Abstract
Purpose
To investigate the association of cumulus cell (CC)-related expression of a selected cluster of key genes (PTGS2, CAMK1D, HAS2, STC1, and EFNB2) with embryo development to blastocyst.
Methods
Exploratory study at a private clinic. Eighteen advanced maternal age patients were enrolled (37.3 ± 4.0 years). Seventy-five cumuli were collected, whose oocytes resulted in either developmental arrest (N = 33) or blastocyst formation (N = 42). The noninvasive CC gene expression was combined with time-lapse morphokinetic parameters and, for blastocysts, with qPCR-based aneuploidy testing on trophectoderm biopsies.
Results
The detection rate was 100% for all transcripts, but STC1 (96%) and CAMK1D (89%). Among amplified assays, CC mean expression levels of CAMK1D, PTGS2, and HAS2 were higher from oocytes that developed to blastocyst. No difference in CC key gene expression was reported between euploid (N = 21) and aneuploid (N = 21) blastocysts. Some timings of early embryo development were faster in embryos developing to blastocyst (time of pronuclei appearance and fading, division to two- and four-cells, first and second cell cycles). However, the generalized linear models outlined increasing CAMK1D expression levels as the strongest parameter associated with oocytes’ developmental potential from both a general (AUC = 0.78 among amplified samples) and an intrapatient perspectives (AUC = 0.9 among patients obtaining ≥ 2 zygotes from the cohort with different developmental outcomes).
Conclusions
CAMK1D level of expression in CCs associated with blastocyst development. If confirmed from larger studies in wider populations of patients, the investigation of CC key gene expression might suit IVF clinics not adopting blastocyst culture. Future investigations should clarify the role of CAMK1D in ovarian physiology and could provide novel insights on how oocytes gain competence during folliculogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The goal of IVF is to maximize couple’s reproductive potential and reduce their clinical risks. In this regard, frozen single-embryo transfer (SET) on an unstimulated endometrium is pivotal to minimize both ovarian hyperstimulation syndrome and multiple pregnancies [1]. The need for novel biomarkers of oocyte and embryo developmental and reproductive competence, pivotal to prioritize the embryo(s) to transfer and adopt a SET strategy, is indeed a central topic in modern IVF. At present, embryo selection is still largely based on morphological scoring, but embryo morphology in day 2 or day 3 of preimplantation development showed a relatively low predictive power upon the reproductive competence [2, 3]. Alternatively, many authors suggested blastocyst culture as a more effective strategy to conduct a better selection of competent embryo(s) [4, 5]. However, blastocyst culture requires an upskilling of the lab, which needs to ensure accurate culture conditions and an appropriate oxygen tension [6]. Time-lapse morphokinetics (TLM) provided new promises for the prediction of embryo competence. Indeed, different studies demonstrated that morphokinetic evaluation at early stage of embryo development could predict blastocyst formation [7, 8]. Other authors proposed some algorithms to use morphokinetic parameters as indicators for improved clinical outcomes during IVF [9]. However, to date, there is no consensus on which algorithm may be clinically relevant, and a systematic review showed no significant advance deriving from TLM [10].
Chromosomal aneuploidies per se represent the most important cause of implantation failure and miscarriage in humans [11, 12]. The aneuploidy rate of the blastocysts increases from a 25–30% baseline level in women younger than 35, up to > 90% in women older than 42 [13, 14]. Therefore, in the last decades, preimplantation genetic testing of aneuploidies (PGT-A) has been theorized and applied to discriminate euploid from aneuploid embryos based on the analysis of a biopsy. Euploid SET after PGT-A then, regardless of woman age, allows a higher implantation rate per transfer (~ 50%) and a lower miscarriage rate with respect to standard IVF, as reported in two meta-analyses [15, 16]. Yet, PGT-A should be conducted at the blastocyst stage on trophectoderm (TE) biopsies through 24-chromosome testing techniques, to obtain a reliable diagnosis and not to affect embryo reproductive competence [17, 18]. To date, reliable and solid noninvasive criteria that might predict chromosomal constitution have not been defined yet [19]. For instance, two studies outlined some TLM criteria as predictive of embryo chromosomal constitution [20, 21], but when tested on a different dataset, neither of them was confirmed [22]. Therefore, novel biomarkers are required in IVF to complement PGT-A and possibly increase our predictive power upon implantation.
Looking more deeply into the microenvironment in which the oocyte acquires its developmental and reproductive competence, cumulus (CC) and granulosa cells (GC) have been proposed as a mirror of oocyte quality, mainly because of the crucial crosstalk established during folliculogenesis between these somatic cells and the oocyte [23]. Additionally, the oocyte orchestrates its own microenvironment through the release of oocyte-secreted factors, like growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15), which in turn regulate CCs’ gene expression during folliculogenesis [24,25,26]. Therefore, many authors focused their interest on CCs’ gene expression to obtain more information upon the oocyte health status. Many papers reported different CCs’ gene expression patterns to be correlated with several IVF outcomes, for instance prostaglandin-endoperoxide synthase 2 (PTGS2) was positively correlated with oocyte nuclear maturation [27]. Similarly, other interesting studies revealed that the upregulation of hyaluronic acid synthase 2 (HAS2), gremlin 1 (GREM1), and PTGS2 is associated to oocyte developmental competence and increased embryo morphological quality [28,29,30]. Other authors analyzed the mRNA expression levels of three genes in the CCs, namely angiogenin (ANG), regulator of G protein signaling 2 (RGS2), and perilipin 2 (PLIN2), as potential predictors of blastocyst development [31]. In general, various studies identified genes expressed in CCs as candidate noninvasive biomarkers of oocyte and embryo quality and/or successful clinical outcomes as recently reviewed by Kordus and LaVoie [32]. All these papers shed light on basic mechanisms of follicle biology, thereby underlining that what happens to the oocyte during folliculogenesis may have an effect later on embryo development. However, the endpoints were often heterogeneous, and this resulted in some discrepancies in the results across the studies. For instance, ephrinB2 (EFNB2) was found upregulated by Wathlet and colleagues in CCs of embryos that resulted in a delivery after blastocyst SET [33]. In contrast, Burnik Papler and colleagues found that EFNB2 was downregulated in the CCs of embryos that resulted in a pregnancy after SET [34], and imputed the discrepancy of the results to the different primary outcome set, namely the live birth in Whatlet’s study, and the clinical pregnancy in theirs. To date, despite the intriguing hypotheses and the promising results achieved, no clear data are yet available to define CCs’ gene expression as a noninvasive tool to assess oocyte competence.
The purpose of this exploratory study was to validate expression patterns of previously described CC genes and to correlate them with oocytes’ developmental competence to blastocyst. Therefore, blastocyst prediction was set as primary outcome of the analysis. Additionally, the main technologies of embryo quality assessment currently applied in our clinic, namely TLM and aneuploidy testing, were combined with target gene expression of CCs to produce an integrated overview of oocyte competence.
