Abstract
Purpose
This paper aims to investigate the feasibility of performing pre-implantation genetic diagnosis (PGD) and pre-implantation genetic screening (PGS) simultaneously by a universal strategy without the requirement of genotyping relevant affected family members or lengthy preliminary work on linkage analysis.
Methods
By utilizing a universal Mutated Allele Revealed by Sequencing with Aneuploidy and Linkage Analyses (MARSALA) strategy based on low depth whole genome sequencing (~3x), not involving specific primers' design nor the enrichment of SNP markers for haplotype construction. Single-sperm cells and trephectoderm cells from in vitro fertilized embryos from a couple carrying HBB mutations were genotyped. Haplotypes of paternal alleles were constructed and investigated in embryos, and the chromosome copy number profiles were simultaneously analyzed.
Results
The universal MARSALA strategy allows the selection of a euploid embryo free of disease mutations for in uterus transfer and successful pregnancy. A follow-up amniocentesis was performed at 17 weeks of gestation to confirm the PGD/PGS results.
Conclusion
We present the first successful PGD procedure based on genotyping multiple single-sperm cells to obtain SNP linkage information. Our improved PGD/PGS procedure does not require genotyping the proband or relevant family members and therefore can be applicable to a wider population of patients when conducting PGD for monogenic disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Monogenic diseases are inherited disorders resulting from mutations in a single gene and have a prevalence of ~1% in all live births [1]. Most monogenic diseases are associated with developmental defects or, more severely, lethality. However, effective medical interventions are currently available but only for a few of those diseases [2]. Pre-implantation genetic diagnosis (PGD) is a genetic testing used to select embryos free of a monogenic mutation before implantation takes place during an in vitro fertilization (IVF) treatment. Currently, the most widely used techniques for PGD generally rely on polymerase chain reaction (PCR), either targeted or by whole-genome amplification of a single-cell or equivalent [3, 4] such as biopsy from D3 cleavage-stage or D5 blastocyst-stage embryos. These samples are then used for subsequent analysis (for instance karyomapping and next-generation sequencing (NGS)). However, the nonuniform allelic amplification due to the allele dropout (ADO) in single-cell analysis, is one of the main causes of misdiagnosis in PGD [5]. Consequently, linkage analysis became highly recommended to increase PGD accuracy [7, 8], which has been reported to decrease misdiagnosis rates from 3–4 to 0.3–0.5% [9]. It relies upon single-nucleotide polymorphism (SNP) [6] or short tandem repeat (STR) markers in combination with the specific mutation.
Linkage analysis starts with selecting informative markers adjacent to the mutation, based on previous analysis of the genotypes from the couple and relevant information from affected and unaffected family members. A haplotype is then constructed accordingly to the identified alleles linked to the mutation. However, the lack of availability of family member, and particularly the proband sample, often limits a wider application of this PGD strategy. Moreover, the design of specific primers as markers in haplotype construction and subsequent amplification are generally required, further increasing the difficulty of the process. Therefore, a universal PGD procedure with less demanding steps, which does not require a proband or multiple family members would conceivably broaden the applicablility of the PGD technique.
Chromosomal aneuploidy [10], such as trisomy or monosomy, is a common cause for miscarriages and congenital malformations [11]. Such chromosomal abnormalities can be identified in IVF embryos via pre-implantation genetic screening (PGS) [12], allowing the selection of chromosomally-normal embryos for implantation [13]. Previous studies have showed that PGS can increase the implantation and live birth rates in certain populations [14,15,16,17]. Therefore, combining PGD and PGS becomes essential to avoid errors when selecting a healthy and transferrable embryo for PGD patients. Mutated allele revealed by sequencing with aneuploidy and linkage analyses (MARSALA) is a recently developed approach for simultaneous detection of single-gene mutations, linked SNPs and chromosomal aneuploidies at a single-cell levels based on NGS platform [18]. It has been applied in IVF and demonstrated to successfully prevent the transmission of causal mutation for hereditary multiple exostoses, hypohidrotic ectodermal dysplasia and spinal muscular atrophy [18, 19].
