Abstract
The modulation in physiological and metabolic attributes associated with colonisation by cyanobacteria in wheat has been little explored. In the present investigation, the performance of six selected cyanobacterial strains was evaluated with wheat (variety HD2687). The fresh weight of plants, measured after 2 weeks, exhibited a 30–60 % increase, while 14–40 % increase in plant dry weight was also recorded, as compared to uninoculated control. The nitrogen-fixing potential (expressed as acetylene-reducing activity or ARA) was 20-fold higher in the treatment involving inoculation of Anabaena laxa RPAN8 as compared to that in the uninoculated control. The inoculation of Calothrix sp. RPC1 brought about a more than 90 % increase in endoglucanase activity and root chlorophyll. Comparison of DNA fingerprints (highly iterated palindrome (HIP)-TG profiles) of wheat roots with those of corresponding pure cultures revealed a high degree of similarity, confirming the colonisation. Significant correlation of plant parameters with nitrogen-fixing potential and growth attributes and fingerprints of cyanobacteria from roots further illustrated the novelty of our results. This represents a first report on understanding hydrolytic enzyme-mediated colonisation of cyanobacteria on roots of wheat plants using plant growth parameters and DNA fingerprints. Such synergistic combinations of cyanobacterium and wheat can lead to savings of nitrogen and increased yields, besides being a prelude to generating nitrogen-independent wheat plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticuma estivum L.) contributes about 33.5 % to the total world food grain production and represents one of the major sources of calories and protein in the human diet. India ranks second in the world in terms of acreage (12.5 %) and production (12.05 %) in intensive cropping systems (www.agricoop.nic.in). Crop productivity has been traditionally enhanced using chemical fertilisers in modern agriculture, but their low efficiency mainly due to losses through denitrification, volatilisation, leaching or immobilisation in soil has necessitated the need for identifying viable alternatives for use as nutrient supplements.
It is generally understood that plant growth promotion is dependent upon nutrient acquisition or stimulation, mainly mediated by associative microorganisms (Dobbelaere et al. 2003). Plant-growth-promoting rhizobacteria (PGPR) represent mainly soil bacteria growing in association with plants, which stimulate growth by increased mobilisation, uptake and enrichment of micronutrients and macronutrients in the plant. As a result, they can indirectly lead to bio-control of pathogenic flora/fauna or other competing native organisms for space or nutrients (Lucas García et al. 2004; Çakmakçi et al. 2006). Nitrogen is unique among the essential mineral elements in the plants, present in both anionic and cationic (NO3 and NH4) forms, and supplying the plant with a mixture of NO3 and NH4 often results in better vegetative growth and enhanced nutrient accumulation than either form separately (Hageman 1984).
Enhancement of rice seed germination, root and shoot growth and the fertilising action of N2 fixed from the atmosphere by free-living cyanobacteria has been illustrated in several studies. This generally leads to the release of synthesized nitrogenous compounds either by decomposition of the cells or excretion (Venkataraman and Neelakantan 1967; Nayak et al. 2004; Prasanna et al. 2008). In symbiotic associations, the nitrogen fixed by the cyanobiont is made available to the system, which in turn is supplied by the host with a niche for its growth and survival (Rai 1990). Due to the difficulty in maintaining specific N ratios in soil and its buffering capacity, much of this evidence has been obtained by growing plants hydroponically. Such studies indicate that vegetative growth of wheat is enhanced when both NO3 and NH4 are present (Weissman 1951) leading to higher total protein content and protein concentration in leaves of young wheat plants grown in solution culture containing NH4 and NO3 than on either form alone. Similarly, an enhancement (74 %) was observed in vegetative dry weight of 16-day-old wheat plants when NH4 was supplied at the rate of 17 % of total N, as compared to plant grown with only NO3 (Cox and Reisenauer 1973).
Inoculation with cyanobacteria can lead to yield improvement of 5–25 % not only in cereals such as rice and wheat but also in solanaceous fruits including tomato and in legumes and cotton (Prasanna et al. 2008, 2009, 2013a; Karthikeyan et al. 2009; Nain et al. 2010). Cyanobacteria represent promising organic inputs in rice-wheat cropping system, as they help in N and C accretion and improve crop productivity and soil health; the colonisation and establishment of inoculated cyanobacteria have been demonstrated using DNA-based markers (Prasanna et al. 2012). Earlier studies have shown that sugars and amino acids may serve as signalling molecules in the development of functional associations between relevant cyanobacteria and wheat seedlings (Jaiswal et al. 2008; Karthikeyan et al. 2009). Kovtunovych et al. (1999) found a high degree of correlation between the activity of cell wall depolymerising enzyme pectate lyase and internal colonisation of wheat roots by Klebsiella spp.; however, the role of endoglucanases, especially in cyanobacteria, is less investigated. Such studies can be particularly useful for developing nitrogen-fixing cyanobacteria associated wheat plants, as current research is being directed for genetic engineering of nif genes in cereals.