Materials and methods
Selection of the five key genes to be analyzed from the cumulus cells
We performed a systematic review of the studies published from 2004 up to September 2015 that outlined putative biomarkers of oocyte and embryo quality and/or pregnancy among the genes expressed in the CCs. Pertinent research articles published in English were acquired and reviewed. The keywords included in the search were “cumulus cells,” “granulosa cells,” “gene expression,” “cumulus cells transcriptome,” “oocyte competence,” “non-invasive biomarker,” “blastocyst stage,” and “oocyte quality.” We focused our investigation on CC gene expression of two genes previously reported in many papers as involved in cumulus expansion and correlated with good-quality embryo development: HAS2 and PTGS2 [27, 28]. Then, we added to our pattern another three candidate genes, namely ephrinB2 (EFNB2), calcium/calmodulin-dependent protein kinase 1D (CAMK1D), and stanniocalcin-1 (STC1). The former two (EFNB2 and CAMK1D) previously found upregulated, the latter (STC1) instead downregulated, in the CCs of reproductively competent oocytes that resulted in a delivery after blastocyst SET [33]. All these genes are involved in the regulation of late folliculogenesis [35,36,37,38], but CAMK1D whose role in the ovary is yet to be unveiled.
The purpose of this study was therefore to correlate the expression of these five key genes in the CCs, to the in vitro embryonic development up to the blastocyst stage.
Study design and patient population
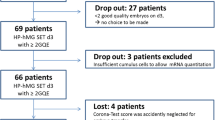
This is an exploratory study, approved by the IRB of Clinica Valle Giulia, G.EN.E.R.A. Center for Reproductive Medicine (Italy). From September 2015 to September 2016, 18 advanced maternal age (AMA) patients (37.3 ± 4.0 years; body mass index 22.2 ± 3.6) provided informed consent and were included in the study. Trying to limit any putative confounder that could affect blastocyst formation, severe male factor (sperm count lower than 1 × 106/mL), surgical sperm extraction, and female pathologies such as endometriosis and premature ovarian failure were set as exclusion criteria. Controlled ovarian stimulation for the present study population was performed only using GnRH antagonist protocol, as previously described [1, 39]. All the enrolled patients underwent ICSI [40]. We included only oocytes that fertilized after insemination.
Collection of CCs and time-lapse embryo culture
The follicular fluid was retrieved 36 h after hCG triggering. It was inspected under a laminar flow hood as described previously [41]. Cumulus oocyte complexes (COCs) were collected and incubated at 37 °C, 6% CO2, and 5% O2 for at least 2 h in continuous single culture medium (CSCM, Irvine Scientific, USA) until denudation and ICSI. Collection of CCs was performed individually for each COC. Each COC was cultured for 10 s into a droplet of Hepes-buffered medium containing 20 IU/μL hyaluronidase (Irvine Scientific, USA) and washed sequentially in two droplets of medium without enzyme. CCs were mechanically removed using stripper pipettes with decreasing diameters (170 μm and 140 μm, respectively; COOK Medical, USA). Soon after dissociation from the oocyte, the whole cumuli were washed in cold PBS, centrifuged for 10 min at 1200 rpm, then further processed for RNA extraction. During ICSI and embryo culture, the oocytes were kept individually to assess the correlation with their respective CCs’ key gene expression. Injected oocytes were transferred in EmbryoSlide microwells, each one in 25 μl of CSCM (Irvine Scientific) at 37 °C, 6% CO2 and 5% O2 in a time-lapse culture system (Embryoscope, Vitrolife, Sweden) up to the fully expanded blastocyst stage (tEB). The blastocysts produced underwent TE biopsy [42] and qPCR-based PGT-A [43, 44].
A total of 75 CC samples were individually collected and analyzed for the expression of the 5 candidate key genes. The primary outcome of this exploratory study was to outline the correlation between CC key gene expression pattern and blastocyst formation. We analyzed the CC key gene expression of the whole cohort of zygotes produced by each patient, regardless of whether the embryos reached the blastocyst stage or arrested their preimplantation development. Therefore, it was possible to cluster the samples into two biological groups: CCs whose oocytes arrested their preimplantation development versus CCs whose oocytes reached the blastocyst stage. Moreover, the blastocysts were subclustered according to their morphology. Blastocyst quality was assessed immediately before TE biopsy, graded according to the criteria defined by Gardner and Schoolcraft in 1999 and categorized in four groups: excellent, group 1 (≥AA); good, group 2 (AB and BA); average, group 3 (BB, AC and CA); poor, group 4 (<BB) based on inner cell mass (ICM) and TE quality score [42, 45, 46].
TLM-based assessment of preimplantation development was performed to determine whether any change in timings could be associated to CC key gene expression and blastocyst development. One embryologist performed the TLM parameter annotations, as previously described [22]. The TLM parameters considered were second polar body extrusion (tPB2), pronuclear appearance (tPNa), pronuclear fading (tPNf), and completion of cleavage to two, three, four, and five cells (t2, t3, t4, and t5, respectively). We also considered the length of cell cycles at the first and second divisions (cc1 and cc2, respectively) and the length of synchronization of cell divisions (s2). All the timings from t8 (cleavage to eight cells) to expanded blastocyst stage (tEB) were annotated but not compared in the two groups. Indeed, the main stage of embryo arrest during preimplantation development in humans corresponds to the embryonic genome activation (EGA), which in our species occurs between the four- and eight-cell stages [47]. Beyond the t5, more than half of the embryos would undergo arrest, thereby preventing a reasonable comparison between the two groups (i.e., “arrested embryos” versus “blastocysts”). Therefore, the purpose of the TLM comparison here conducted was to outline the predictivity on blastocyst formation derived from embryo behavior during the very first divisions.
RNA extraction, cDNA synthesis, and qPCR
Total RNA was extracted using TaqMan Gene Expression Cell-to-Ct Kit (Thermo Fisher Scientific, USA). The extraction protocol was performed using lysis solution with DNase I (Thermo Fisher Scientific) following manufacturer’s instruction. Each CC sample was processed individually adding 50 μL of lysis solution with 0.5 μL of DNase I, for 5 min at room temperature. Soon after, the reaction was stopped adding 5 μL of stop solution to each CC sample, to avoid mRNA degradation. The RNA extraction was concluded after the incubation at room temperature for 2 min. Each sample was then laid on ice until reverse transcription, which was performed according to the manufacturer’s protocol in a final 100 μL reaction volume for each CC sample. The thermocycler program used was 37 °C for 60 min, 95 °C for 5 min, and 4 °C ∞. For each experiment, we generated two negative controls by omitting the reverse transcriptase enzyme mix or the RNA in the reaction, respectively.