In the present study, we performed PGD/PGS to a couple carrying Beta Thalassemia causal mutations. We used single-sperm genotyping instead of proband or family members for linkage analysis of MARSALA. Different from previous MARSALA, this new approach utilized direct low-depth whole genome sequencing to genotype SNP markers to avoid the PCR enrichment of the targets. The conventional MARSALA was conducted in parallel to verify the results prior to the embryo transfer. A follow-up amniocentesis further confirmed our PGD/PGS results.
Materials and methods
Patient information
A couple visited the Reproductive Medicine Center, First Affiliated Hospital of Sun Yat-sen University in 2014 for PGD. The 22-year-old male was a carrier of the − 28A > G (rs33931746) mutation for HBB (NM_000518.4) and the 22-year-old female was a carrier of the CD41–42 mutation (c.126-129delCTTT, rs281864900) of the same gene, which were confirmed by Sanger sequencing. The female had a previous pregnancy in July 2013, and the fetus was identified to harbor compound heterozygote mutations by chorionic villus biopsy at the 16th gestational week, so the couple chose to terminate the pregnancy at the 19th week and prepared for IVF-PGD treatment. We obtained written informed consent from the patients prior to the PGD/PGS procedure. The present study was approved by the Research Ethics Committee of the First Hospital of Sun Yat-sen University [2014]134.
Single-sperm cell collection, embryo biopsy, and genomic DNA extraction
Semen was collected by masturbation and was diluted by a factor of ~1000 in PBS. Next, each single-sperm cell was isolated by mouth pipetting under the microscope and placed into a PCR tube containing 5 μl of the single-cell lysis reaction mix for subsequent MALBAC whole genome amplification (WGA) (Yikon Genomics, China). In vitro fertilization and trophectoderm (TE) biopsy of blastocysts were performed using prevously described protocols [20]. All embryos intended for PGD/PGS purposes were inseminated by intracytoplasmic sperm injection (ICSI) and cultured following a standard blastocyst culture procedure. Approximately three to five TE cells were biopsied from each blastocyst on day 5 and transferred into lysis buffer for WGA. The genomic DNA was also extracted from the peripheral blood of the couple.
Whole-genome amplification
The MALBAC single-cell WGA method [21] was used to amplify individual sperm cells from the male patient, the biopsied TE cells, as well as the couple's extracted genomic DNA by following the standard amplification protocol provided by the manufacturer (Yikon Genomics, China).
The sequencing strategy
The WGA products were used for direct whole genome sequencing on Illumina HiSeq 2500 platform with a ~3× mean genome coverage. The mutation sites and SNPs were then genotyped and the CNV profiles were investigated as described before [21].
Additionally, to verify this MARSALA result, a targeted sequencing strategy was also employed for single sperms, TE cells, and genomic DNA which involved the enrichment and sequencing of the two parental mutations together with 60 selected SNP markers. The PCR amplicons of the SNPs and mutations were mixed with corresponding WGA products and subjected to library construction for NGS. By doing so, mutation detection, linked SNP detection and chromosomal aneuploidy screening was accomplished in a single NGS process with an average genome coverage of ~0.1× as previously described [18].
The Sanger sequencing method was used to confirm the mutation sites in all samples.
SNP selection for linkage analysis
Sixty SNP markers link to the HBB mutations were chosen according to the following criteria: (a) the minor allele frequency (MAF) > 0.1 in the Asian population, (b) coverage > 10× in MALBAC-WGA sequencing results under the 3× average genome coverage, and (c) SNPs should be no more than 1.5 Mb upstream or downstream of the mutations.
Results
Analysis of the coverage of HBB gene and adjacent SNPs in the MALBAC WGA products
In order to evaluate the feasibility of genotyping HBB mutations and adjacent SNPs for PGD by the universal MARSALA approach based on low-depth, whole genome sequencing, we retrospectively analyzed the whole genome sequencing data obtained from previous MALBAC products. Among the SNPs 1.5 Mb upstream and downstream of the HBB gene, the minor allele frequencies (MAF), of which in the Eastern Asian population are greater than 0.1 (in 1000Genome), more than 90 SNPs reveal a relative coverage greater than 5× compared to the average genome coverage, indicating a sufficient number of SNP markers suitable for subsequent linkage analysis.
