Abstract
Purpose
To evaluate the overall expression patterns of imprinted genes in human embryonic stem cells following long term culture and differentiation.
Materials and methods
Expression levels of 65 imprinted genes determined by PCR array were analyzed in one human embryonic stem cell line (cHES1) following prolonged passaging and differentiation.
Results
Transcripts of 63 imprinted genes were detected in cHES1 cells. Expression levels of all but 5 imprinted genes did not correlate with passage numbers or differ in cells after passage 50 compared with those before passage 50. SLC22A2, SLC22A3, CPA, H19, COPG2IT1 and IGF2 expression were significantly increased in embryoid bodies compared with undifferentiated cells.
Conclusions
The global expression profiles of imprinted genes are generally stable in human embryonic stem cells after prolonged passaging and differentiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human embryonic stem (hES) cells carry the potential to differentiate into all known types of somatic cells in living organisms and capacity to self renew indefinitely [1, 2]. Thus hES hold great promise for cell replacement therapies. Embryonic stem (ES) cells are derived from pre-implantation embryos, a stage characterized by global epigenetic remodeling [3]. Genomic imprinting is a unique epigenetic phenomenon, which refers to silencing of one parental allele in somatic nucleus depending upon the parent of origin, eventually resulting in allele specific expression. Although imprinted genes comprise a small subset of the human genome, they have been shown to have profound effects on cell behavior and play important roles in fetal and behavioral development [4]. Differential epigenetic/chromatin modifications occur during gametogenesis and preimplantation development. Thus the epigenetic status of ES cells may be disturbed due to embryo manipulation by assisted reproductive technology (ART) or derivation and in vitro culture of ES cells. Inappropriate expression of imprinted genes correlates with several human pathologies, including cancers [5–7]. Therefore investigation on imprinting status of hES cells is crucial for their use in regenerative medicine.
It has been reported that ART actually destroys the imprinting status of some genes in mammals such as human oocytes [8], sheep [9] and mouse [10] embryos. Furthermore, imprinted genes in mouse and rhesus monkey ES cells were documented to be perturbed [11, 12]. Research on a small number of imprinted genes showed that hES cells have a more stable imprinting status compared with mouse ES cells [13]. Kim examined allele specific expression of 22 imprinted genes in hES cells and found that 14 imprinted genes showed monoallelic while 8 genes showed biallelic expression in the majority of hES cell lines. This result suggests that there are specific genes which are vulnerable to imprinting variability in hES cells [14]. However, the dynamic alteration ofexpression of these genes during prolonged passage was not recorded in Kim’s study.
ES cells could form embryoid bodies (EBs) in vitro, attachment of EBs to the adhesive substrate is accompanied by subsequent differentiation into various somatic tissues [15]. Since EBs represent the common initial stage of lineage specific differentiation in hES cells, the investigation on imprinted genes in EBs would be essential to the potential use of hES cells in cell therapies. Additionally, ES cells are derived from inner cell mass of blastocyst, their differentiation into EBs, which comprise three embryonic germ layers resembles events that occur in vivo shortly before and after the embryonic implantation [16]. Thus, study on imprinted genes of EBs will also help to understand the epigenetic regulatory mechanisms in early human development. Whereas little is known about epigenetic factors accompanying proliferation and differentiation of hES cells, a recent data reported expression levels of a limited amount of imprinted genes in hES cells derived EBs [17], However, the mRNA levels of the majority of imprinted genes in EBs still remain obscure.
Our study selected 65 genes (27 maternally and 38 paternally expressed genes) which cover the majority of currently known imprinted genes in humans. Expression levels of imprinted genes were examined simultaneously by real-time quantitative reverse transcriptase-polymerase chain reaction (Q-RT-PCR) using PCR array in the human embryonic stem cell line cHES1. Changes in dynamic expression of imprinted genes were tested during long-term culture of hES cells to evaluate the disruption of imprinted genes by in vitro culture. Expression profiles of imprinted genes were also determined in EBs at day 14 to investigate the epigenetic status in the early stage of hES cells differentiation.
Materials and methods
Culture and in vitro differentiation of hES cells
One hES cell line, which was derived in our center was cHES1, 46XX. Methods of in vitro passage and differentiation into EBs at day 14 were performed as previously reported [18]. Undifferentiated cells at different passages (p27, p37, p47, p57, p67 and p77) and hES (p27, p37 and p47) derived EBs at day 14 were collected for mRNA level examination by Q-RT-PCR using PCR array.