Quantitative PCR for the selected key genes was performed using ViiA7 Real-Time PCR System (Thermo Fisher Scientific) in a 20 μL reaction, of which 10 μL of gene expression master mix, 5 μL of pure water, 1 μL of Taqman primers for CAMK1D, STC1, EFNB2, PTGS2, HAS2 (the five target key genes), B2M or UBC (the two selected endogenous controls) (Supplementary Table 1), and 4 μL of cDNA for each gene. The program was 50 °C for 2 min, 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Each reaction was performed in triplicate. The geometric mean of the Ct values of B2M and UBC was adopted to normalize the gene expression. Both these genes were already suggested to be stable in human CCs through the geNorm program and adopted from previous studies [33, 48, 49]. PCR no-template control was included for each experiment. The levels of gene expression are reported as 2−dCT [50]. In case of failure of detection of a given gene, the Ct value was neither replaced with an arbitrary value (e.g., “40”), nor estimated based on the other transcripts detected from the same CCs sample, since both these strategies might result in a bias greater than excluding them [51].
Statistical analysis
Continuous data are presented as mean ± SD and range. Shapiro–Wilk tests were conducted to investigate a normal distribution of the data. Mann–Whitney U or Kruskal–Wallis tests were conducted to assess heterogeneity and homogeneity. Categorical variables are presented as absolute and percentage. Fisher’s exact or chi-squared tests were used to assess differences between categorical variables. A p value ≤ 0.05 was considered significant. The software G*Power was used to define the effect sizes and for post hoc power analyses. The software R was used for statistics, generalized linear models, and receiver operating characteristic (ROC) curve analyses.
Results
Embryo development and morphokinetic analysis
Figure 1a shows the developmental outcome of the 75 oocytes whose cumuli were isolated to be included in this study from 18 consenting patients. During in vitro preimplantation development, 33 embryos arrested (44.0%), while 42 developed to the blastocyst stage (56.0%). The chromosomal status of the embryos that reached blastocyst stage was assessed by qPCR. Twenty-one embryos were euploid (50.0%) and 21 aneuploid (50.0%). No difference was reported in terms of timing of CC sampling from the trigger in the two study groups (4.7 ± 0.7 h, 2.8–6.3 h vs 4.6 ± 0.7 h, 2.8–6.1 h). Among the 33 arrested embryos, only 4 arrested before the t8 (12.1%), most of them arrested their development at the t8 (N = 13, 39.4%), the rest at the time of morula formation (tM; N = 7, 21.2%) or of starting and full blastulation (tSB and tB, N = 9, 27.3%) (Fig. 1b). We compared the recorded timings from the embryos that arrested versus the embryos that developed to blastocyst up to t5 (i.e., five-cell stage). Significant differences, namely slower kinetics, were reported in embryos that arrested their development with respect to embryos that reached the blastocyst stage for tPNa, tPNf, t2, cc1, t4, and cc2 (Table 1; Supplementary Table 2).
a Characteristics of the patients and related oocytes whose cumulus cells were included in the study. b Timings of developmental arrest for the 33 arrested embryos. Second polar body extrusion (tPB2), completion of cleavage to two, three, four, five and eight cells (t2, t3, t4, t5, and t8, respectively), time of morula formation (tM), time of starting blastulation (tSB), time of full blastulation (tB)
Cumulus cell expression of EFNB2, PTGS2, HAS2, STC1, CAMK1D versus oocyte developmental competence and chromosomal constitution
The qPCR analysis outlined comparable mean Ct values in the two groups for B2M and UBC (Supplementary Fig. 1). The expression of STC1, EFNB2, CAMK1D, PTGS2, and HAS2 was therefore analyzed and correlated to embryo developmental competence. EFNB2, PTGS2, and HAS2 were amplified from all the tested samples; STC1 was amplified in 97% (n = 32/33) of the cumuli from arrested embryos and 95.2% (n = 40/42; p = 1.0) of those from blastocysts; CAMK1D was detected in 94% (n = 31/33) of the cumuli from arrested embryos and 87% (n = 36/42; p = 0.45) of those from blastocysts. For both of these key genes, the geometric mean of B2M and UBC Ct values was significantly lower from the samples that showed amplification than from the samples resulting in no detection, thereby suggesting either a lower efficiency of the qPCR reaction or a lower number of cells collected (Supplementary Fig. 2).
The levels of CAMK1D (mean 2−dCT in arrested embryos versus blastocysts: 0.025 ± 0.018, 0.003–0.066 versus 0.07 ± 0.056, 0.006–0.241; p = 0.01), PTGS2 (0.22 ± 0.21, 0.008–0.954 versus 0.59 ± 0.78, 0.027–4.336; p = 0.05) and HAS2 (0.24 ± 0.29, 0.015–1.304 versus 0.53 ± 0.73, 0.056–3.575; p = 0.03) were significantly lower in the CCs isolated from oocytes that resulted into embryo developmental arrest rather than reaching the blastocyst stage (Fig. 2a–c). No statistically significant difference was instead found in the expression levels of STC1 and EFNB2 in the two study groups (Supplementary Fig. 3).
Relative expression levels of CAMK1D (a), PTGS2 (b), and HAS2 (c) from the cumulus cells of developmentally incompetent versus competent oocytes. Total RNA was extracted from cumulus cells obtained from oocytes that resulted in either arrested embryos or blastocysts after IVF. Quantitative-PCR was conducted using Taqman primers. Each reaction was performed in triplicate. Each sample was normalized to its B2M and UBC mRNA content. For each key gene, the boxplots of the 2−dCT data are shown and the median of their distribution among all the samples analyzed is highlighted by a dotted line. Mann–Whitney U tests were conducted to assess statistically significant differences. Circles and stars represent outlier values
A subanalysis of CAMK1D, PTGS2, and HAS2 data among CCs related to oocyte developing as euploid or aneuploid blastocysts was conducted, but no significant difference was reported (Supplementary Fig. 4). Nonetheless, the samples size was limited; therefore, it cannot be excluded an effect to be reported from future studies targeting blastocysts’ chromosomal constitution as primary outcome.
In a further subanalysis, we clustered the CC samples, whose oocytes developed to blastocyst, according to embryo morphological quality (i.e., “high” defined as morphological classes 1 and 2, “low” defined as morphological classes 3 and 4): (i) all the three key genes showed significant differences between high-quality blastocysts and arrested embryos (p < 0.01), (ii) only CAMK1D showed significant differences between low-quality blastocysts and arrested embryos (p = 0.02), and (iii) only PTG2 showed a significant difference between high- and low-quality blastocysts (p = 0.02) (Supplementary Fig. 5).