It is very well understood that considerable expertise is required to identify cyanobacteria, and studies on Anabaena species have revealed information that both morphological and developmental characters can vary with the growth conditions, particularly the shape and size of the cell/thalli, which are the basis for the classification and taxonomy of Anabaena (Hiroki et al. 1998). However, the use of DNA-based attributes is getting significant popularity in the identification of cyanobacteria at both generic and species levels. Filamentous heterocystous cyanobacteria, including Anabaena, are known to have several distinct families of highly conserved repetitive sequences, such as short tandemly repeated repetitive (STRR) sequences and long tandemly repeated repetitive (LTRR) sequences, which have been used in combination with restriction fragment length polymorphism (RFLP) and polymerase chain reaction (PCR) typing (Rasmussen and Svenning 1998; Nayak et al. 2009) to distinguish the isolates at both inter- and intra-generic levels. Another set of interspersed repeated sequences, namely highly iterated palindrome (HIP) sequences, have proved valuable in the identification of differentiation of strains (Gupta et al. 2012). Such repeat/palindrome sequences, as a result of their conserved status, represent methodologically important tools for DNA fingerprinting as well as for diversity studies among related cyanobacteria.
In order to understand the chemical and biological nature of interactions of cyanobacterial strains with wheat seedlings, a hydroponics experiment was taken up at the National Phytotron Facility, Indian Agricultural Research Institute, New Delhi. The strains used in the experiment had been earlier characterised for their nitrogen-fixing and PGP abilities (Nain et al. 2010; Prasanna et al. 2008, 2013b), and in the present study, efforts were undertaken to analyse the plant-cyanobacterium associations using biochemical/physiological attributes and employing molecular tools to illustrate colonisation of inoculated organisms in root tissues based on HIP-TG profiles and 16S ribosomal DNA (rDNA) sequences.
Materials and methods
Cultivation and maintenance of selected cyanobacterial strains
Cyanobacterial cultures used in study—Anabaena torulosa BF1, Anabaena laxa C11, Anabaena azollae C16, Anabaena oscillarioides C20, Calothrix sp. RPC1 and A. laxa RPAN8—were maintained by using standard protocols and cultivated in Haffkine flasks, in nitrogen-free BG11 medium (Stanier et al. 1971) at 27 ± 1 °C and light/dark cycles (L/D) of 16:8 and white light (50–55 μmol photons m−2 s−1) as stationary cultures.
Experimental setup
An experiment with wheat and selected cyanobacterial strains was undertaken under controlled conditions, including five replicates of each treatment in the National Phytotron Facility (NPF), IARI, New Delhi. The experiment was carried out in 4-in pots containing 300 mL of autoclaved nitrogen-free BG11 liquid medium. Seeds (T. aestivum; variety HD2687) were sterilised with 0.1 % mercuric chloride for 1 min followed by washing with sterile water. Seeds were imbibed overnight in sterile water in dark at room temperature and kept for germination for 24 h. A set of pots containing 300 mL of autoclaved chemically defined BG11 liquid medium was used as control. A thermocol plate with four 1.5-mL Eppendorf tubes containing overnight 24-h soaked seeds was adjusted on each pot individually. The base of each tube was cut to permit the emergence and growth of seedlings in pots. According to designed treatments for this study, the selected strains were inoculated with 1 mL of packed cell volume of each strains individually (approximately 10 μg chlorophyll mL−1 media) and grown for 14 days at 22 ± 1 °C (day) and 18 ± 1 °C (night), light intensities of 570 μmol photons m−2 s−1 for 3 h during peak day hours and 320 μmol photons m−2 s−1 in the beginning and end of the day for 1 h, with prominent red and far-red spectrum ratio 5:2, 0.6 % m s−1 air flow (bottom to top) and humidities of 45 ± 1 % (day) and 55 ± 1 % (night).
The seedlings were transferred to the pots, and after 15 days, the plant samples from each treatment were harvested for the analyses.
