Abstract
Purpose
By comparing the chromosomal constitution among the arrested cleavage-stage embryos, blastocysts and human embryonic stem cells (hESCs) which are all derived from monopronuclear (1PN) zygotes, it is aimed to determine whether chromosomally normal embryos can be reliably selected by blastocyst culture.
Methods
After 1PN zygotes are sequentially cultured for 5 days, the blastocysts and arrested cleavage-stage embryos were analyzed by fluorescence in situ hybridization (FISH) with probes for chromosomes 18, X and Y; G-banding analysis was adopted to analyze the karyotype of 1PN hESCs.
Results
The diploid rate of blastocysts was 74.6%, which was significantly (P < 0.001) higher than that of arrested cleavage-stage embryos (31.6%), and the diploid rate of hESCs was 97.0%, which was significantly (P < 0.01) higher than that of blastocysts; the haploid embryos were excluded by blastocyst culture; nevertheless, there still existed such chromosomal abnormalities as mosaic and monosomic in blastocysts and trisomy in hESCs.
Conclusions
Blastocyst culture is an effective method to select against chromosomal abnormalities, especially the haploids in 1PN embryos; however, development to the blastocyst stage is not a reliable marker for mosaicism or aneuploidy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In conventional in-vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) treatments, normal fertilization is determined by the presence of two distinct pronuclei (2PN) at 16–20 h after insemination. Occasionally, only one pronucleus (1PN) is observed in oocytes. The frequencies of the occurrence of 1PN zygotes ranged from 1.6% to 7.7% [1]. The mechanism of 1PN zygotes has been extensively studied as: a) asynchrony of the pronuclear appearance [2–4] or b) the male and female pronuclear fusion together [5, 6], which will result in chromosomally normal embryos; while c) androgenetic or gynogenetic parthenogenesis, which will lead to haploid embryos [7, 8]. The embryos from normal fertilized 1PN zygotes and those chromosomally abnormal ones look so similar in morphological appearance that it is hard to distinguish between them [9], which blocks their clinical application. Establishing a method to select the chromosomally normal ones from the 1PN-derived embryos will improve the efficiency of ART treatment.
As a routine method in clinical IVF, blastocyst culture and transfer has proved to bring in a significantly increase in implantation [10], which helps to select embryos with high developmental potential and chromosomally normal embryos [11]. Although there have been several case reports about transferring 1PN blastocysts and achieving normal live-birth, the chromosomal constitution of 1PN blastocyst still remains uncertain.
The human embryonic stem cells (hESCs), which are derived from the inner cell mass (ICM) of blastocysts and have the ability to differentiate into the three germ layers of our body [12, 13], are similar to the fetus in the gestation. Thus, it is believed that the cytogenetic analysis of hESCs may helps to estimate whether the 1PN-derived embryos selected by blastocyst culture will be chromosomally normal after implantation. To our knowledge, so far, there has not been any systemic cytogenetic analysis of hESCs derived from 1PN zygotes.
Our study is aimed to determine whether chromosomally normal embryos can be reliably selected by blastocyst culture through comparing the chromosomal constitution among the arrested cleavage-stage embryos, blastocysts and hESCs which are all derived from 1PN zygotes by using FISH and G-banding analysis.
Materials and methods
The experiment was approved by the ethical committee of the CITIC-Xiangya Reproductive and Genetic Hospital, and the 1PN zygotes used in this study were donated by couples who underwent IVF/ICSI and had signed consent forms.
IVF and ICSI treatment
Ovarian stimulation was performed by our routine down-regulation protocol using the gonadotrophin-releasing hormone agonist (GnRHa): gonadotrophin was given from day 2–3 of menstrual cycle to stimulate follicle growing; h-HCG was injected when there were at least 3 dominant follicles with diameter ≥ 18 mm. 34–36 h after administration of h-HCG, oocyte retrieval was performed by ultrasound-guiding transvaginal aspiration; sperm preparation was performed employing swim-up or density-gradient centrifugation method. Based on sperm concentration and vitality [?], routine IVF or ICSI were applied for insemination.