Generalized linear models and ROC curve analyses
Supplementary Table 2 shows the results of the generalized linear models performed to test the association between early time-lapse parameters and selected key gene expression in the cumulus cells with the primary outcome (i.e., developmental arrest/blastocyst formation). The data confirmed that tPNa, tPNf, t2, cc1, t4, cc2, as well as CAMK1D, PTGS2, and HAS2 expression associate with the chance of a zygote to reach the blastocyst stage.
Importantly, the Kruskal–Wallis tests highlighted that the CC key gene expression was patient-specific; indeed, the mean values were significantly different across the 18 women included in the study (p < 0.01 for all the key genes analyzed; Supplementary Fig. 6). Figure 3a–c shows the expression values for CAMK1D, PTGS2, and HAS2 from each CC sample analyzed clustered according to the “patient” and by defining its related final outcome (arrested development/blastocyst). For each of these three key genes, but especially for CAMK1D, it is evident that higher gene expression levels characterize the cumuli of oocytes developing to the blastocyst stage from both an overall perspective and a patient-specific one. Standing this evidence and since the value of each putative predictor of oocyte developmental competence (including the TLM parameters) had to be assessed within each cohort, we included the “patient” variable in the generalized linear models. Due to the low number of patients enrolled in this study, we could not evaluate if and which specific woman’s characteristic (e.g., maternal age, body mass index, FSH) [52] might modulate CC key gene expressions; therefore, a deeper insight is eagerly required from future studies. Only t2, as well as CAMK1D and PTGS2 expression maintained a significant association within each cohort (data not shown). Therefore, we created a model that included all these three parameters and the “patient” variable. Table 2 shows this model that outlined CAMK1D as the only significant putative predictor of oocyte developmental competence. Indeed, the partial eta squared, the power and the R2 parameters of a model based only on the expression level of this transcript corrected per “patient” (0.16, 0.86, and 0.44, respectively) testify its promising value.
CAMK1D (a), HAS2 (b), and PTGS2 (c) 2−dCT data of each cumulus cell (CC) sample clustered according to its related patient and developmental outcome (arrested embryos represented by gray stars; blastocysts represented by orange circles). For CAMK1D, patients 1 and 18 showed no amplification from a CCs sample whose oocyte resulted into developmental arrest, while patients 3, 7, 12, 13, 16, and 17 showed no amplification from a CC sample whose oocyte resulted into blastocyst development. For HAS2 and PTGS2, all CC samples were amplified. The median general expression value has been reported for each key gene as a continuous black line. Overall, the CC samples whose oocytes developed to the blastocyst stage mostly clustered above the median, but in general, they showed higher expression than sibling arrested embryos within each patient-specific cohort of zygotes
To conclude, the overall predictive power of CAMK1D upon oocyte developmental competence was therefore estimated through a ROC curve analysis that showed an area under the curve (AUC) 0.78, 95% CI 0.67–0.89, p < 0.01 among the samples expressing this key gene (Supplementary Fig. 7a). The same analysis was conducted by adding the “patient” variable, which outlined an AUC 0.9, 95% CI 0.83–0.97, p < 0.01, among the cohorts of zygotes where a comparison between arrested embryos and blastocysts was doable (Supplementary Fig. 7b; Fig. 3a).
Discussion
In the present study, we analyzed the association between the expression pattern of selected key genes in the CCs and the developmental potential of the oocytes they surrounded during folliculogenesis. Since the blastocyst population is not homogeneous, we also analyzed the expression of our five target key genes in the CCs according to their corresponding blastocyst morphological quality and chromosomal constitution. All the embryos were cultured individually in an undisturbed time-lapse incubator with the same continuous media, thereby preventing any putative stress deriving from the environment and/or the manipulation. To further improve the objectivity in the assessment of embryo development, we also correlated the CC gene expression to the main morphokinetic parameters of early embryo preimplantation development. Three of the five key genes we analyzed showed a higher mean expression level in the CCs whose oocytes developed to the blastocyst stage, namely CAMK1D, PTGS2, and HAS2. While EFNB2 and STC1 showed no correlation.
Importantly, since mRNA levels of PTGS2, CAMK1D, and HAS2 increase a few hours before ovulation, the timing between hCG administration and COC denudation is crucial to exclude stochastic or biased results. However, this was not the case here, since similar timings between triggering and CCs sampling were reported for all the patients included in the analysis.
The results on CAMK1D, PTGS2, and HAS2 expression levels are in line with the previous literature [28, 29, 33], while the data related with EFNB2 and STC1 do not confirm what reported by Wathlet and colleagues [33]. Probably the reason is the different endpoints among ours and Wathlet’s studies, since we limited the investigation to the developmental rather than reproductive competence.
In our clinical setting, PGT-A allows the identification of aneuploid blastocysts, which are not transferred. Such strategy, by preventing the impact of chromosomal aneuploidies on pregnancy outcomes, ensures per se a good predictivity upon embryo reproductive competence [1, 15]. For this reason, we set the endpoint of this study as the blastocyst development rather than embryo implantation, since the former is expected to be more probably associated with the dialog between CCs and the oocytes during folliculogenesis. In fact, recently, Green and collaborators set the endpoint as live birth and analyzed the CCs’ whole transcriptome by RNA-seq in sibling oocytes searching for biomarkers of embryo reproductive competence, but could not find any significant association [53].
Importantly, the levels of these key gene expression from CCs was shown to be patient-specific. Therefore, we conducted the analyses also within each cohort of zygotes. PTGS2, but especially CAMK1D, showed high association with blastocyst development also from these analyses. Clearly, the low number of patients here included limits the general value of such finding. Nonetheless, it supports the evidence previously reported by Adriaenssens and colleagues [54] and encourages future investigation to address which woman’s characteristic(s) influence the expression of these key genes in the CCs.
In the present study, only the expression of CAMK1D in the CCs was confirmed as a significant predictor of blastocyst formation from both an overall perspective (AUC = 0.79) and a patient-specific one (AUC = 0.9). Clearly, the former evidence is limited to the samples that resulted in the detection of this transcript, while the latter one is limited to the patients enrolled from this cohort of selected women and showing at least two zygotes with different developmental outcomes. Yet, the effect size and post hoc statistical power achieved are sufficiently high to support future investigation on a larger sample size and a wider patient population. Additionally, the level of expression of CAMK1D seems to be lower with respect to the other transcripts; indeed, it might go undetected if less CCs are collected or in case of a lower qPCR efficiency. Future studies should therefore consider its preamplification after the retrotranscription step and before conducting qPCR.