Quantitative real time reverse transcription-PCR
Samples were derived into 3 groups (Undifferentiated hES cells before passage 50, undifferentiated hES cells after passage 50, hES cells derived EBs at day 14). Each group has 3 independent samples (biological replicates). Each sample was repeated twice. For undifferentiated cHES1 cells and cHES1 derived EBs, total RNA was extracted and Dnase digested using the RNeasy Mini Kit (Qiagen). 1.5 μg of total RNA was reverse transcribed (RT) into cDNA using SuperScript III Reverse Transcriptase (Invitrogen), and oligo(dT) 18(Promega). For quantitative real-time PCR (qPCR, we used RT2 Profiler Custom PCR Array from SuperArray Bioscience to simultaneously assay the mRNA levels of 69 genes including four housekeeping genes (B2M, RPL13A, GAPDH and HPRT1) according to the recommendations of the manufacturer. The gene list focused on those with affirmative imprinting status (27 maternally and 38 paternally expressed genes), excluding predicted, conflicting, provisional and unknown data (listed at least on one of the two web sites: http://WWW.geneimprint.com; http://WWW.otago.ac.nz, Table 1). The selected genes cover the majority of currently known imprinted genes in humans. Thermal cycling was carried out with a 10 min denaturation step at 95°C, followed by 40 two-step cycles: 15 s at 95°C and 60 s at 60°C. Each reaction included 40 ng of total RNA and the proper negative controls (no reverse transcription, no template), positive PCR control and genomic DNA control. Amplification data was collected by the ABI PRISM 7900 and analyzed by the Sequence Detection System 2.0 software (ABI). After completion of these cycles, melting–curve data were then collected to verify PCR specificity. Array results showed that expressions of 4 candidate housekeeping genes (HKG) did not change significantly in hES cells under the experimental conditions (in vitro culture and differentiation); see supplementary Fig. 1. Thus, GAPDH, B2M, HPRT1 and RPL13A were all chosen as suitable reference genes for quantitative gene expression analysis. Threshold cycle numbers (Ct) for each imprinted gene from different samples were determined and standardized to the average HKG value (△Ct = Gene Ct- average HKG Ct), The values were then compared between groups using the 2-△△Ctmethod [19]. RT-qPCR was also carried out on total RNA extracted from the mouse feeders to confirm the absence of reactivity with mouse cDNA.
Statistical analysis
Pearson’s or Spearman’s correlation coefficient test was used to analyze the correlations among levels of gene expression and passage numbers. One way ANOVA or Student’s t test was used for the analysis of gene expression. A P value of <0.05 was considered significant. All statistical analyses were performed with Excel X and SPSS15.0.
Results
Imprinted genes detected in undifferentiated hES cells
We showed that paternally expressed genes INS and MKRN3 were not detected in all the passages of hES cells. The other 63 (96.9%) imprinted genes were present in hES cells (Table 2). 27(100%) maternally and 36 (94.7%) paternally expressed genes were detected in hES cells. All the examined genes were not detected in MEFs, confirming no reaction of feeder cDNA (Data not shown).
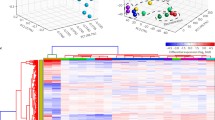
a, b, c High correlation between relative expression level of imprinted genes (INPP5F, KCNQ1OT1, NNAT) and passage numbers for hES cells (n = 6). d Comparison of relative expression of imprinted genes between hES cells before and after passage 50. Y-value is expressed as relative fold change in mRNA levels in cells after passage 50 when compared with those in cells before passage 50, defined as 1. Bars indicate mean ± SEM (n = 3). *P < 0.05, **P < 0.01
Transcriptional levels of imprinted genes in undifferentiated hES cells during long term culture
Transcriptional levels of INPP5F (r 2 = 0.8506, P < 0.01) and KCNQ1OT1 (r 2 = 0.9504, P < 0.01) showed negative correlation with passage numbers, while NNAT (r 2 = 0.6995, P < 0.05) showed positive correlation with passage numbers (Fig. 1a, b, c). The other 62 imprinted genes did not correlate with passage numbers of hES cells (Table 2). Furthermore, expression levels of five genes including INPP5F, KCNQ1OT1 and NNAT changed dramatically with no alteration of other 60 genes in hES cells after passage 50 compared with those before passage 50 (Fig. 1d). INPP5F expression levels decreased 2.9 fold (P < 0.05); KCNQ1OT1 expression levels decreased 2.9 fold (P < 0.05); NNAT levels increased 6.7 fold (P < 0.01). Additionally, expression of CALCR and PEG10 levels decreased 2.5 and 3.6 fold respectively.
