Abstract
Preimplantation genetic testing has been used widely in recent years as a part of assisted reproductive technology (ART) owing to the breakthrough development of deoxyribonucleic acid (DNA) sequencing. With the advancement of technology and increased resolution of next generation sequencing (NGS), extensive comprehensive chromosome screening along with small clinically significant deletions and duplications can possibly be performed simultaneously. Here, we present a case of rare chromosomal aberrations: 46,XY,dup(15)(q11.2q13),t(16;18)(q23;p11.2), which resulted in a normally developed adult but abnormal gametes leading to recurrent pregnancy loss (RPL). To our best knowledge, this is the first report of t(16;18) translocation with such a small exchanged segment detected by NGS platform of MiSeq system in simultaneous 24-chromosome aneuploidy screening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Preimplantation genetic testing has become a promising practice of assisted reproductive technology (ART) in recent years. For known genetic abnormalities such as single-gene diseases or small chromosome segmental imbalances, preimplantation genetic diagnosis (PGD) can stop the transmission of chromosome abnormalities from the affected couples to their offspring [1]. Preimplantation genetic screening (PGS), on the other hand, is an evolving technique, which enables the assessment of numerical chromosomal constitution of embryos before transfer during the in vitro fertilization (IVF) treatment. PGS has been proposed to be beneficial for patients with advanced maternal age, RPL, and repeated implantation failure [2]. Aneuploidy is the most common cause of reproductive failure and may occur in any of the 24 chromosomes, which indicates that comprehensive chromosome screening for aneuploidy may augment embryo selection [3].
Case presentation
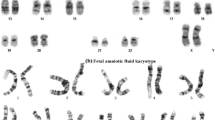
A 36-year-old Taiwanese woman suffering from five consecutive pregnancy losses was referred to our infertility clinic for ART because of an abnormal chromosomal analysis of her husband, which revealed a proximal 15q euchromatic variant and a reciprocal translocation between the long arm of chromosome 16 and the short arm of chromosome 18 by means of standard Giemsa-trypsin banding: 46,XY,dup(15)(q11.2q13),t(16;18)(q23;p11.2) (Fig. 1). Her obstetric history was remarkable for five times spontaneous abortion including one missed abortion at around the 7–8th week of gestation, blighted ovum twice at around the 6–7th week of gestation, and chemical pregnancy twice. Surveys for her RPL including infection (such as genital tract chlamydia and bacterial vaginosis), serum hormones (such as prolactin, thyroid function, anti-thyroid peroxidase antibody), uterine anatomy, and autoimmunity (such as anti-phospholipid antibodies, lupus anticoagulant, anti-cardiolipin antibody) were all negative. The husband was normal in development and intelligence without any known disease or neurodevelopment delay.
In the aid of Chromosome Analysis Suite (ChAS) software, we analyzed the sequencing data and aligned the reads to the human reference genome hg19. The size of the translocated segment of 16q23->qter was estimated to be 6~16 megabase (Mb). Conventionally, such a small segmental imbalance must be identified by PGD technology. To perform PGD testing, a unique probe customized for each couple is required in advance. Building the probe takes time, and multiple detection approaches are required to ensure the most reliable results. Moreover, not only the translocated segment but also the aneuploidy could lead to RPL, which is indicating the use of PGS. Performing both PGS and PGD requires an additional embryo biopsy and frozen-thaw process which may harm the embryos, not to mention the heavy economic burden on the couple. Therefore, we offered an option to detect the small segmental imbalances with NGS-based PGS based on our in-house data that the Illumina VeriSeq kit was able to detect small segmental imbalance as 5–6 Mb in particular position of the chromosome. Although it was possible that a euploid result of NGS was actually a carrier of balanced reciprocal translocation or failure to identify the very small segmental imbalances, the couple decided to undergo an IVF treatment with PGS.
She had antral follicle counts of 10 (right 4 and left 6) on the third day of menstruation. She received controlled ovarian hyperstimulation with gonadotropin-releasing hormone (GnRH) antagonist protocol using agonist triggering ovulation. The gonadotropin doses were 225 IU per day for 9 days (human menopausal gonadotropin, Menopur@). Nine oocytes were retrieved including seven metaphase II (MII) oocytes and two immature germinal vesicle (GV) oocytes. Five of the seven fertilized embryos reached blastocyst stage and were biopsied for PGS using the VeriSeq PGS Kit on the Illumina MiSeq@ System, including the DNA amplification of the samples with the SurePlex DNA Amplification Kit followed by sequencing of the amplified samples with the VeriSeq DNA Library Kit-PGS and the MiSeq Reagent Kit v3-PGS. The MiSeq Reporter Software analyzed the sequencing data and aligned the reads to the human reference genome hg19. After the bioinformatics from the MiSeq System was processed by the BlueFuse Multi Analysis Software (Illumina, Inc.), whole chromosome aneuploidy was called automatically while segmental gain or loss less than 20 Mb was called manually [4]. Among the five biopsied blastocysts, only two embryos were euploid (labeled as No. 2 and No. 5; Fig. 2), and the detected translocated segments size of del/dup(16)(q23.1q24.3) was 11~12.1 Mb. Single embryo transfer with the frozen-thawed No. 2 embryo was performed. The level of serum beta-human chorionic gonadotropin (b-hCG) 10 days after embryo transfer was 245.4 mIU/ml, which became 709 mIU/ml 2 days later. An intrauterine gestational sac with positive fetal heartbeat was confirmed by ultrasound at the sixth week of gestation. Amniocentesis was performed in the second trimester for karyotyping and array comparative genomic hybridization (aCGH) (Cytoscan 750 K Array, Affymetrix, Santa Clara, CA, USA) in order to identify possible balanced translocation and small 15q11-13 duplication which could not be detected by PGS. Standard Giemsa-trypsin banding with 550-band level revealed a balanced reciprocal translocation 46,XX,t(16;18)(q23; p11.2), and aCGH showed normal result. No 15q11-13 duplication was identified. The pregnancy was uneventful, and a healthy female baby was delivered at term.