CAMK1D is a member of the calcium/calmodulin-dependent protein kinase 1 family and has been associated with multiple functions, e.g., regulatory or gene activation ones, regulation of granulocyte function, aldosterone synthesis, differentiation and activation of neutrophil cells, and apoptosis of erythroleukemia cells. To date, no ovarian function has yet been reported for CAMK1D; only a higher expression in CCs has been reported by Wathlet and colleagues in competent oocytes resulting in positive pregnancy outcomes after transfer [33]. In their paper, the authors suggested that CAMK1D might be involved in the regulation of steroidogenesis during folliculogenesis. Qin and collaborators recently described a role for CAMK1D in trophoblast cells. Intriguingly, these authors demonstrated a crosstalk between eukaryotic initiation factor 5A (eIF5A), a highly conserved protein which is involved in various cell functions (i.e., protein translation, migration, proliferation, apoptosis, cell cycle progression, inflammation, and tumorigenesis) [55, 56], and CAMK1D aimed at the activation in trophoblast cells of specific pathways of cell proliferation, migration, and invasion [57]. Furthermore, they demonstrated, through RNA interference, that CAMK1D overexpression acts also as an inhibitor of apoptosis in vitro [57]. In human hepatocytes, CAMK1D is instead involved in the regulation of glucose metabolism in an insulin-independent way [58]. A similar mechanism might be crucial also in the oocyte microenvironment during folliculogenesis, where the regulation of glucose or pyruvate uptake during maturation could affect subsequent embryo development. CAMK1D overexpression is also associated to breast cancer proliferation, where it induces loss of cell–cell adhesion, a characteristic typical of cells undergoing epithelial to mesenchymal transition [59]. Our study strongly suggests further investigations of CAMK1D in the acquisition of oocyte developmental competence to blastocyst. In this regard, its putative role in the mediation of glucose uptake in the oocyte during the latest phases of the folliculogenesis, as well as its antiapoptotic function should be both assessed.
To provide an even more comprehensive insight, we further inspected the data according to the morphological quality of the blastocysts obtained (i.e., good- or low-quality). Besides CAMK1D, also PTGS2 and HAS2 showed significantly higher level of expression in CCs associated to good-quality blastocysts, thereby confirming the results produced from previous investigations [28, 33]. PTGS2 is an enzyme involved in prostaglandin biosynthesis, that when is produced by CCs covers a crucial role in the cumulus expansion and oocyte maturation during meiotic resumption [60, 61]. Indeed, the mice knock-out for this gene (PTGS2−/−) showed a failure in the cumulus expansion and extrusion of the first polar body [62,63,64]. HAS2 instead is crucial for the production of hyaluronic acid (HA), the key component of the matrix of the expanded mature cumulus [65,66,67]. An overexpression for both of these key genes in the last phases of folliculogenesis might therefore entail beneficial effects to the oocyte quality lasting up to full blastulation.
A correlation of CAMK1D, PTGS2, and HAS2 CC gene expression with blastocyst morphology did not correspond to an equal correlation with blastocyst chromosomal constitution. In fact, these last two variables are just mildly correlated among each other [42, 68]. To date, only the introduction of PGT-A allowed the identification of euploid blastocysts for transfer from a cohort produced by a couple during IVF, thereby improving the efficiency of embryo selection (i.e., higher implantation success per transfer and lower miscarriage rate were achieved with respect to non-PGT-A cycles) [15, 16]. However, an invasive TE biopsy is needed that, even if reported safe for the blastocyst [17, 18], still requires specific devices (e.g., laser-equipped micromanipulator) and skills (e.g., blastocyst culture, biopsy, and vitrification). Therefore, a quest of widely applicable noninvasive parameters to conduct embryo selection has started, which is still ongoing. For instance, Fragouli and colleagues analyzed the CCs’ gene expression of oocytes that underwent also polar body biopsy; the authors found tumor protein P53 inducible protein 3 (TP53I3) and SplA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) to be underexpressed in aneuploid oocytes [69]. Since then, other groups started analyzing the gene expression patterns in CCs, but the data were not consistent and reproducible among the studies, therefore requiring further investigation [70]. Nonetheless, we still cannot exclude that an association exists between CCs’ gene expression and chromosomal aneuploidies. Indeed, if meiotic chromosomal impairments (i.e., the only ones identifiable by qPCR on a TE biopsy in this study) seem not be correlated with the expression level of our selected key genes, mitotic ones instead could be. In fact, this latter kind of aneuploidies have been proposed as a significant cause of embryo arrest during development, especially after the EGA [71,72,73], which in humans occurs between the four- and eight-cell transitions [47]. In particular in the first 3 days of development, the embryo is in fact under the exclusive control of maternally derived transcripts and no cell cycle checkpoint is in place. Therefore, a role especially for CAMK1D in the prevention of chromosomal missegregation during this delicate phase of embryo development cannot be disregarded and deserves investigation from future studies.
All the embryos included in the analysis were cultured in a time-lapse system and the morphokinetic parameters up to t5 were compared between arrested embryos and blastocysts. We confirmed that faster kinetics of early embryo development could predict blastocyst formation as suggested in many previous papers [8, 9, 21, 74]. Namely, tPNa, tPNf, t2, cc1, t4, and cc2 were significantly faster in oocytes that developed to the blastocyst stage, but in general, all the early timings up to t5 were associated, if slower, with the risk of developmental arrest and degeneration. The t2 was the only early TLM parameter that maintained a significant association with blastocyst development also when corrected per each specific “patient” (i.e., within each cohort of zygotes) and suggests that cellular impairments already in the very first karyokinesis and cell division might be detrimental for the developing embryo. Yet, embryo arrest before t5 is minimal [75]. Mainly the t8, and to some extent the following stages up to the tEB, represent the graveyard of developmentally incompetent embryos, possibly for the same reasons described above.
In this study, we combined morphokinetic parameters with the CC key gene expression aiming at providing an integrated noninvasive evaluation of oocyte developmental competence. To the best of our knowledge, only Hammond and colleagues [76] proposed a comparable approach. The authors reported that the embryos undergoing an earlier division to the four-cell stage have a higher chance to develop into high-quality blastocysts on day 5, and the predictivity was boosted if they included also the CC expression of several genes in a multivariate model. In our setting, however, when combining the t2 with PTGS2 and CAMK1D CCs’ expression corrected per “patient,” only CAMK1D maintained a significant association with the primary outcome. Still, the limited sample size of this study might have hindered the additional effect of those variables, which needs to be addressed from future analyses.
The main strength of our study was that CC key gene expression was corrected across each “patient.” If on the one hand this correction prevented us from discriminating which woman’s specific characteristic might modulate such expression, on the other hand, it implicitly encompassed all the putative confounding factors, such as maternal age, BMI, number of zygotes obtained per cohort, male factor, and timing of CC sampling. As stated previously, future studies from a wider patient population might provide a more detailed insight to address this issue. From a laboratory perspective, embryo culture was here conducted in a controlled undisturbed incubator and with a continuous media to limit any putative stress derived from the environment and/or embryo manipulation. All these characteristics increased the objectivity of the endpoint, which was set as blastocyst development. The morphological evaluation of blastocyst quality and the qPCR-based definition of the chromosomal constitution represented further subanalyses that provided a comprehensive overview upon oocyte developmental competence.