Plant-growth-promoting attributes
Fresh and dry weights of plants were measured as growth indices. Chlorophyll content of plants (shoot and root) and BG11 medium inoculated with selected cyanobacterial strains were estimated following standard methods (Mackinney 1941; Daizy and Kohli 1991) using 95 % methanol. Nitrogenase activity of plants was measured by acetylene reduction activity (ARA) by using Bruker 450-gas chromatograph (GC) with FID detector. For the analysis of nitrogenase activity, plants were placed in glass test tubes (55 mL) and sealed with airtight rubber stoppers followed by replacement of 10 % air space (v/v) with acetylene. The tubes were incubated for 24 h. A set of tubes without acetylene gas injection served as control. All experiments were carried out in triplicates. After incubation, 0.1 mL of gaseous sample was taken tubes and injected into GC for analysis. The ethylene produced by the reduction of acetylene was estimated and expressed as nanomoles of ethylene produced per plant per hour.
Activity of hydrolytic and defence enzymes in seedlings
Root tissues were cleaned by running tap water and homogenised with pre-chilled 50 mM (5 mL) Tris-HCl buffer in a mortar pestle. The homogenous extracts were centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatant was collected and stored at −20 °C. Polyphenol oxidase (PPO) and peroxidase (PO) activities were analysed using catechol and 1 % guaiacol (molar extinction coefficient 26.6 mM cm−1) as substrate by minor modifications (Prasanna et al. 2013a). In reaction mixture of PPO activity, 1 mL of 0.02 M citrate phosphate buffer, pH 6.0; proline (5 mg mL−1), 0.5 mL; catechol (2 mg mL−1), 0.5 mL; and 500 μL of appropriate diluted plant extract were used and read at 546 nm by time scan method for 3 min with 30-s intervals of readings. The mixture was aerated before addition of catechol which served as blank. One unit of enzyme is defined as the change in absorbance of 0.01 unit min−1. In the reaction mixture of PO activity, 1 % (v/v) guaiacol in 0.01 M sodium phosphate buffer (pH 6.0) and 0.1 M H2O2 were added to the extract at the end to initiate the reaction followed by reading absorbance at 470 nm using time scan method for 3 min with 30-s intervals. One unit of enzyme is defined as the change in absorbance of 0.01 unit min−1 g−1 fresh weight of extract. Phenylalanine ammonia lyase (PAL) activity was estimated in root extracts (100 μL) using 2.5 mL of 0.2 % L-phenylalanine in 50 mM Tris-HCl (pH 5 to 6) buffer solution. The reaction mixture was incubated at 40 °C for 60 min. The amount of released trans-cinnamic acid from L-phenylalanine was measured at 290 nm. Distilled water instead of cyanobacterial cultures served as blank, and the details are given in Prasanna et al. (2013a). One unit of enzyme is defined as change in absorbance min−1 g−1 fresh tissue weight.
Chitosanase, endoglucanase (β-1,3-glucanase) and carboxy methyl cellulase (CMCase/β-1,4-endoglucanase) activities were assayed spectrophotometrically using glycol chitosan, laminarin and carboxy methyl cellulose respectively, as substrate using the methodology optimised earlier (Prasanna et al. 2013a). One unit of endoglucanase, CMCase and chitosanase activities was defined as 1 μmol of glucose and glucosamine (chitosanase) released per minute per millilitre culture filtrate, under the assay conditions respectively.
DNA fingerprinting and analyses
Anabaena filaments (approximately 50-mg wet weight) were collected by centrifugation and washed four times with sterile Milli-Q water. The samples were vortexed and homogenised using mini pestles, in order to rupture the filaments. The pellet was resuspended in 5 μL of Milli-Q water and used directly as a template in PCR reaction mixture for HIP-TG pattern analysis (Nayak et al. 2009). Amplification of HIP-TG sequence was done by using the following primer sequence (5′- to 3′-GCGATCACTG) in PCR reaction mixture. The primers were synthesized by Bangalore Genei Merck (India). The final concentration of constituents in PCR reaction mixture was 50 pmol of each primer, 2 mM dNTP, 2.5 % (v/w) BSA (bovine serum albumin), 10 % (v/v) DMSO (dimethyl sulphoxide), 1 U of Taq DNA polymerase, Taq buffer 2 μL and 5 μL of cyanobacterial filaments. The amplification was carried out for 35 cycles in a PEQLAB QB96 thermocycler following the protocol as pre-denaturation at 95 °C for 6 min, denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, extension at 65 °C for 5 min, and final extension at 65 °C for 16 min. After amplification, the products were analysed by running in 1.5 % agarose gel prepared using TAE (1×) buffer at 80 V for 2 h. The PCR amplification for HIP-TG was repeated at least three times for each to ascertain the reproducibility of the band patterns. The size of bands in HIP-TG profiles were measured with the help of AlphaImager software by running 1-kb DNA ladder obtained from Bangalore Genei Merck (India).