Linkage analysis of parental disease mutations and PGD/PGS by universal MARSALA approach on single-sperm cells and embryos
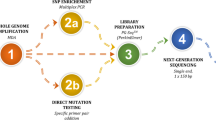
A proband sample is usually difficult to obtain, therefore we developed a novel strategy to avoid the need of a proband and instead incorporated sequencing of single-sperm cells and embryos (Fig. 1). Whole genome sequencing was performed on WGA products of seven single-sperm cells, TE cells of six embryos, and extracted gDNA from parents peripheral blood, with an average genome coverage of ~3× for each samples. Genotypes of mutation sites in single-sperm cells and embryos were illustrated in Fig. 2a, b. SNPs with coverage > 10× (or > 5× in single-sperm cells) located within 1.5 Mb upstream or downstream of the mutation sites were genotyped. Those genotypes that were heterozygous in the father and homozygous in the mother were used for paternal linkage analysis and screening for paternal mutation in embryos, as demonstrated in Fig. 3a. A total of 48 paternal heterozygous sites met these criteria, and 74% of them were detected in each single-sperm cell on average. In TE cell samples, approximately 22 sites (~46%) are detectable. By genotyping these SNPs and the paternal mutation in sperms, we constructed haplotypes of two paternal alleles and then deduced the defective inherited allele from the father for each embryos. As illustrated in Figs. 2a and 3c, two out of seven sperm cells were identified with the paternal − 28A > G mutation and the haplotype linked was then determined accordingly. Four embryos were identified with the same paternal mutation, which wasfurther confirmed by the linked haplotype, hencewere not recommended for transfer (Table 1). However, heterozygotes embryos might be used for transfer when no normal homozygotes are available. Similarly, the construction of the maternal allele haplotype was performed by investigating the SNPs with heterozygous genotypes in the mother but homozygous in the father as illustrated in Fig. 3b. There were 106 maternal heterozygous sites and on average ~45% of them were detectable in TE cell samples. Only one embryo was identified with the maternal mutation (Fig. 2b and Table 1), which was also confirmed by the corresponding haplotype (Fig. 3d). The HBB causal mutations were also confirmed by Sanger sequencing using specific primers (data not shown).
The workflow of MARSALA-PGD/PGS coupled with single-sperm genotyping. The DNA from single-sperm cells and embryo biopsy samples is amplified using the MALBAC technique. MARSALA-PGD/PGS is then performed to identify target mutation, linked SNPs, and chromosomal abnormalities in individual single-sperm cells and embryo biopsies. Based on the SNP information obtained in single sperms and embryos, the haplotypes of the causal mutations are constructed, which can further confirm the alleles identified in embryos. Euploid embryos free of target mutations then can be selected for implantation. A follow-up amniocentesis is performed to confirm the PGD/PGS results
The results of mutation carrying status and CNV profiles obtained from universal MARSALA analysis on single sperm cells and embryos. a The fraction of NGS reads for paternal mutation allele (red) in the couple, embryos (E01–E06), and single-sperm cells (S1–S7). b The fraction of NGS reads for maternal mutation allele (red) in the couple and embryos (E01–E06). c The CNV profiles of the embryos. A deletion of the p of chromosome 17 was identified in embryo E04
Linkage analysis and haplotype construction via MARSALA. a Schematic representation of MARSALA for linkage analysis to confirm the carrying status of paternal mutation allele in embryos. The amplified genomes of the peripheral blood samples, the sperms, and the embryos are sequenced. By analyzing the paternal mutation and SNPs which are heterozygous in father but homozygous in mother, we could deduce the inherited allele of the embryos under screening. The allele carrying the paternal mutation is indicated in black. The red asterisk indicates the paternal mutation − 28 A > G (HBB gene is reverse to the genome; therefore, the genome sequence on the corresponding site is T > C). And the blue asterisk indicates the maternal mutation. b Schematic representation of MARSALA for linkage analysis to confirm the carrying status of maternal mutation allele in embryos. The amplified genomes of the peripheral blood samples and the embryos are sequenced. By analyzing maternal mutation and SNPs which are heterozygous in mother but homozygous in father, we could deduce the inherited allele of the embryos under screening. The allele carrying the maternal mutation was indicated in black. The red asterisk indicates the maternal mutation c.126–129delCTTT (HBB gene is reverse to the genome; therefore, the genome sequence on the corresponding site is delAAAG), and the blue asterisk indicates the paternal mutation. c Linkage analysis of paternal mutation allele by SNPs. Ten SNP markers were selected to construct the haplotype of the paternal disease-causing allele, by which four embryos (E01, E02, E03, E04) were identified to carry the mutation allele which is consistent with the direct sequencing result of the mutation site. d Linkage analysis of maternal mutation allele by SNPs. Nine SNP markers were selected to construct the haplotype of the maternal disease-causing allele, by which one embryo (E02) was identified to carry the mutation allele which is consistent with the direct sequencing result of the mutation site
Chromosome copy number variations were investigated for all embryos revealing normal euploid profiles, with the exception of one embryo (E04), which displayed a missing p arm of the chromosome 17 karyotype (46, XN, − 17p) (Fig. 2c).