To conclude, the interesting finding that CAMK1D level of expression is associated with blastocyst development should encourage future investigations to understand how oocytes gain competence during folliculogenesis. Concurrently, the improvement of our knowledge about the role of CAMK1D in CCs during the last crucial steps of folliculogenesis could conduct to a better fine-tuning of follicular stimulation, or to new advances in in vitro maturation protocols.
References
Ubaldi FM, Capalbo A, Colamaria S, Ferrero S, Maggiulli R, Vajta G, et al. Reduction of multiple pregnancies in the advanced maternal age population after implementation of an elective single embryo transfer policy coupled with enhanced embryo selection: pre- and post-intervention study. Hum Reprod. 2015;30(9):2097–106. https://doi.org/10.1093/humrep/dev159.
Guerif F, Le Gouge A, Giraudeau B, Poindron J, Bidault R, Gasnier O, et al. Limited value of morphological assessment at days 1 and 2 to predict blastocyst development potential: a prospective study based on 4042 embryos. Hum Reprod. 2007;22(7):1973–81. https://doi.org/10.1093/humrep/dem100.
Racowsky C, Ohno-Machado L, Kim J, Biggers JD. Is there an advantage in scoring early embryos on more than one day? Hum Reprod. 2009;24(9):2104–13. https://doi.org/10.1093/humrep/dep198.
Glujovsky D, Farquhar C. Cleavage-stage or blastocyst transfer: what are the benefits and harms? Fertil Steril. 2016;106(2):244–50. https://doi.org/10.1016/j.fertnstert.2016.06.029.
Gardner DK. Blastocyst culture: toward single embryo transfers. Hum Fertil (Camb). 2000;3(4):229–37.
Gardner DK. The impact of physiological oxygen during culture, and vitrification for cryopreservation, on the outcome of extended culture in human IVF. Reprod BioMed Online. 2016;32(2):137–41. https://doi.org/10.1016/j.rbmo.2015.11.008.
Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, et al. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100(2):412–9 e5. https://doi.org/10.1016/j.fertnstert.2013.04.021.
Kirkegaard K, Kesmodel US, Hindkjaer JJ, Ingerslev HJ. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod. 2013;28(10):2643–51. https://doi.org/10.1093/humrep/det300.
Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26(10):2658–71. https://doi.org/10.1093/humrep/der256.
Armstrong S, Bhide P, Jordan V, Pacey A, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev. 2018;5:CD011320. https://doi.org/10.1002/14651858.CD011320.pub3.
Heffner LJ. Advanced maternal age—how old is too old? N Engl J Med. 2004;351(19):1927–9. https://doi.org/10.1056/NEJMp048087.
Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. https://doi.org/10.1038/35066065.
Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101(3):656–63 e1. https://doi.org/10.1016/j.fertnstert.2013.11.004.
Capalbo A, Hoffmann ER, Cimadomo D, Maria Ubaldi F, Rienzi L. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum Reprod Update. 2017;23:1–17. https://doi.org/10.1093/humupd/dmx026.
Dahdouh EM, Balayla J, Garcia-Velasco JA. Comprehensive chromosome screening improves embryo selection: a meta-analysis. Fertil Steril. 2015;104(6):1503–12. https://doi.org/10.1016/j.fertnstert.2015.08.038.
Chen M, Wei S, Hu J, Quan S. Can comprehensive chromosome screening technology improve IVF/ICSI outcomes? A meta-analysis. PLoS One. 2015;10(10):e0140779. https://doi.org/10.1371/journal.pone.0140779.
Scott RT Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013;100(3):624–30. https://doi.org/10.1016/j.fertnstert.2013.04.039.
Cimadomo D, Capalbo A, Ubaldi FM, Scarica C, Palagiano A, Canipari R, et al. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis. Biomed Res Int. 2016;2016:7193075. https://doi.org/10.1155/2016/7193075.
Pennetta F, Lagalla C, Borini A. Embryo morphokinetic characteristics and euploidy. Curr Opin Obstet Gynecol. 2018;30(3):185–96. https://doi.org/10.1097/GCO.0000000000000453.
Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod BioMed Online. 2013;26(5):477–85. https://doi.org/10.1016/j.rbmo.2013.02.006.
Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101(3):699–704. https://doi.org/10.1016/j.fertnstert.2013.12.005.
Rienzi L, Capalbo A, Stoppa M, Romano S, Maggiulli R, Albricci L, et al. No evidence of association between blastocyst aneuploidy and morphokinetic assessment in a selected population of poor-prognosis patients: a longitudinal cohort study. Reprod BioMed Online. 2015;30(1):57–66. https://doi.org/10.1016/j.rbmo.2014.09.012.
Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122(6):829–38.
Paulini F, Melo EO. The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod Domest Anim. 2011;46(2):354–61. https://doi.org/10.1111/j.1439-0531.2010.01739.x.
Trombly DJ, Woodruff TK, Mayo KE. Roles for transforming growth factor beta superfamily proteins in early folliculogenesis. Semin Reprod Med. 2009;27(1):14–23. https://doi.org/10.1055/s-0028-1108006.
Kedem A, Fisch B, Garor R, Ben-Zaken A, Gizunterman T, Felz C, et al. Growth differentiating factor 9 (GDF9) and bone morphogenetic protein 15 both activate development of human primordial follicles in vitro, with seemingly more beneficial effects of GDF9. J Clin Endocrinol Metab. 2011;96(8):E1246–54. https://doi.org/10.1210/jc.2011-0410.
Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22(12):3069–77. https://doi.org/10.1093/humrep/dem336.
McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19(12):2869–74. https://doi.org/10.1093/humrep/deh535.
Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007;134(5):645–50. https://doi.org/10.1530/REP-07-0182.
Anderson RA, Sciorio R, Kinnell H, Bayne RA, Thong KJ, de Sousa PA, et al. Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction. 2009;138(4):629–37. https://doi.org/10.1530/REP-09-0144.
Feuerstein P, Puard V, Chevalier C, Teusan R, Cadoret V, Guerif F, et al. Genomic assessment of human cumulus cell marker genes as predictors of oocyte developmental competence: impact of various experimental factors. PLoS One. 2012;7(7):e40449. https://doi.org/10.1371/journal.pone.0040449.
Kordus RJ, LaVoie HA. Granulosa cell biomarkers to predict pregnancy in ART: pieces to solve the puzzle. Reproduction. 2017;153(2):R69–83. https://doi.org/10.1530/REP-16-0500.
Wathlet S, Adriaenssens T, Segers I, Verheyen G, Janssens R, Coucke W, et al. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil Steril. 2012;98(2):432–9 e1–4. https://doi.org/10.1016/j.fertnstert.2012.05.007.