DNA extraction, sequencing of 16S rDNA gene and phylogenetic analysis
DNA from plant roots of each treatment was extracted by using UltraClean plant DNA isolation kit (MoBio Inc., USA). Amplification of 16S rDNA gene was performed using primers fD1 and rD1 to isolate the nearly complete 16S rDNA gene sequence (Weisburg et al. 1991). The reaction mixture contained 1× Taq buffer, 2.5 mM MgCl2, 0.3 mM each of the deoxynucleotide triphosphate, 10 pmol of each primer, 1 U Taq DNA polymerase, 50 ng of template DNA and Milli-Q to give final volume of 20 μL. The reaction mixture was incubated in a PEQLAB Primus 96 thermal cycler using the optimised program as described by Hisbergues et al. (2003), with annealing temperature of 50 °C. All PCR-related chemicals were purchased from Bangalore Genei Merck Pvt. Ltd., India. PCR-amplified products were purified and sequenced using both fD1 and rD1 primers for forward and reverse reactions, respectively. Sequencing of amplified gene was carried out by using Applied Biosystems ABI prism automated DNA sequencer (Applied Biosystems, USA). The 16S ribosomal RNA (rRNA) gene sequences were compared with sequences available in the NCBI database. Isolates were identified to species level on the basis of 16S rRNA gene sequence similarity of ≥97 % with the sequences in GenBank. Sequence alignment and comparison used the multiple sequence alignment tool CLUSTALW (Thompson et al. 2002) with default parameters. The phylogenetic tree was constructed on aligned data sets using the neighbour-joining (NJ) method (Saitou and Nei 1987) and the program MEGA 4.0.2 (Tamura et al. 2007). Bootstrap analysis was performed on 1,000 random samples taken from multiple alignments (Felsenstein 1981).
Statistical analysis
The experiment was based on a completely randomised design (CRD), which included seven treatments, along with control. The data obtained in triplicates for the various parameters were analysed by ANOVA (analysis of variance) using MSTAT-C statistical package, and critical differences values were calculated at probability level of 0.05 %. Standard deviations (SD) are denoted as error bars in graphs. Correlations were analysed using Pearson’s coefficient in the Microsoft Excel package. Path coefficient analyses were carried out by using Windostat version 8.5 and the procedure outlined by Singh and Chaudhary (1979).
Results
Growth parameters and nitrogen fixation
In the present investigation, all the cyanobacterial strains were able to form a close association with wheat roots in hydroponics experiment. The colonisation by these cyanobacteria was reflected by the presence of blue-green colonies on roots, which were visible to the naked eye and could not be dislodged even after gentle washing using distilled water. In order to study the degree of colonisation by cyanobacteria, the chlorophyll content of media, roots and shoots (Fig. 1 and Supplementary Table 1) was estimated, which revealed a significant increase in the values of media and plants in cyanobacteria-inoculated treatments. The chlorophyll values ranged from 0.065 to 2.904 mg mL−1 in the medium, and the highest value was recorded in treatment involving inoculation of A. oscillarioides CW3, followed by treatments A. azollae CW2 and A. laxa CW1 with values of 1.91 and 0.68 mg mL−1 chlorophyll. A significant increase of 80 to 90 % in chlorophyll accumulation in medium (Fig. 1a) over the control was recorded. Root chlorophyll values were highest in the treatment involving A. laxa RPAN8 with a value of 0.094 mg g−1 fresh weight, followed by treatment with A. oscillarioides CW3 and A. azollae CW2 with values of 0.085 and 0.045 mg g−1. The treatment involving RPAN8 A. laxa and A. oscillarioides CW3 showed the highest root chlorophyll, with an increase of 20 to 80 % over control (Fig. 1b). An increase of 28 to 70 % in shoot chlorophyll, as compared to control, was observed, with the treatment involving inoculation of A. laxa RPAN8 recording the highest increase in chlorophyll (an index of photosynthesis). Shoot chlorophyll ranged from 0.30 to 1.07 mg g−1; the highest chlorophyll was recorded in treatment involving inoculation of A. laxa RPAN8 with values of 1.075 mg g−1, followed by treatment with Calothrix sp. and A. oscillarioides CW3.