Blastocyst culture
16–18 h after insemination (Day 1), fertilization status was evaluated using an inverted microscope. 1PN zygotes were moved to G1.3 (vitrolife, Sweden), cultured to day 3 in 37.5°C, 6%CO2 incubator, then they were moved to G2.3 (vitrolife, Sweden), cultured to day 5 in 37.5°C, 6%CO2 incubator.
hESC derivation and cultivation
The hESCs analyzed in this study come from our stem cell bank. The hESC derivation method has been described previously[14] as: the isolated ICM from blastocyst was plated on mitotically inactivated human embryonic fibroblasts (HEFs), which were cultured in our laboratory from an aborted fetus of 2–3 months of age. The cells were first cultured in DFSR medium containing DMEM/F12 supplemented with 15% knockout serum replacement, 2 mM nonessential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol and 50 ng/ml of basic fibroblast growth factor (bFGF) (all from Invitrogen, Carlsbad, CA). The medium was changed every two days and the cells were passaged every 7 days by mechanical cutting. Once established and propagated through several passages, the cells were transferred back to DFSR medium containing 4 ng/ml bFGF for long-term propagation.
Cytogenetics analysis
FISH: On day 5, the embryos that developed to blastocyst stage and the embryos that arrested in cleavage stage were put into acid Tyrode’s solution for 1–2 min in order to get the zona pellucida removed. Then all blastomeres were analyzed by FISH. Blastomeres were put in hypotonic solution (6 mg/ml BSA + 1% NaAc) for 5 min, and then moved to slides. Being added with 0.01 N HCL + 0.01% Tween, observed until the disappearing of blastomeres under stereoscope, slides were kept roasted at 60°C for 2 h, and digested using ribonuclease for 1 h; then slides were rinsed in 2 × SSC, 70% ethanol, 90% ethanol and absolute ethanol respectively for 2 min. After air-dry, slides were put in denaturation solution (70% formamide, 2 × SSC, 0.1 mM EDTA) at 75°C for 5 min, and then rinsed in 70% ethanol, 90% ethanol and absolute ethanol respectively for 2 min. Being denatured at 75°C for 5 min, three chromosome specific probes (D18Z1, CEPX and CEPY, Abbott Molecular Inc.) were hybridized with nuclei at room temperature overnight. Slides were rinsed three times in WS-I (50% formamide, 10% 20 × SSC, 40% distilled water) at 45°C, each for 5 min, later another three times in WS-I (4 × SSC/0.05% Tween20) at room temperature, each for 5 min, and then in WS-III (4 × SSC), 70% ethanol, 90% ethanol and absolute ethanol respectively for 2 min, followed by air-dry. Nuclei were counterstained by DAPI, and then examined using Olympus fluorescence microscope with DAPI, FITC and Texas fluorescence filter.
G-banding analysis: hESCs were treated with 100 ng/ml demecolcine (Sigma, St Louis, MO) for 4 h and dissociated with 0.05% trypsin/EDTA (Invitrogen) into single cells for standard G-banding analysis. Usually more than 20 metaphases were analyzed for every sample.
Statistics
χ2 tests were performed to analyze the differences between the results of each group. P < 0.05 was considered significant.
Results
After five days of blastocyst culture, there were 62 1PN embryos developing to the blastocyst stage, and 169 embryos arresting in the cleavage stage. The blastocyst formation rate was 26.9% (62/231). Among those embryos, 95 arrested cleavage-stage embryos and 59 blastocysts were analyzed successfully by FISH (Table 1).
According to the signal number of autosome chromosome 18 and sex chromosome X and Y, five kinds of chromosomal constitution of the arrested cleavage-stage embryos were examined: diploid, haploid, mosaic, aneuploid and polyploid. The diploid rate of the arrested embryos was 31.6% (30/95), which was significantly (P < 0.001) lower than that of blastocysts (74.6%, 44/59) and hESCs (97.0%, 32/33). There were 30.5% (29/95) of haploid and 28.4% (27/95) mosaic among arrested embryos. There were 7 aneuploid embryos, containing the numeral error of autosome and/or sex chromosome (one 1818XO, two 1818XYY, one 1818XXY, two 18XY and one 18YY). Besides, there were two polyploid embryos: one tetraploid (18181818XXXX) and the other octoploid (1818181818181818XXXXYYYY).
