted in azoospermia gene family in the context of frequent interloci gene conversion. PLoS One. 2016;11:1–28.
Giacco DL, Chianese C, Sanchez-Curbelo J, Bassas L, Ruiz P, Rajmil O, et al. Clinical relevance of Y-linked CNV screening in male infertility: new insights based on the 8-year experience of a diagnostic genetic laboratory. Eur J Hum Genet. 2014;22:754–61.
Bunyan DJ, Callaway JLA, Laddach N. Detection of partial deletions of Y-chromosome AZFc in infertile men using the multiplex ligation-dependent probe amplification assay. J Reprod Infertil Avicenna Research Institute. 2012;13:174–8.
Saito K, Miyado M, Kobori Y, Tanaka Y, Ishikawa H, Yoshida A, et al. Copy-number variations in Y-chromosomal azoospermia factor regions identified by multiplex ligation-dependent probe amplification. J Hum Genet. 2015;60:127–31.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57 1–13.
Barratt CLR, Mortimer D, Jouannet P. ‘ How to count sperm properly ’: checklist for acceptability of studies based on human semen analysis. Hum Reprod. 2016;31:227–32.
WHO laboratory manual for the examination and processing of human semen, 5th ed. World Health Organization; 2010.
Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, Oates RD, et al. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 y chromosomes. Am J Hum Genet. 2012 Nov 2;91(5):890-6.
Patsalis PC, Skordis N, Sismani C, Kousoulidou L, Koumbaris G, Eftychi C, et al. Identification of high frequency of Y chromosome deletions in patients with sex chromosome mosaicism and correlation with the clinical phenotype and Y-chromosome instability. Am J Med Genet. 2005;135(A):145–9.
Jiang Y, Wang W, Guo Q, Sha Y, Ouyang H, Zhou Y. Multiplex ligation-dependent probe amplification for detecting AZF microdeletions on the Y chromosome in infertile men with azoospermia or severe oligozoospermia. Zhonghua Nan Ke Xue. 2012;18:115–21.
Martinez MC, Bernabés MJ, Gómez E, Ballesteros A, Landeras J, Glover G, et al. Screening for AZF deletion in a large series of severely impaired spermatogenesis patients. J Androl. 2000;21:651–5.
Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb, and AZFc regions. Hum Reprod. 2003;18:1660–5.
Choi JM, Chung P, Veeck L, Mielnik A, Palermo GD, Schlegel PN. AZF microdeletions of the Y chromosome and in vitro fertilization outcome. Fertil Steril. 2004;81:337–41.
Kleiman SE, Yogev L, Lehavi O, Hauser R, Botchan A, Paz G, et al. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature (1994-2010). Fertil Steril. 2011;95:2005–12.
Liu XY, Wang RX, Fu Y, Luo LL, Guo W, Liu RZ. Outcomes of intracytoplasmic sperm injection in oligozoospermic men with Y chromosome AZFb or AZFc microdeletions. Andrologia. 2017;49:1–6.
Stouffs K, Vloeberghs V, Gheldof A, Tournaye H, Seneca S. Are AZFb deletions always incompatible with sperm production? Andrology. 2017;5:691–4.
Zhang YS, Li LL, Xue LT, Zhang H, Zhu YY, Liu RZ. Complete azoospermia factor b deletion of Y chromosome in an infertile male with severe oligoasthenozoospermia: case report and literature review. Urology. 2017;102:111–5.
Kozlowski P, Jasinska AJ, Kwiatkowski DJ. New applications and developments in the use of multiplex ligation-dependent probe amplification. Electrophoresis. 2008;29:4627–36.
Lin YW, Hsu LCL, Kuo PL, Huang WJ, Chiang HS, Der Yeh S, et al. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28:486–94.
Noordam MJ, Westerveld GH, Hovingh SE, van Daalen SKM, Korver CM, van der Veen F, et al. Gene copy number reduction in the azoospermia factor c (AZFc) region and its effect on total motile sperm count. Hum Mol Genet. 2011;20:2457–63.
Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124:399–410.
Vogt PH. Genomic heterogeneity and instability of the AZF locus on the human Y chromosome. Mol Cell Endocrinol. 2004;224:1–9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Research Ethics Committee of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo (HC-FMUSP) approved this study, and written informed consent for publication was obtained from the patients (CAPPesq # 535.321).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Franchim, C.S., Soares-Junior, J.M., Serafini, P. et al. Efficacy of MLPA for detection of Y-chromosome microdeletions in infertile Brazilian patients. J Assist Reprod Genet 37, 1251–1259 (2020). https://doi.org/10.1007/s10815-020-01777-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-020-01777-8