Discussion
We report a case of a 36-year-old female presented with five consecutive pregnancy losses, and her husband carried a rare chromosomal aberration with proximal 15q euchromatic variant and a balanced reciprocal translocation, designated as 46,XY,dup(15)(q11.2q13),t(16;18)(q23;p11.2). The translocated segment 16q23->qter was estimated as small as 6~16 Mb which must be identified by PGD technology in the past. In the published literature, no other case with the same breakpoints of chromosomes 16 and 18 had been reported. However, Baptista et al. [5] had reported a case of recurrent miscarriage whose chromosome analysis was ascertained as 46,XX,t(16;18)(q24;q21.1)mat, which was similar to our patient. As for the duplication, the 15q11-13 locus harbors several genes that regulate genomic imprinting and have been reported in association with developmental delay, intellectual disability, and autism [6]. The diagnosis of dup15q is established by detection of at least one extra maternally derived copy of the Prader-Willi/Angelman critical region (PWACR), a region approximately 5 Mb long within chromosome 15q11.2-q13.1 [7]. Cytogenetically detectable elongation of the 15q proximal region can be associated with PWACR interstitial duplications or with inherited juxtacentromeric euchromatic variants [8]. The former as we mentioned above are pathogenic recurrent duplications while the latter euchromatic variants reflect polymorphic copy number variations of segments containing genes and pseudogenes which are polymorphic in normal population and reach a cytogenetically detectable level only when multiple copies are present. Euchromatic variants usually segregate without apparent phenotypic consequence.
In this case, the husband was apparently a normal adult without any known diseases or neurodevelopment disorders. We consequently speculated that the husband to be a euchromatic variant judged by the phenotype. The pathogenic relevant 15q11-13 duplications are not distinguishable from the innocuous euchromatic variant with conventional cytogenetic methods, but only with molecular analysis. The aberration can be detected with PGD but not with NGS-based PGS since the locus contains mostly pseudogenes and the size is beyond the resolution, though it is apparently an innocuous euchromatic variant in this case. Besides, considering there are approximately less than 60% euploid embryos in the wife’s age [9], concomitant aneuploidy screening may be beneficial by reducing pregnancy loss related to aneuploidy in chromosomes not involved in the segmental imbalance. However, performing both PGD and PGS not only increased the potential harm to the embryos but also formed a heavy economic burden on the couple. Taking all together, we suggested NGS-based comprehensive chromosome screening for aneuploidy and the reciprocal translocation simultaneously based on our in-house data that the Illumina VeriSeq kit was able to detect small segmental imbalance as 5–6 Mb in particular position of the chromosome. Intra-partum amniocentesis was arranged for karyotyping and aCGH to validate the pre-implantation NGS result as well as to identify possible balanced translocation and small 15q11-13 duplication which could not be detected by pre-implantation NGS.
Over the past decades, DNA sequencing had a breakthrough development. By means of massive parallel sequencing, NGS enables analysis of massive sequencing in a single run with a hugely reduced sequencing cost per megabase [5]. NGS-based PGS has become the mainstream in IVF practice owing to its efficiency yet affordable cost. With the advancement of technology, chances are extensive comprehensive chromosome screening and single-gene disorder/clinical significant deletions and duplications can be performed simultaneously by improving the resolution of NGS-based PGD [10].
According to the manufacture’s declaration, the effective resolution of the MiSeq@ system for PGS used in our case is 20 Mb.1 Fiorentino et al. had demonstrated that the NGS-based comprehensive chromosome aneuploidy screening protocol using the MiSeq@ from Illumina had a high resolution and allowed accurate detection of segmental imbalances as small as 14 Mb [11]. To compare, the NGS-based PGS which readily detected the segmental imbalance of 16q23->16qter in our case was even smaller as 11 Mb. We had shown that the resolution of NGS-based PGS was further improved and made it possible using a single methodology to detect small chromosome segmental imbalances with simultaneous 24-chromosome aneuploidy screening.