The diploid rate was 74.6% (44/59) in embryos developing to the blastocyst stage, which was significantly (P < 0.001) higher than that of arrested cleavage-stage embryos. Two kinds of chromosomal abnormalities in blastocysts were observed: mosaic (22.0%, 13/59) and monosomy X (3.4%, 2/59), and the percentages were similar to that of arrested cleavage-stage embryos. No haploid, aneuploid of autosome chromosome or polyploid embryos was found in the blatocysts.
Our lab has established 33 hESC lines from 1PN embryos by the time of this study. The karyotypes of these hESC lines were examined by G-banding analysis. The diploid rate of hESCs was 97.0% (32/33), which was significantly (P < 0.01) higher than that of blastocysts(74.6%, 44/59), and significantly (P < 0.001) higher than that of arrested cleavage-stage embryos (31.6%, 30/95). The only kind of abnormal karyotype found in hESCs was trisomy 16 (47, XY, + 16) (Table 1, Fig. 1).
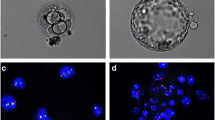
a FISH analysis of an arrested cleavage-stage 1PN embryo showing haploid: one 18 chromosome signal (blue) and one X chromosome signal (green) in each nuclear; b FISH analysis of a 1PN blastocyst showing diploid: two 18 chromosome signals (blue), one X chromosome signal (green) and one Y chromosome signal (red) in each nuclear; c G-banding analysis of chHES-22-P2 cells: 46, XY; d G-banding analysis of chHES-22-P75 cells: 46, XY
Discussion
In this study, 31.6% of arrested cleavage-stage embryos resulted from 1PN zygotes were diploid, which was significantly lower than that of blastocysts and hESCs. Four kinds of chromosomal abnormalities were found among arrested cleavage-stage embryos: haploid, mosaic, aneuploid and polyploid. The haploid (30.6%) was one of the main chromosomal abnormalities in 1PN cleavage embryos, and it might be the result of those 1PN embryos originating from parthenogenetic activation [7]; the mosaic(28.4%) was the other main chromosomal abnormality, in accordance with the observation that mosaic is a common feature occurring in embryos both from normal and abnormal fertilization [15, 16]; the aneuploid was found in 7.4% 1PN cleavage embryos, containing the numeral error of autosome and/or sex chromosome, which suggests that aneuploid embryos can survive through early cleavage stage; besides, there were two polyploid (2.1%) embryos, which may be due to the fact that the chromosome were duplicated several times while the cells were not divided. It is indicated in the result that the chromosomal constitutions of arrested 1PN cleavage embryos are complicated and have a variety of sources [2, 7, 9]. The observation that there were 81.3% (65/80) chromosomally abnormal embryos arresting in the cleavage stage leads to the conclusion that the chromosomal abnormality of 1PN embryos is a crucial cause for their arrested development.
The diploid rate of the embryos developing to the blascyst stage increased to 74.6% in the present study, which was significantly higher than that of 1PN embryos arresting in the cleavage stage (31.6%); and such chromosomal abnormalities as haploid, aneuploid of autosome chromosome and polyploid were eliminated in blastocysts. From the above result, it could be demonstrated that the formation of a blastocyst is a useful morphology indicator for normal fertilization and chromosomal constitution, which agrees with the study of Sandalinas et al. that embryos with haploid, aneuploid excepted monosomy X and 21 never develop to blastocyst [17]. In addition, blastocyst culture is an effective method to select against chromosomally abnormal embryos, especially the haploid, which was the main chromosomal abnormality in 1PN embryos. In previous studies, the mechanism of selection is not clear, probably attributed to the developmental block against chromosome abnormal embryos at compaction [17] or cavitation stage [18]. Besides, it was still observed that there existed mosaic (22.0%) and aneuploid (3.4%) in 1PN-derived blastocyst, and the percentages were similar to that of arrested cleavage-stage embryos, which suggests that blastocyst culture can not select against these chromosomal abnormalities.