Thus, when combining the mutation site sequencing with linkage analysis, and CNV profiles, two embryos (E05 and E06; Table 1) were identified to be euploid and free of either disease mutation, and recommended for transfer.
Comparison between targeted sequencing and universal MARSALA strategies
In order to validate the results obtained with the universal MARSALA strategy, we also performed the previously reported MARSALA approach based on targeted sequencing [18]. Briefly, the mutation sites of HBB gene and 60 adjacent SNPs were re-amplified with specific primers and then the PCR products were mixed with the MALBAC product for NGS. Following this procedure, targeted mutations and aneuploidy were detected simultaneously in one NGS run with only ~0.1× average genome coverage, and > 100× coverage for mutation sites and SNPs were obtained for linkage analysis. Comparable with the result obtained by the universal MASALAR method(Table 1), E05 and E06 were found to be euploid and disease allele-free, which were suitable for transfer. E06 was eventually chosen for transfer and a successful pregnancy was achieved.
Validation of the PGD/PGS results by prenatal diagnosis
A follow-up amniocentesis was performed at 17-weeks of gestation to confirm the previously obtained PGD/PGS results. DNA was extracted from the amniotic fluid to examine the mutation sites and comprehensive chromosomal screening using Sanger sequencing and NGS, respectively. The results of Sanger sequencing (Fig. 4a) demonstrated that the fetus was free of the − 28A > G and CD41–42 mutations. Additionally, no chromosomal aneuploidy was detected in fetus by NGS (Fig. 4b). These results demonstrate that our newly developed universal method is accurate and reliable, and that single-sperm cells can be used instead of proband and other family members's samples to select embryos free of monogenic mutations and chromosome abnormalities.
Detection of the mutation sites and chromosome ploidy of the fetus by amniocentesis. a The Sanger sequencing results for the − 28A > G mutation (left) and the CD41–42 mutation (right). The result of the reverse-strand sequencing of the − 28 locus was a T base, showing no mutation at the locus; the result of the CD41–42 also showed that this locus is free of mutation. b The CNV profile of the fetus indicated a euploidy with 46 chromosomes
Discussion
Use of Single-sperm cell genotyping by MARSALA as a universal method to obtain linkage information in the standard practice of PGD for paternally inherited diseases
Single-sperm cell genotyping has been previously used for studying the mechanism of meiotic recombination [22, 23]. However, due to a lack of effective WGA and genotyping techniques in the past, single-sperm cell genotyping had not been used to obtain linkage information in clinical practice. Progress in the development of NGS [24,25,26,27] and WGA [21] in recent years has enabled comprehensive and accurate genotyping of a single-cell, thereby opening the possibility of using single-cell genotyping to obtain linkage information. In previous studies [23, 25], single-sperm cell genotyping by SNP array or NGS was used to study meiotic recombination in healthy individuals. In the present study, we conducted a universal MARSALA strategy, based on the MALBAC-NGS technique, using single-sperm cells in order to deduce the haplotype for linkage analysis in pre-implanted embryos. Subjecting our samples to one reaction, we could simultaneously perform whole genome sequencing, check mutation status and obtain linked SNPs. The result obtained were comparable with the targeted sequencing-based MARSALA approach established previously [18]. Using the linkage analysis results from single sperm, targeted mutation detected in in vitro fertilized embryos can be further validated, increasing the accuracy of embryo selection for implantation. This universal procedure eliminates the requirement of proband or multiple family members, which sometimes can be very difficult or nearly impossible to obtain, and thus making it more applicable to a wider population of patients where the conventional PGD is not suitable. Such situations include, but are not limited to: (a) certain severe genetic disorders, where the mutation-carrying proband is often deceased, and (b) the couple intended for PGD treatment have undergone carrier screening before conception, such as for alpha and beta thalassemia in southern China [28].