There are other NGS methods reporting even higher resolution. Based on a semiconductor ion torrent platform, Ion PGM™ Sequencing (Life Technologies), the NGS method validated for PGD of translocations reported a resolution of 5 Mb for segmental imbalances [12]. However, the ion torrent platform has some disadvantages. It has more hands-on time and fewer reads at higher cost per megabase relative to MiSeq@ System (Illumina, Inc.) and also smaller user community [13, 14]. Another quantitative NGS method termed copy number variation sequencing (CNV-Seq) reported a precisely 0.8 Mb imbalance detection [15]. However, this new NGS method requires higher depth of sequencing, which would add extra costs of the PGS/PGD test. It is also still an experimental technology and not commercialized yet. With scattered cases, the NGS methods mentioned accordingly are neither justified to diagnose the chromosome segmental imbalances generally nor superior to other NGS methods. However, they do demonstrate promising results and lead the way for further investigation. It remains to be determined what is the optimal number of reads to strike a balance between detection of small imbalance chromosomal segments and the cost per megabase. How to distinguish the signal noise from chromosome aberrations is also of concern in the quality control.
We acknowledge that there is large heterogeneity among chromosomes. The sensitivity for detecting a segmental imbalance is not only related to technology improvement but also to the position where the segmental imbalance resides. Some regions have higher density than the others, and the probes in NGS do not evenly cover the whole chromosome. However, we would like to share our experience by this case report that it was feasible to detect a small segmental imbalance located at 16q23 by NGS-based PGS instead of PGD even though the segmental imbalance was smaller than the officially claimed resolution. In this case, there was good coverage in the particular area which facilitated calls in that region and came with good results. Nevertheless, we had limitations of no validation for the remaining embryos with different methodology.
Another aspect regarding this case was that the karyotyping obtained from the amniocentesis revealed balanced reciprocal translocation. Array CGH and ordinary NGS methods available in market are unable to detect balanced translocations and inversions. Therefore, the combination of karyotyping with genotyping data is necessary to provide complete information, especially in cases of known parental chromosome aberrations as in our case.
In conclusion, we present a case of a phenotypically normal adult suffering from RPL possibly related to a rare chromosomal balanced reciprocal translocation. By using the Illumina NGS platform, we performed PGS only to select and transfer a euploid embryo which resulted in a successful pregnancy outcome. To the best of our knowledge, this is the first report of such an unusual breakpoints and extremely small chromosome segmental imbalance detected by NGS platform of MiSeq system in simultaneous 24-chromosome aneuploidy screening.
References
Handyside AH. Preimplantation genetic diagnosis after 20 years. Reprod BioMed Online. 2010;21(3):280–2.
Liss J, et al. Current methods for preimplantation genetic diagnosis. Ginekol Pol. 2016;87(7):522–6.
Fiorentino F, et al. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29(12):2802–13.
Illumina product information sheet: VeriSeq PGS Kit-MiSeq System. Accessed on 10 December 2016; Available from: http://www.illumina.cin/centent/dam/illumina-marketing/documents/products/product_information_sheets/product-info-ceriseq.pdf.
Baptista J, et al. Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur J Hum Genet. 2005;13(11):1205–12.
Kalsner L, Chamberlain SJ. Prader-Willi, Angelman, and 15q11-q13 duplication syndromes. Pediatr Clin N Am. 2015;62(3):587–606.
Finucane BM, Lusk L, Arkilo D, et al. 15q duplication syndrome and related disorders. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 2016. p. 1993–2017.
Carelle-Calmels N, et al. Proximal 15q familial euchromatic variant and PWS/AS critical region duplication in the same patient: a cytogenetic pitfall. Eur J Med Genet. 2008;51(6):547–57.
Demko ZP, et al. Effects of maternal age on euploidy rates in a large cohort of embryos analyzed with 24-chromosome single-nucleotide polymorphism–based preimplantation genetic screening. Fertil Steril. 2016;105(5):1307–13.
Brezina PR, et al. Preimplantation genetic testing for aneuploidy: what technology should you use and what are the differences? J Assist Reprod Genet. 2016;33(7):823–32.
Fiorentino F, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014;101(5):1375–82.
Bono S, et al. Validation of a semiconductor next-generation sequencing-based protocol for preimplantation genetic diagnosis of reciprocal translocations. Prenat Diagn. 2015;35(10):938–44.
Glenn TC. Field guide to next-generation DNA sequencers, 2016 update. NGS Field Guide-Table 4—Advantages & Disadvantages; Available from: http://www.molecularecologist.com/next-gen-table-4-2016/.
Quail MA, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13(1):341.
Zhang W, et al. Clinical application of next-generation sequencing in preimplantation genetic diagnosis cycles for Robertsonian and reciprocal translocations. J Assist Reprod Genet. 2016;33(7):899–906.
Acknowledgements
The authors thank all the members of the Genetics Generation Advancement Corp. (GGA Corp.) for supporting this study in various forms, including sample preparation and interpretation, technical support, and extremely helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, YX., Chen, CW., Lin, YH. et al. Prediction of a rare chromosomal aberration simultaneously with next generation sequencing-based comprehensive chromosome screening in human preimplantation embryos for recurrent pregnancy loss. J Assist Reprod Genet 35, 171–176 (2018). https://doi.org/10.1007/s10815-017-1044-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-017-1044-x