Transcriptional levels of imprinted genes in differentiated hES cells
We examined the expression levels of imprinted genes in EBs at day 14 from hES cells of different passages (passage 27, 37 and 47) compared with their undifferentiated counterparts by RT-qPCR and found that in hES cells derived EBs, expression levels of 5 maternally (SLC22A2, SLC22A3, COPG2IT1, H19 and CPA4) and 1 paternally (IGF2) expressed genes were markedly increased (Fig. 2). SLC22A2 levels increased 4.7 fold, P < 0.01; SLC22A3 levels increased 3.7 fold, P < 0.01; CPA4 levels increased 3.7 fold; P < 0.01; H19 levels increased 2.2 fold, P < 0.05; COPG2IT1 levels increased 3.0 fold, P < 0.05; IGF2 levels increased 2.3 fold P < 0.05. The expression levels of other 59 imprinted genes did not change significantly following EB formation in hES cells (Table 3).
Discussion
This study assayed the mRNA levels of imprinted genes in undifferentiated and differentiated hES cells to describe the global transcriptional status of imprinted genes comprehensively and investigate the dynamic transcript alteration of imprinted genes in hES cells following prolonged passages and upon differentiation. To our knowledge, the widest range of imprinted genes in hES cells was investigated in this research to date. Sun [17] and Abeyta [20] detected 19, 28, 22 and 32 imprinted genes in 4 different hES cell lines by Affymetrix Oligo micro-array. Actually, we found that not only 19 ∼ 32 but nearly all the imprinted genes (63/65, 96.9%) were detected in cHES1 cells. The fact that there were more imprinted genes detected in our study than previously reported might be due to the different methods used to examine imprinted genes. We used PCR array which is more sensitive and accurate than Affymetrix Oligo micro-array used by previous studies. INS and MKRN3 were not detected in all the passages of cHES1 cell line, while somatic cells showed significant expression of INS and MKRN3 tested on the same arrays (unpublished data from our lab), indicating that these genes are still silenced in human blastocysts and hES cells. So far, the expression status of 22 imprinted genes in hES cells have been reported previously. Most of them have been identified as monoallelic expression (establishment of imprint). Our study expanded the scope of imprinted genes and demonstrated that 96.9% imprinted genes were expressed in hES cells, suggesting that the majority of imprinted genes have been activated at least at blastocyst stage or during derivation of hES cells, and hES cells offer an excellent model for studying epigenetic regulation of imprinting.
Recent work focused on monoallelic or biallelic expression status of imprinted genes in hES cells [13, 14, 17]. However, the transcriptional levels of imprinted genes are largely unknown. Taking into account that the mRNA levels influence the amount of protein directly, resulting in different biological effects, we first analyzed the expression levels of 65 imprinted genes in hES cells from passage 27 to passage 77. We showed that transcriptional levels of INPP5F, KCNQ1OT1 and NNAT gradually reduced or increased with passage numbers, indicating that transcript of these three imprinted genes might closely relate with duration of in vitro culture of hES cells. This study also showed that under the same culture conditions, the variation trends of expression levels for imprinted genes were different. INPP5F and KCNQ1OT1 showed a decrease while NNAT showed an increase. The reason for these results is not understood and may relate with epigenetic property of individual imprinted genes. Expression of 5 genes (CALCR, PEG10, INPP5F, KCNQ1OT1 and NNAT) varied greatly in hES cells after passage 50 compared with those before passage 50, suggesting that in vitro culture actually affect the expression of a few imprinted genes in hES cells.
Rugg-Gunn revealed that H19 was biallelically expressed with elevated expression levels after prolonged passages (passage66 ∼ passage101) in H9 line, while this phenomenon did not exist in three other hES cell lines [13]. In our study, H19 did not vary significantly in cHES1 cells line after extended culture, which might suggest that imprinted genes which are vulnerable to disruption varied among hES cell lines. All the 65 imprinted genes except CALCR, PEG10, INPP5F, KCNQ1OT1 and NNAT did not change dramatically after long term culture of cHES1, indicating that mRNA levels of the majority of imprinted genes are quite stable in hES cells, in accordance with previous data that imprinted genes could generally maintain monoallelic expression after long term culture of hES cells [13, 14, 17]. Since many more imprinted genes were included in our study, we provide more evidence to show that hES cells posses substantial imprinting stability during prolonged passaging at least in the cell line we examined. This is an encouraging fact when considering the safety of transplanting hES cell derived tissues. However, differential expression of a few imprinted genes was identified in subculture of cHES1. Evidence has been presented that prenatal growth retardation may be due to deletion of PEG10 [21]. Defects of KCNQ1OT1 were found to be associated with children born small for gestational age when they are conceived by intracytoplasmic sperm injection [22]. NNAT is frequently overexpressed in a variety of human cancers [23–25]. Thus whether these aberrantly expressed genes influence the cellular function, cause phenotypic features and eventually affect the therapeutic use of hES cells needs further investigation.