In addition, this newly reported universal MARSALA approach employs MALBAC for WGA, which has been demonstrated in numerous published articles to have good coverage and high reproducibility, as well as a low ADO rate [21, 29]. We achieved sufficient coverage (> 10×) for SNP analysis under a ~3× average genome coverage sequencing without additional targeted enrichment. This strategy simplifies the PGD procedure by reducing the amount of work involved in specific primer design and PCR reactions.
Comparison with other techniques that can perform PGD/PGS simultaneously
In addition to NGS, a karyomapping approach utilizing SNP array has also been developed and applied to perform simultaneous PGD and PGS [6,7,8]. The ADO derived from WGA can be largely corrected by the haplotype construction via this genome-wide computing of SNPs. By comparison of the haplotypes between the tested embryos and the parents or affected siblings chromosome imbalance can also be deduced. More recently, a method called haplarithmesis which allows for both, copy number calling and genotype calling, has been invented and validated on monogenic cases as well as on translocation cases [30]. This method also relies on whole genome SNP array and mainly employs an algorithm called iChilds, which infers haplotypes of single cell from parental haplotypes. Nevertheless, both SNP-array based methods are incapable of detecting a mutation locus directly, and moreover, they require information from at least one additional family member, limiting their applicability.
As discussed above, the universal MARSALA strategy we reported in this study eliminates lengthy preliminary work-ups on linkage analysis of specific mutations and SNP markers and the need of additional information from relevant family members is. Based on the NGS platform, this approach also possesses some advantages over other technologies: (a) it detects the mutation sites directly allowing the discovery of de novo mutations; (b) the detection of chromosomal mosaicism in the blastocysts can be achieved with greater sensitivity and at a finer resolution than array-based tests [31]; (c) the analyses on mitochondrion genome (for instance copy number or mutation site) are also achievable [32].
Relevant to mention, the sole application of this newly proposed universal MARSALA strategy is only feasible in PGD for monogenetic diseasescaused by genes with relative high coverage in WGA products, due to its low-depth whole-genome sequencing coverage strategy. For genes without sufficient coverage, a MARSALA approach combined with the enrichments of specific SNP markers is recommendable to be employed. Nevertheless, the quick falling in the cost of NGS will soon make the whole genome deep sequencing affordable. We present here a new universal MARSALA PGD method for monogenic diseases which does not requir proband or additional family members studies to perform linkage analysis, it is cheaper and demonstrated to be reliable. We believe this method will conceivably broaden the applicable situations of the PGD technique.
References
Genes and human disease: Human Genetic Programmes. 2015. http://www.who.int/genomics/public/geneticdiseases/en/index2.html
Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15(suppl 1):R67–74.
Handyside AH, Pattinson JK, Penketh RJ, Delhanty JD, Winston RM, Tuddenham EG. Biopsy of human preimplantation embryos and sexing by DNA amplification. Lancet. 1989;333(8634):347–9.
Hellani A, Coskun S, Benkhalifa M, Tbakhi A, Sakati N, Al-Odaib A, et al. Multiple displacement amplification on single cell and possible PGD applications. Mol Hum Reprod. 2004;10(11):847–52.
Wells D, Sherlock JK. Strategies for preimplantation genetic diagnosis of single gene disorders by DNA amplification. Prenat Diagn. 1998;18(13):1389–401.
Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014;16(11):838–45.
Thornhill AR, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, et al. Karyomapping—a comprehensive means of simultaneous monogenic and cytogenetic PGD: comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–56.