Burnik Papler T, Vrtacnik Bokal E, Maver A, Lovrecic L. Specific gene expression differences in cumulus cells as potential biomarkers of pregnancy. Reprod BioMed Online. 2015;30(4):426–33. https://doi.org/10.1016/j.rbmo.2014.12.011.
Sanchez F, Smitz J. Molecular control of oogenesis. Biochim Biophys Acta. 2012;1822(12):1896–912. https://doi.org/10.1016/j.bbadis.2012.05.013.
Wissing ML, Kristensen SG, Andersen CY, Mikkelsen AL, Host T, Borup R, et al. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum Reprod. 2014;29(5):997–1010. https://doi.org/10.1093/humrep/deu008.
Luo CW, Kawamura K, Klein C, Hsueh AJ. Paracrine regulation of ovarian granulosa cell differentiation by stanniocalcin (STC) 1: mediation through specific STC1 receptors. Mol Endocrinol. 2004;18(8):2085–96. https://doi.org/10.1210/me.2004-0066.
Deol HK, Varghese R, Wagner GF, Dimattia GE. Dynamic regulation of mouse ovarian stanniocalcin expression during gestation and lactation. Endocrinology. 2000;141(9):3412–21. https://doi.org/10.1210/endo.141.9.7658.
Vaiarelli A, Cimadomo D, Patrizio P, Venturella R, Orlando G, Soscia D, et al. Biochemical pregnancy loss after frozen embryo transfer seems independent of embryo developmental stage and chromosomal status. Reprod BioMed Online. 2018;37(3):349–57. https://doi.org/10.1016/j.rbmo.2018.05.019.
Rienzi L, Ubaldi F, Anniballo R, Cerulo G, Greco E. Preincubation of human oocytes may improve fertilization and embryo quality after intracytoplasmic sperm injection. Hum Reprod. 1998;13(4):1014–9.
Ubaldi F, Anniballo R, Romano S, Baroni E, Albricci L, Colamaria S, et al. Cumulative ongoing pregnancy rate achieved with oocyte vitrification and cleavage stage transfer without embryo selection in a standard infertility program. Hum Reprod. 2010;25(5):1199–205. https://doi.org/10.1093/humrep/deq046.
Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, et al. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29(6):1173–81. https://doi.org/10.1093/humrep/deu033.
Capalbo A, Treff NR, Cimadomo D, Tao X, Upham K, Ubaldi FM, et al. Comparison of array comparative genomic hybridization and quantitative real-time PCR-based aneuploidy screening of blastocyst biopsies. Eur J Hum Genet. 2015;23(7):901–6. https://doi.org/10.1038/ejhg.2014.222.
Treff NR, Tao X, Ferry KM, Su J, Taylor D, Scott RT Jr. Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril. 2012;97(4):819–24. https://doi.org/10.1016/j.fertnstert.2012.01.115.
Gardner DK, Schoolcraft B. In vitro culture of human blastocyst. In: Jansen R, Mortimer D, editors. Towards reproductive certainty: infertility and genetics beyond. Carnforth: Parthenon Press; 1999. p. 377–88.
Cimadomo D, Capalbo A, Levi-Setti PE, Soscia D, Orlando G, Albani E, et al. Associations of blastocyst features, trophectoderm biopsy and other laboratory practice with post-warming behavior and implantation. Hum Reprod. 2018;33:1992–2001. https://doi.org/10.1093/humrep/dey291.
Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–61. https://doi.org/10.1038/332459a0.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034.
Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26(5):1035–51. https://doi.org/10.1093/humrep/der036.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8.
McCall MN, McMurray HR, Land H, Almudevar A. On non-detects in qPCR data. Bioinformatics. 2014;30(16):2310–6. https://doi.org/10.1093/bioinformatics/btu239.
Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25(5):1259–70. https://doi.org/10.1093/humrep/deq049.
Green KA, Franasiak JM, Werner MD, Tao X, Landis JN, Scott RT Jr, et al. Cumulus cell transcriptome profiling is not predictive of live birth after in vitro fertilization: a paired analysis of euploid sibling blastocysts. Fertil Steril. 2018;109(3):460–6 e2. https://doi.org/10.1016/j.fertnstert.2017.11.002.
Adriaenssens T, Segers I, Wathlet S, Smitz J. The cumulus cell gene expression profile of oocytes with different nuclear maturity and potential for blastocyst formation. J Assist Reprod Genet. 2011;28(1):31–40. https://doi.org/10.1007/s10815-010-9481-9.
Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38(2):491–500. https://doi.org/10.1007/s00726-009-0408-7.
Fujimura K, Choi S, Wyse M, Strnadel J, Wright T, Klemke R. Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and Rho-associated kinase (ROCK) protein expression levels. J Biol Chem. 2015;290(50):29907–19. https://doi.org/10.1074/jbc.
Qin X, Liang Y, Guo Y, Liu X, Zeng W, Wu F, et al. Eukaryotic initiation factor 5A and Ca(2+) /calmodulin-dependent protein kinase 1D modulate trophoblast cell function. Am J Reprod Immunol. 2018;80(1):e12845. https://doi.org/10.1111/aji.12845.
Haney S, Zhao J, Tiwari S, Eng K, Guey LT, Tien E. RNAi screening in primary human hepatocytes of genes implicated in genome-wide association studies for roles in type 2 diabetes identifies roles for CAMK1D and CDKAL1, among others, in hepatic glucose regulation. PLoS One. 2013;8(6):e64946. https://doi.org/10.1371/journal.pone.0064946.
Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, Langerod A, et al. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2(4):327–39. https://doi.org/10.1016/j.molonc.2008.09.004.
Eppig JJ. Prostaglandin E2 stimulates cumulus expansion and hyaluronic acid synthesis by cumuli oophori isolated from mice. Biol Reprod. 1981;25(1):191–5.
Hizaki H, Segi E, Sugimoto Y, Hirose M, Saji T, Ushikubi F, et al. Abortive expansion of the cumulus and impaired fertility in mice lacking the prostaglandin E receptor subtype EP(2). Proc Natl Acad Sci U S A. 1999;96(18):10501–6.
Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91(2):197–208.
Davis BJ, Lennard DE, Lee CA, Tiano HF, Morham SG, Wetsel WC, et al. Anovulation in cyclooxygenase-2-deficient mice is restored by prostaglandin E2 and interleukin-1beta. Endocrinology. 1999;140(6):2685–95. https://doi.org/10.1210/endo.140.6.6715.
Takahashi T, Morrow JD, Wang H, Dey SK. Cyclooxygenase-2-derived prostaglandin E(2) directs oocyte maturation by differentially influencing multiple signaling pathways. J Biol Chem. 2006;281(48):37117–29. https://doi.org/10.1074/jbc.M608202200.