Influence of cyanobacterial inoculation on chlorophyll content after 14 days of growth. a Percent change in chlorophyll content in root and medium over control; b percent change in chlorophyll of shoot and medium over control; c Nitrogen-fixing potential of the associations measured as acetylene-reducing activity (ARA) and expressed in terms of nanomoles of N fixed per plant per hour
The ARA values were significantly higher (20-fold increase over uninoculated control) in treatment involving inoculation of A. laxa RPAN8, with a value of 2.34 μmol C2H4 plant−1 h−1 (Table 1). Nitrogen-fixing potential was recorded for whole plants which revealed values ranging from 0.11 in control plants (uninoculated) to 2.34 μmol ethylene plant−1 h−1 (Fig. 1c), equivalent to 7 nmoles N plant−1 h−1 (obtained by converting these values in terms of N fixed, using a factor of 3, as suggested by Dommergues et al. (1973)). The treatments involving inoculation of Calothrix sp. and A. oscillarioides C20 exhibited at par values of 1.75 and 1.48 μmol C2H4 plant−1 h−1 respectively. ARA showed a positive correlation with root chlorophyll (r = 0.70) and shoot chlorophyll (r = 0.75).
The fresh weight of plants measured after 2 weeks (Table 1) revealed an increase of 35–60 %, as compared to that in control. The fresh weight values ranged from 0.21 to 0.51 g fresh weight, with the highest plant fresh weight recorded in treatment involving inoculation of A. laxa RPAN8 followed by treatment Calothrix sp. and A. oscillarioides CW3. A similar trend of 15–40 % increase in plant dry weight (Supplementary Table 1) was also recorded with respect to control.
Activity of hydrolytic and plant defense enzymes
The CMCase (β-1,4-endoglucanase) activity (Table 1) in root tissues was very high, as compared to the activity of other enzymes, ranging from 12.20 IU g−1 fresh weight (in uninoculated control ) to 17 IU g−1 fresh weight in treatment involving inoculation of Calothrix sp. CMCase activity showed a significant positive correlation with endoglucanase (r = 0.87), shoot chlorophyll (r = 0.70), PPO (r = 0.92), ARA (r = 0.88), and plant fresh weight (r = 0.83). CMCase activity recorded the highest percent increase in T6 (28 %), T5 (25 %) and T7 (25 %) over control. β-1,3-endoglucanase showed a significant enhancement of activity in the inoculated treatments—A. torulosa BF1 and A. laxa RPAN8—as compared to that in the control. β-1,3 endoglucanase activity (Supplementary Table 1 and Fig. 2a) in root tissues ranged from 0.001 to 0.056 IU g−1 fresh weight, with the highest endoglucanase activity being recorded in treatment involving inoculation of Calothrix sp. with a value of 0.056 IU g−1 fresh weight. This was followed by A. oscillarioides CW3 and A. laxa RP8 recording 0.043 and 0.038 IU g−1 fresh weight, which were statically at par with control. A significant (25 to 28 %) increase in CMCase activity of treatment involving A. oscillarioides CW3, Calothrix sp. and A. laxa RPAN8 was recorded as compared to that in control. Chitosanase activity ranged from 0.066 to 0.103 IU g−1 fresh weight. The highest activity was recorded in treatment involving inoculation of A. laxa CW1, followed by treatment with A. azollae CW2 and uninoculated control (Table 1). A positive correlation of CMCase (r = 0.88), β-1,3-endoglucanase (r = 0.78) and PPO (r = 0.88) with ARA was recorded. A positive correlation (r = 0.78) was observed between β-1,3-endoglucanase activity and ARA.