It was reported that Expression of IGF2 and H19 increased significantly in SHhES1 and HUES-7 derived EBs [17]. We showed that mRNA levels of H19 and IGF2 increased remarkably in cHES1 derived EBs at day 14. H19 and IGF2 are known to be involved in embryonic growth and development [26]. Additionally, we found that expression levels of 4 imprinted genes (COPG2IT1, CPA4, SLC22A2 and SLC22A3) were elevated in EBs. SLC22A2, SLC22A3 and SLC22A18 are highly expressed in tissues with metabolite transport functions [27–29]. Dosage regulation of the metabolite transporter genes by imprinting may regulate placental and fetal growth [29]. CPA4 has a potential role in cell proliferation and differentiation [30]. Our study is the first to note that expression levels of SLC22A2, SLC22A3 and CPA4 increased during EB formation of hES cells. This suggests that these genes are developmentally regulated. SLC22A2/SLC22A3, COPG2IT1/CPA4 and H19/IGF2 are located nearby on chromosome 6q26 [31], 7q32.2 [32] and 11p15.5 [33] respectively. Therefore these results raise important questions about mechanistic and functional relationships between imprinted genes in a cluster. Previous evidence showed that in the imprinted IGF2/H19 domain. IGF2 and H19 are co-expressed in endoderm- and mesoderm-derived tissues during embryogenesis, co-regulate embryonic growth and development. H19 influences growth by way of a cis control on IGF2 expression [34]. Transcripts of both genes are controlled by a common imprinted control region, (ICR) [35]. However, such interactions and mechanisms remain unclear in the imprinted domain 6q26 and 7q32. Our study showed that imprinted domain at 6q26, 7q32.2 and 11p15 might be involved in formation of the 3 germ layers with differentiation of hES cells, and might play an important role in human early development. Though how these physically linked imprinted genes interact with each other to exert their effects needs to be further elucidated.
The other 59 imprinted genes did not differ significantly in EBs compared with undifferentiated cells This may indicate that only a small amount of imprinted genes are involved in early differentiation of hES cells, while the transcriptional levels of other imprinted genes, which are irrelevant to differentiation, remain stable.
During the differentially expressed genes identified in our study, PEG10 and KCNQ1OT1 were reported to show monoallelic expression in most examined hES cells lines [14]. H19 showed monoallelic expression when its expression increased dramatically in EBs [17]. Combined with previous reports, these phenomenons might suggest that expression level is not always consistent with mono or biallelic expression status in imprinted genes. Transcriptional level might reflect imprinting stability of imprinted genes more sensitively. However, the relationship between mRNA level and allelic expression of imprinted genes and their regulatory mechanism need to be further studied.
Conclusions
hES cells expressed the majority of imprinted genes (63/65,96.9%). Long-term culture in vitro did not affect transcriptional levels of the majority of (60/65, 92.3%) imprinted genes. H19, IGF2, COPG2IT1, CPA4, SLC22A and SLC22A3 might play a role in early differentiation of hES cells. Whether the differential expression of a few imprinted genes identified in subculture of hES cells affect the cellular function and cause phenotypic consequences needs to be established.
References
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7.
Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708.
Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14.
Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93.
Wu J, Qin Y, Li B, He WZ, Sun ZL. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics. 2008;91:443–50.
Wang CC, Xiao Y, Hu ZH, Chen YB, Liu N, Hu GX. PEG10 directly regulated by E2Fs might have a role in the development of hepatocellular carcinoma. FEBS Lett. 2008;582:2793–8.
Yu WQ, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–10.
Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22:26–35.
Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–4.
Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–26.
Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout 3rd WM, Biniszkiewicz D, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–7.
Fujimoto A, Mitalipov SM, Kuo HC, Wolf DP. Aberrant genomic imprinting in rhesus monkey embryonic stem cells. Stem Cells. 2006;24:595–603.
Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–7.
Kim KP, Thurston A, Mummery C, Oostwaard DWV, Priddle H, Allegrucci C, et al. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–42.
Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–55.
Poirier F, Chan CT, Timmons PM, Robertson EJ, Evans MJ, Rigby PW. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113:1105–14.