Since the human embryonic stem cells (hESCs) are derived from the ICM of blastocysts which will develop to the fetus in the gestation, the cytogenetic analysis of hESCs may helps to answer the question that whether the 1PN-derived embryos selected by blastocyst culture will be chromosomally normal after implantation or not. Our lab has established 33 hESC lines derived from 1PN embryos. Most of these hESCs were diploid (97.0%), which was significantly higher than the blastocysts (74.6%); besides, no mosaic was found in the hESCs. Such a result indicates that mosaic blastocysts could further self-correct to be chromosomally normal after implantation, which may be explained by the hypothesis that embryos can possibly self-correct by moving the mosaic cells to trophoblast during development process [19]. Another possible mechanism of self-correction is apoptosis. Apoptosis is a common feature in human embryos to eliminate abnormal cells[20]; the chromosomally abnormal embryos or cells probably undergo apoptosis, causing blstocysts to show an increased normal diploid rate, which is even higher in hES cells generated from those blastocysts. Suss-Toby et al. also reported a hESC coming from 1PN embryos with normal diploid karyotype (46, XX) [21]. However, the embryo development maybe differ from hESCs in-vitro, as suggested by Lavon et al. that the mosaic could be corrected to normal karyotype in hESCs by faster proliferating of normal cells than abnormal cells during the growth of the hESCs, and then took over the culture, whereas the mosaic ICM of blastocyst only divided a few times in-vivo, which may lead to a mosaic fetus [22]. Although most of the 1PN hESCs were normal diploid, one aneuploid hESC line with trisomy 16 (47, XY, + 16) was still observed. As a common phenominon in human, aneuploid is mostly derived from errors in maternal meiosis I. No fewer than 5% of the clinically recognized pregnancies are aneuploid, which is the leading known cause of miscarriage and congenital birth defects and mental retardation [23]. Trisomy 16 accounts for at least 1–2% of all first-trimester miscarriages [24]. The frequency of aneuploid occurring in hESCs still remains unknown. Maybe it is very rare, but there have already been several reports about trisomic in hESCs [25, 26]. The presence of trisomic in the hESCs suggests that the aneuploid may not be eliminated by further development after embryo implantation. Moreover, besides the trisomic, the interesting thing is that we had found a homozygous hESC line in these diploid hESCs [27], which maintained a stable karyotype of 46, XX, but the expression of five paternally expressed imprinted genes were almost undetectable and their HLA genotype were identical with the oocyte donor. Staessen et al. also observed a possible diploid, androgenetic 1PN-derived cleavage stage embryo with two Y and two 18 signals [7]. It indicates that spontaneous diploidization may be another possible reason for the diploid embryos resulted from the 1PN zygotes which are actually haploid, and that the homozygous diploid embryo can not be excluded by blastocyst culture.
In conclusion, it is demonstrated in our study that blastocyst culture is an effective method to select against chromosomal abnormalities, especially the haploid which is the main chromosomal abnormality in 1PN embryos. It should also be noted that the ability to the blastocyst stage is not a reliable marker against the mosaic or aneuploid embryos, which could lead to miscarriage and congenital birth defects after implantation. Blastocyst culture and transfer of 1PN embryos may be employed in clinical, but in order to exclude the possible mosaic and aneuploid anomalies, it’s necessary to do the prenatal diagnosis for the pregnancies after 1PN blastocysts transfer.
References
Dasig D, Lyon J, Behr B, Milki AA. Monozygotic twin birth after the transfer of a cleavage stage embryo resulting from a single pronucleated oocyte. Journal of Assisted Reproduction and Genetics. 2004;21:427–9.
Staessen C, Janssenswillen C, Devroey P, Van Steirteghem AC. Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod. 1993;8:221–3.
Nagy ZP, Liu J, Joris H. Time-course of oocyte activation, pronucleus formation and cleavage in human oocytes fertilized by intracytoplasmic sperm injection. Hum Reprod. 1994;9:1743–8.
Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod. 1997;12:532–41.
Levron J, Munne S, Willadsen S, Rosenwaks Z, Cohen J. Male and female genomes associated in a single pronucleus in human zygotes. Biol Reprod. 1995;52:653–7.