Plant defense enzyme activity, in terms of peroxidase (Table 1), polyphenol oxidase and phenylalanine ammonia lyase enzyme (Supplementary Table 1), was analysed in the root samples of the wheat seedlings. Peroxidase activity in root samples ranged from 29.00 to 141.00 IU g−1 fresh weight (Fig. 2b). The highest peroxidase activity of 141 IU g−1 fresh weight was recorded in treatment involving inoculation of A. laxa RPAN8, followed by Calothrix sp. and Anabaena sp. CW2 with values of 123 and 117 IU g−1 fresh weight. However, these values were statistically at par with control (29 IU g−1 fresh weight). Interestingly, a significant positive correlation of (r = 0.44) with chitosanase, ARA (r = 0.88), plant fresh weight (r = 0.75), PAL (r = 0.81), activity of CMCase (r = 0.92) and endoglucanase (r = 0.91), as also with root chlorophyll (r = 0.61) and shoot chlorophyll (r = 0.78) were also recorded. PPO activity in roots ranged from 8 to 66.66 IU g−1 fresh weight. The treatment involving inoculation of A. laxa RPAN8 showed the highest value of 66.66 IU g−1 fresh weight followed by Calothrix sp. and A. oscillarioides, with values of 66.65 and 45.33 IU g−1 fresh weight, which were statistically at par with control. A significant increase of 60 to 88 % in PPO activity was recorded in these treatments as compared to that in control. Phenylalanine ammonia lyase activity in root ranged from 80.99 to 172.18 IU g−1 fresh weight. The highest activity was recorded in microbial inoculation involving A. laxa RPAN8 with a value of 172.18 IU g−1 fresh weight, followed by treatment with Calothrix sp. and A. oscillarioides. The observations on A. laxa (CW1) and A. azollae (CW2) were also found to be statistically at par with those in the uninoculated control (80.99 IU g−1 fresh weight). PAL shows a significant positive correlation (r = 0.81) with peroxidase; significant 52.9 % increase in PAL activity was recorded in T6 (Calothrix sp.) as compared to that in control. The plant dry weight values ranged from 0.06 to 0.10 g dry weight, and the highest values were recorded in A.-laxa-RPAN8-inoculated treatment (Fig. 2c).
PCR-based DNA fingerprinting and their 16S-rDNA-based phylogenetic analyses
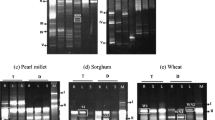
Comparison of DNA fingerprints of wheat root DNA with those of the treatments involving inoculation with cyanobacterial strains (Fig. 3) showed that the common bands were in the range of 300–4,000 bp, with the maximum number of bands in the 500 to 3,500 bp region. The DNA fingerprints of rice roots of control (uninoculated) treatment seedlings did not show any bands. The treatment inoculated with A. torulosa BF1 showed a distinct band pattern, in which four unique bands were observed of sizes 2,690 and 1,523 bp and duplet of 1,016 and 824 bp, which were common to both pure cultures and treatment. In the sample from A. laxa (CW1), one band of 1,372 bp was common. The profile of treatment with A. azollae(CW2) revealed a pattern comprising a triplet of 3,854, 3,570 and 2,944 bp and duplet of 1,513 and 1,372 bp, which were similar to that of pure culture used for inoculation. A.-oscillarioides-inoculated treatment showed a band pattern of 1,372, 1,229, 604 and 464, which matched with the fingerprint of the pure culture. T6 showed a band pattern, made up of bands of 3,272, 2,831, 1,478 and 1,280 bp, identical with that of the inoculated pure culture fingerprint. A. laxa (RPAN8) showed only one band of 2,005 bp, also present in the DNA profile of the pure culture. The details of the common bands present in both root DNAs of all the treatments vis-a-vis pure cultures (Supplementary Table 2).
HIP-TG PCR-based DNA fingerprints of wheat plant roots and pure cyanobacterial samples. Lane 1 is the 1-kb marker; lanes 2–8 are root samples from uninoculated control, Anabaena torulosa BF1 inoculated, Anabaena laxa CW1 inoculated, Anabaena azollae CW2 inoculated, Anabaena oscillarioides CW3 inoculated, Calothrix sp. RPC1 inoculated, and A. laxa RPAN8 inoculated. Lanes 9–14 are pure culture samples of A. torulosa BF1, A. laxa CW1, A. azollae CW2, A. oscillarioides CW3, Calothrix sp.RPC1, and A. laxa RPAN8
The 16S rDNA sequences generated from plant root DNA of all the samples were compared with those of the inoculated cyanobacterial strains (available in NCBI database), and a phylogram was generated, which revealed two major clusters (Fig. 4). Both clusters were 83 % similar. The first cluster comprised two subclusters, in which A. laxa CW1 showed 100 % similarity with the sequence of A. laxa RPAN8, available in the database. The treatment involving inoculation of A. azollae(CW2) showed the nearest similarity with Anabaena flos-aquae and 99 % similar to A. laxa (CW1) and the sequence of RPAN18 A. laxa available in the database. In the second subcluster, A. oscillarioides showed 99 % similarity with the NCBI sequence of RPAN69 A. oscillarioides. A. laxa RPAN8 showed 100 % similarity with that of Anabaena variabilis (AB016520), while A. torulosa BF1 and NCBI sequence GU396091 (A. torulosa BF1) exhibited 100 % similarity. The treatment inoculated with Calothrix sp. showed 100 % similarity with the deposited sequence of this strain in the NCBI.