Sun BW, Yang AC, Feng Y, Sun YJ, Zhu YF, Zhang Y, et al. Temporal and parental-specific expression of imprinted genes in a newly derived Chinese human embryonic stem cell line and embryoid bodies. Hum Mol Genet. 2006;15:65–75.
Li T, Zhou CQ, Mai QY, Zhuang GL. Establishment of human embryonic stem cell line from gamete donors. Chin Med J (Engl). 2005;118:116–22.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–8.
Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–6.
Kanber D, Buiting K, Zeschnigk M, Ludwig M, Horsthemke B. Low frequency of imprinting defects in ICSI children born small for gestational age. Eur J Hum Genet. 2009;17:22–9.
Schulz R, McCole RB, Woodfine K, Wood AJ, Chahal M, Monk D, et al. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum Mol Genet. 2009;18:118–27.
Siu IM, Bai R, Gallia GL, Edwards JB, Tyler BM, Eberhart CG, et al. Coexpression of neuronatin splice forms promotes medulloblastoma growth. Neuro Oncol. 2008;10:716–24.
Uchihara T, Okubo C, Tanaka R, Minami Y, Inadome Y, Iijima T, et al. Neuronatin expression and its clinicopathological significance in pulmonary non-small cell carcinoma. J Thorac Oncol. 2007;2:796–801.
Morison IM, Becroft DM, Taniguchi T, Woods CG, Reeve AE. Somatic overgrowth associated with overexpression of insulin-like growth factor II. Nat Med. 1996;2:311–6.
Lee WK, Wolff NA, Thevenod F. Organic cation transporters: physiology, toxicology and special focus on ethidium as a novel substrate. Curr Drug Metab. 2009;10:617–31.
Bourdet DL, Pritchard JB, Thakker DR. Differential substrate and inhibitory activities of ranitidine and famotidine toward human organic cation transporter 1 (hOCT1; SLC22A1), hOCT2 (SLC22A2), and hOCT3 (SLC22A3). J Pharmacol Exp Ther. 2005;315:1288–97.
Dao D, Frank D, Qian N, O’Keefe D, Vosatka RJ, Walsh CP, et al. IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes. Hum Mol Genet. 1998;7:597–608.
Bentley L, Nakabayashi K, Monk D, Beechey C, Peters J, Birjandi Z, et al. The imprinted region on human chromosome 7q32 extends to the carboxypeptidase A gene cluster: an imprinted candidate for Silver-Russell syndrome. J Med Genet. 2003;40:249–56.
Verhaagh S, Schweifer N, Barlow DP, Zwart R. Cloning of the mouse and human solute carrier 22a3 (Slc22a3/SLC22A3) identifies a conserved cluster of three organic cation transporters on mouse chromosome 17 and human 6q26-q27. Genomics. 1999;55:209–18.
Yamada T, Mitsuya K, Kayashima T, Yamasaki K, Ohta T, Yoshiura K, et al. Imprinting analysis of 10 genes and/or transcripts in a 1.5-Mb MEST-flanking region at human chromosome 7q32. Genomics. 2004;83:402–12.
Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65 Suppl 3:50–8.
Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188–93.
Viville S, Surani MA. Towards unravelling the Igf2/H19 imprinted domain. Bioessays. 1995;17:835–8.
Acknowledgments
This work was supported in part by grants from the Doctoral Station Foundation, Ministry of Education (20050558097); National 863 program (20060102A1022, 2006AA02A102, 2006AA02A101); Guangdong provincial science and technology foundation (2008A030201028); National 973 program (2007CB948100); Chinese National Nature Foundation (30801239); The PhD programs foundation of ministry of China (200805581164). 2009 Sun yat-sen university sponsored programs (09YKP436).
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiuyun Mai and Qingyun Mai contributed equally to this work.
Capsule
Transcriptional levels of imprinted genes are generally stable in human embryonic stem cells following extended culture and upon differentiation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Comparison of housekeeping genes (HKG) expression among 3 groups (undifferentiated hES cells before passage 50, after passage 50 and differentiated hES cells). No change of expression level in 4 HKG (B2M, HPRT1, RPL13A and GAPDH) was observed. Y-value is expressed as relative fold change in mRNA levels when compared with those in cells before passage 50 defined as 1. Bars indicate mean ± SEM (n = 3). The P value were determined by ANOVA, P > 0.05 (DOC 70 kb)
Rights and permissions
About this article
Cite this article
Mai, X., Mai, Q., Li, T. et al. Dynamic expression patterns of imprinted genes in human embryonic stem cells following prolonged passaging and differentiation. J Assist Reprod Genet 28, 315–323 (2011). https://doi.org/10.1007/s10815-010-9524-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-010-9524-2