Natesan SA, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, et al. Live birth after PGD with confirmation by a comprehensive approach (karyomapping) for simultaneous detection of monogenic and chromosomal disorders. Reprod BioMed Online. 2014;29(5):600–5.
Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod. 2009;24(5):1221–8.
Wang, L, Cram DS, Shen J, Wang X, Zhang J, Song Z, Xu G, Li N, Fan J, Wang S, Luo Y, Wang J, Yu L, Liu J, Yao Y, Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biology of reproduction, 2014: p. biolreprod. 114.120576.
Korenberg J, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci. 1994;91(11):4997–5001.
Treff NR, Northrop LE, Kasabwala K, Su J, Levy B, Scott RT Jr. Single nucleotide polymorphism microarray–based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil and Steril. 2011;95(5):1606–12. e2.
Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88(3):69.
Yang Z, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5(1):1–8.
Schoolcraft W, Surrey E, Minjarez D, Gustofson RL, Scott RT Jr, Katz-Jaffe MG. Comprehensive chromosome screening (CCS) with vitrification results in improved clinical outcome in women > 35 years: a randomized control trial. Fertil Steril. 2012;98(3):S1.
Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fert Steril. 2013;100(1):100–7. e1.
Scott RT Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100(3):697–703.
Yan L, Huang L, Xu L, Huang J, Ma F, Zhu X, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112(52):15964–9.
Ren Y, Zhi X, Zhu X, Huang J, Lian Y, Li R, et al. Clinical applications of MARSALA for preimplantation genetic diagnosis of spinal muscular atrophy. J Genet Genomics. 2016;43(9):541–7.
Jiao Z, Zhou C, Li J, Shu Y, Liang X, Zhang M, et al. Birth of healthy children after preimplantation diagnosis of β-thalassemia by whole-genome amplification. Prenat Diagn. 2003;23(8):646–51.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–6.
Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci. 1992;89(13):5847–51.
Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–30.
Zhou X, Ren L, Meng Q, Li Y, Yu Y, Yu J. The next-generation sequencing technology and application. Protein Cell. 2010;1(6):520–36.
Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150(2):402–12.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4.
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–85.
Xu X, Zhou YQ, Luo GX, Liao C, Zhou M, Chen PY, et al. The prevalence and spectrum of α and β thalassaemia in Guangdong Province: implications for the future health burden and population screening. J Clin Pathol. 2004;57(5):517–22.
Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102.
Zamani Esteki M, Dimitriadou E, Mateiu L, Melotte C, Van der Aa N, Kumar P, et al. Concurrent whole-genome haplotyping and copy-number profiling of single cells. Am J Hum Genet. 2015;96(6):894–912.
Sermon K. Novel technologies emerging for preimplantation genetic diagnosis and preimplantation genetic testing for aneuploidy, expert review of molecular diagnostics. Expert Rev Mol Diagn. 2017;17(1):71–82.
Shang W, Zhang Y, Shu M, Wang W, Li R, Chen F, et al. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reprod BioMed Online. 2017;36:67–74. https://doi.org/10.1016/j.rbmo.2017.10.110.
Funding
The work was supported by grants from the National Natural Science Foundation of China (81370765), Guangdong Provincial Key Laboratory of Reproductive Medicine (2012A06140003), Guangzhou Science and Technology Foundation (201300000097), National Key Technologies Research and Development program (2016YFC0900100 to L.H.), Beijing Municipal Science & Technology Commission Grants (D1511000024150002 to X.S.X.), the Pilot Construction of Reproductive Clinical Research and Transformation Center of Guangzhou (155700011/201508020006), and the funding from Beijing Advanced Innovation Center for Genomics at Peking University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
X.S.X. and S.L. are cofounders of Yikon Genomics Company, Ltd.
Rights and permissions
About this article
Cite this article
Wu, H., Shen, X., Huang, L. et al. Genotyping single-sperm cells by universal MARSALA enables the acquisition of linkage information for combined pre-implantation genetic diagnosis and genome screening. J Assist Reprod Genet 35, 1071–1078 (2018). https://doi.org/10.1007/s10815-018-1158-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-018-1158-9