Path coefficient analyses
The influence of ARA and fresh weight with other parameters was selected for path coefficient analyses, as a significant effect was observed. Positive direct effects of chlorophyll content in roots (0.91) and PPO (0.762) besides plant fresh weight (0.58) and endoglucanase activity (0.46) on ARA were recorded (Table 2 and Fig. 5). The correlation between the activity of PPO, CMCase, PAL and endoglucanase with ARA was very high (0.77–0.85). The residual effect was 0.148, indicative that all the parameters taken into consideration played a major role (84.2 % or 0.842) in the enhancement of ARA, brought about by cyanobacterial inoculation. The fresh weight of plants was positively correlated (0.7–0.8) with all the parameters, except for a negative correlation with endoglucanase activity. The positive direct effect of PAL (0.86) on fresh weight of plants was very high, followed by the effect of chlorophyll content of growth medium, CMCase and ARA on fresh weight. The residual effect was 0.145, which shows that the increase in fresh weight is due to the attributes investigated as influenced by cyanobacterial inoculation (Table 2).
Discussion
Cyanobacteria represent a highly heterogenous assemblage of photosynthetic prokaryotes, which are employed as inoculants for improving crop productivity and soil health in several crops, including rice, wheat, legumes, fruits and vegetables (Venkataraman 1972; Kaushik 2004; Nayak et al. 2004; Karthikeyan et al. 2007; Prasanna et al. 2008, 2009, 2013b, c; Nain et al. 2010;). They are known to produce phytohormones, provide fixed nitrogen and make available other nutrients to plants. Reports are also available on their potential as bio-control agents, especially against soil-borne phytopathogenic fungi (Manjunath et al. 2009; Chaudhary et al. 2012; Prasanna et al. 2013a). In order to increase the benefits of cyanobacterial nitrogen fixation or its plant-growth-promoting properties, there is a need to develop tighter associations, especially as alternatives to obviate repeated inoculation or genetic engineering to mediate transfer of nif genes into plants. The present study was undertaken to evaluate a set of cyanobacterial strains, in terms of their nutritional and biochemical interactions with wheat roots, as a prelude to developing effective associations for use in integrated nutrient management practices in wheat.
In the present investigation, the cyanobacterial strains formed a close association and enhanced the chlorophyll content of plants and fresh/dry weight. A similar observation was recorded earlier for a different set of strains by (Gantar et al. 1991). Some of these cyanobacterial isolates (originally from wheat rhizosphere) used in this investigation are known to produce growth-promoting substances and remain in association with roots, both extra- and intra-cellularly (Jaiswal et al. 2008; Karthikeyan et al. 2007, 2009; Prasanna et al. 2008, 2013a), but the mode of entry or biochemical changes mediating the colonisation could not be elucidated. Cultivation of wheat with cyanobacteria was found to increase root dry weight and chlorophyll (Obreht et al. 1993), as observed in the present investigation too. Prasanna et al. (2013c) recorded improvement in fresh and dry biomass of plants by 12–25 %, through the use of cyanobacterial formulations. The strains used in this study are known to produce IAA (Prasanna et al. 2008), which may be responsible for the increase in plant biomass and chlorophyll. Despite the availability of published research in related areas, the exact mechanisms involved and employed by cyanobacterial cells to enter the plant is not clear; root hair exudates seem to attract the filaments and may be involved in recognition of some lectins (McCowen et al. 1986; Knight and Adams 1996).
Anabaena or Nostoc strains are known to colonise the roots of wheat and exhibit associative nitrogen fixation (Gantar et al. 1991, 1995; Spiller et al. 1993; Gantar and Elhai 1999; Gantar 2000). The cyanobacterial strains in the present study also enhanced plant-growth-promoting traits such as N-fixing potential, measured using the acetylene-reducing assay/ARA. The positive correlation of ARA with root chlorophyll (r = 0.70) and shoot chlorophyll (r = 0.75) revealed that the increase in ARA is growth-linked and indirectly colonisation, as has been observed in other studies (Gantar et al. 1991, 1995; Obreht et al. 1993). This shows the synergistic effect of association of cyanobacteria with wheat, which leads to improved associative N fixation. Path coefficient analysis for the ARA and fresh weight with other parameters was undertaken to partition the correlations into direct and indirect effects. The analyses revealed the parameters taken into consideration, including the activity of defense, and hydrolytic enzymes were significant in their role in modulating ARA and fresh weight.