Tesarik J, Mendoza C. Spermatid injection into human oocytes. I. Laboratory techniques and special features of zygote development. Hum Reprod. 1996;11:772–9.
Staessen C, Van Steirteghem AC. The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod. 1997;12:321–7.
Tesarik J, Mendoza C. In vitro fertilization by intracytoplasmic sperm injection. Bioessays. 1999;21:791–801.
Munne S, Cohen J. Chromosome abnormalities in human embryos. Human Reproduction Update. 1998;4:842–55.
Gardner DK, Vella P, Lane M, Wagley L, Schlenker T, Schoolcraft WB. Culture and transfer of human blastocysts increases implantation rates and reduces the need for multiple embryo transfers. Fertil Steril. 1998;69:84–8.
Jones GM, Trounson AO, Gardner DK, Kausche A, Lolatgis N, Wood C. Evolution of a culture protocol for successful blastocyst development and pregnancy. Hum Reprod. 1998;13:169–77.
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7.
Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404.
Zhou J, Ou-Yang Q, Li J, Zhou XY, Lin G, Lu GX. Human feeder cells support establishment and definitive endoderm differentiation of human embryonic stem cells. Stem Cells Dev. 2008;17:737–49.
Coonen E, Hopman AHN, Geraedts JPM, Ramaekers FCS. Application of in-situ hybridization techniques to study human preimplantation embryos: a review. Human Reproduction Update. 1998;4:135–52.
Almeida PA, Bolton VN. The relationship between chromosomal abnormality in the human preimplantation embryo and development in vitro. Reprod Fertil Dev. 1996;8:235–41.
Sandalinas M, Sadowy S, Alikani M, Calderon G, Cohen J, Munne S. Developmental ability of chromosomally abnormal human embryos to develop to the blastocyst stage. Hum Reprod. 2001;16:1954–8.
Janny L, Menezo YJ. Maternal age effect on early human embryonic development and blastocyst formation. Mol Reprod Dev. 1996;45:31–7.
James RM, West JD. A chimaeric animal model for confined placental mosaicism. Hum Genet. 1994;93:603–4.
Hardy K. Cell death in the mammalian blastocyst. Mol Hum Reprod. 1997;3:919–25.
Suss-Toby E, Gerecht-Nir S, Amit M, Manor D, Itskovitz-Eldor J. Derivation of a diploid human embryonic stem cell line from a mononuclear zygote. Human Reproduction. 2004;19:670–5.
Lavon N, Narwani K, Golan-Lev T, Buehler N, Hill D, Benvenisty N. Derivation of euploid human embryonic stem cells from aneuploid embryos. Stem Cells. 2008;26:1874–82.
Terry Hassold HHaPH. The origin of human aneuploidy: where we have been, where we are going. Human Molecular Genetics. 2007;16:R203–R8.
Menasha JLB, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: New insights from a 12-year study. Genet Med. 2005;7:251–63.
Seol HW, Oh SK, Park YB, Kim HS, Baek JA, Seo J, et al. Separation and maintenance of normal cells from human embryonic stem cells with trisomy 12 mosaicism. Chromosome Res. 2008;16:1075–84.
Catalina P, Montes R, Ligero G, Sanchez L, de la Cueva T, Bueno C, et al. Human ESCs predisposition to karyotypic instability: Is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol Cancer. 2008;7:76.
Lin G, OuYang Q, Zhou XY, Gu YF, Yuan D, Li W, et al. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Research. 2007;17:999–1007.
Acknowledgement
This work was supported by the Ministry of Science and Technology of China (863 program 2006AA02A102), the National Basic Research Program of China (973 program 2007CB948103).
Author information
Authors and Affiliations
Corresponding author
Additional information
Capsule
Blastocyst culture is an effective method to select against the haploid in 1PN embryos, though not against the mosaic or aneuploid.
Rights and permissions
About this article
Cite this article
Liao, H., Zhang, S., Cheng, D. et al. Cytogenetic analysis of human embryos and embryonic stem cells derived from monopronuclear zygotes. J Assist Reprod Genet 26, 583–589 (2009). https://doi.org/10.1007/s10815-009-9355-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-009-9355-1