Fulop C, Salustri A, Hascall VC. Coding sequence of a hyaluronan synthase homologue expressed during expansion of the mouse cumulus-oocyte complex. Arch Biochem Biophys. 1997;337(2):261–6. https://doi.org/10.1006/abbi.1996.9793.
Salustri A, Yanagishita M, Hascall VC. Synthesis and accumulation of hyaluronic acid and proteoglycans in the mouse cumulus cell-oocyte complex during follicle-stimulating hormone-induced mucification. J Biol Chem. 1989;264(23):13840–7.
Salustri A, Ulisse S, Yanagishita M, Hascall VC. Hyaluronic acid synthesis by mural granulosa cells and cumulus cells in vitro is selectively stimulated by a factor produced by oocytes and by transforming growth factor-beta. J Biol Chem. 1990;265(32):19517–23.
Alfarawati S, Fragouli E, Colls P, Stevens J, Gutierrez-Mateo C, Schoolcraft WB, et al. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil Steril. 2011;95(2):520–4. https://doi.org/10.1016/j.fertnstert.2010.04.003.
Fragouli E, Wells D, Iager AE, Kayisli UA, Patrizio P. Alteration of gene expression in human cumulus cells as a potential indicator of oocyte aneuploidy. Hum Reprod. 2012;27(8):2559–68. https://doi.org/10.1093/humrep/des170.
Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 2014;20(1):1–11. https://doi.org/10.1093/humupd/dmt044.
McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, et al. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348(6231):235–8. https://doi.org/10.1126/science.aaa3337.
McCoy RC, Demko ZP, Ryan A, Banjevic M, Hill M, Sigurjonsson S, et al. Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genet. 2015;11(10):e1005601. https://doi.org/10.1371/journal.pgen.1005601.
McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER, Chatzimeletiou K, Cornejo OE, et al. Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Hum Mol Genet. 2018;27(14):2573–85. https://doi.org/10.1093/hmg/ddy147.
Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, et al. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod. 2015;30(2):276–83. https://doi.org/10.1093/humrep/deu331.
Betts DH, Madan P. Permanent embryo arrest: molecular and cellular concepts. Mol Hum Reprod. 2008;14(8):445–53. https://doi.org/10.1093/molehr/gan035.
Hammond ER, Stewart B, Peek JC, Shelling AN, Cree LM. Assessing embryo quality by combining non-invasive markers: early time-lapse parameters reflect gene expression in associated cumulus cells. Hum Reprod. 2015;30(8):1850–60. https://doi.org/10.1093/humrep/dev121.
Author information
Authors and Affiliations
Contributions
CS, DC, AC, LR, and RC designed the study. CS and DC collected and analyzed the samples. CS, DC, and RC drafted the manuscript. All authors contributed to the interpretation and discussion of the data.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
Mean Ct values forB2MandUBCin the cumulus cells from oocytes that underwent developmental arrest versus blastocyst formation. Mann-Whitney U tests were conducted that did not show any statistically significant difference. (PNG 62 kb)
Supplementary Figure 2
Geometric mean ofB2MandUBCCt values in the samples whereSTC1andCAMK1Dwere detected versus the samples where these key-genes were not detected. Mann-Whitney U tests were conducted. Effect size d and post-hoc power analyses were also reported. (PNG 84 kb)
Supplementary Figure 3
Relative expression levels ofSTC1andEFNB2from the cumulus cells of developmentally incompetent versus competent oocytes. Total RNA was extracted from cumulus cells obtained from oocytes that resulted in either arrested embryos or blastocysts after IVF. Quantitative-PCR was conducted using Taqman primers. Each reaction was performed in triplicate. Each sample was normalized to its B2M and UBC mRNA content. The mean 2^-dCT values with SD and range are reported next to each boxplot. Mann-Whitney U tests were conducted to assess statistically-significant differences. No statistically-significant difference was defined. Circles and stars represent outliers values (PNG 67 kb)
Supplementary Figure 4
Relative expression levels ofCAMK1D,PTGS2andHAS2from the cumulus cells whose oocytes developed as aneuploid versus euploid blastocysts (sub-analysis). Total RNA was extracted from cumulus cells obtained from oocytes that resulted in either aneuploid or euploid blastocysts after IVF. Quantitative-PCR was conducted using Taqman primers. Each reaction was performed in triplicate. Each sample was normalized to its B2M and UBC mRNA content. The mean 2^-dCT values with SD and range are reported next to each boxplot. Mann-Whitney U tests were conducted to assess statistically-significant differences. No statistically-significant difference was reported. Effect size d and post-hoc power analyses were also calculated. Circles and stars represent outliers values (PNG 76 kb)
Supplementary Figure 5
Relative expression levels ofCAMK1D,PTGS2andHAS2from the cumulus cells of oocytes that developed as high quality (class 1&2) and low quality (class 3&4) blastocysts or underwent developmental arrest (sub-analysis). Total RNA was extracted from cumulus cells obtained from oocytes that resulted in either arrested embryos or blastocysts after IVF. Quantitative-PCR was conducted using Taqman primers. Each reaction was performed in triplicate. Each sample was normalized to its B2M and UBC mRNA content. The mean 2^-dCT values with SD and range are reported next to each boxplot. Mann-Whitney U tests were conducted to assess statistically-significant differences. Effect size d and post-hoc power analyses were also reported. Circles and stars represent outliers values (PNG 125 kb)
Supplementary Figure 6
Patient-specific mean CCs’ expression of selected key-genes with 95%CI entailing all the samples that were amplified within each cohort of zygotes. Kruskal-Wallis tests were conducted to assess statistically significant differences (PNG 89 kb)
Supplementary Figure 7
Receiver Operating Characteristic (ROC) curve analysis for the predictive power ofCAMK1D2^-dCT upon oocyte developmental competence in vitro from both a general (a) and a patient-specific (b) analyses conducted from this dataset. The former analysis entailed 67 of the 75 cumuli included in this study (89%), namely the samples where this transcript was detected; the latter analysis instead included the patients (N = 11) who obtained at least two oocytes from the cohort with opposite outcomes (arrested embryo and blastocyst development) and whose cumulus cells resulted in CAMK1D detection. AUC, area under the curve. (PNG 115 kb)
Supplementary Table 1
(DOCX 17 kb)
Supplementary Table 2
(DOCX 31 kb)
Rights and permissions
About this article
Cite this article
Scarica, C., Cimadomo, D., Dovere, L. et al. An integrated investigation of oocyte developmental competence: expression of key genes in human cumulus cells, morphokinetics of early divisions, blastulation, and euploidy. J Assist Reprod Genet 36, 875–887 (2019). https://doi.org/10.1007/s10815-019-01410-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-019-01410-3