The regular use of agricultural land for the cultivation of major crops has led to the gradual exhaustion of minerals and nutrient imbalances, emphasizing an urgent need for employing multifunctional microbial consortia/bio-fertilisers which can promote the growth of plants by producing auxins/bioactive molecules and helping to sustain the soil fertility. The role of plant-growth-promoting rhizobacteria (PGPR) in enhancing plant biometric parameters/yield and biological control of soil-borne pathogens has been intensively investigated by several workers (Mäder et al. 2011; Piromyou et al. 2011). Cyanobacteria are known to produce antibacterial and antifungal metabolites/enzymes which help in controlling diseases, besides secreting mucilage and polysaccharides which can improve soil structure and porosity (Prasanna et al. 2008; Gupta et al. 2013). The plant cell wall is a complex mixture of polysaccharides, proteins and other defense compounds, which hinder the invasion of any foreign organism. One of the predicted routes of bacterial penetration into plant tissue besides mechanical/physical modes is through the secretion of an extensive repertoire of glycoside hydrolases, lyases and esterases which can target the numerous linkages as a means of cell wall degradation, especially for pathogen. In the present investigation, the cyanobacterial strains used are known to exhibit hydrolytic enzyme activity such as glucanases or chitosanases (Prasanna et al. 2009; Gupta et al. 2012). The major role of β-1,4-endoglucanases in plants is to degrade the cell wall, but this enzymatic activity can also contribute significantly in the biosynthesis of cellulose, polysaccharide modifications and loosening of cell wall during elongation of cells. Therefore, such type of enzymatic activity has been reported during normal growth and development, as well as in growing shoot, root and germination of grains (Buchanan et al. 2012). The enhanced activity of cell-wall-hydrolysing enzymes in the root tissues of inoculated plants, with increase of 3.1–18.7 % over uninoculated control, can be attributed to the stimulation of the plant-related enzymes, as a result of the cyanobacterial inoculation. The positive correlation (r = 0.78) observed between β-1,3-endoglucanase activity and ARA is in congruence with the earlier reports of Kovtunovych et al. (1999) using Klebsiella spp., who reported an increase in pectate lyase activity (another cell-wall-hydrolysing enzyme) showing positive correlation and concomitant with a tenfold higher rate of internal colonisation of wheat roots and higher ARA. It needs to be emphasized here that despite the enhanced values of these enzymes, they are not potent enough for complete plant cell wall degradation.
The inoculated cyanobacteria also elicited pathogenesis-related (PR) proteins, which serve as plant protection strategies against pathogens and pests. The phenomenon of induced systemic resistance (ISR) and systemic acquired resistance (SAR) is well investigated in terms of bio-control mediated by endophytic bacteria or plant-associated bacteria (Loon et al. 1998; Kloepper and Ryu 2006), but very few reports exist on cyanobacteria (Prasanna et al. 2013a; Singh et al. 2011). A differential systemic accumulation of phenylpropanoids in plant leaves, when inoculated with different cyanobacterial strains, emphasizes that cyanobacterial inoculation correlates positively with plant growth promotion and stress tolerance in rice (Singh et al. 2011). These findings support the conclusions of this study on the significant role of defense/pathogenesis enzymes not only in improving colonisation but also in enhancing plant fitness. Several workers (Karthikeyan et al. 2006; Patil et al. 2011) observed that Actinomycetes help in protecting tomato plants against Rhizoctonia solani, and such plants showed better growth in terms of high chlorophyll and disease reduction. In an earlier study, Prasanna et al. (2013a) investigated the cyanobacteria—phytopathogenic fungi—tomato plant interactions, as a prelude towards developing suitable biological options for combating biotic stress (Fusarium wilt) and enhancing plant vigour.
Cyanobacteria have been mainly identified with the help of morphological characters like shape, size of cells, place and position of heterocyst, or nature of growth in liquid/agar media (Nayak et al. 2009). These prokaryotes have highly conserved repetitive sequences in their genome such as highly iterated palindrome (HIP-TG) sequences, which are specific to these organisms. Such sequences can be an important tool for DNA fingerprinting, besides helping in studying the presence and association with their hosts (Rasmussen and Svenning 1998; Rai et al. 2000; Prasanna et al. 2012). In order to study the colonisation and association of cyanobacterial strains, analyses of the PCR-based profiles of wheat root DNA using repeat sequences were undertaken. DNA fingerprints from the samples revealed that specific bands could be distinguished for monitoring the presence of the inoculated strain and can be valuable for confirming colonisation. Earlier reports on evaluating the role of cyanobacteria on soil microbiological and plant parameters employing agronomic and molecular tools (Singh et al. 2011; Prasanna et al. 2013b) further emphasize the significance of our findings.
In the present investigation, path coefficient analyses helped to correlate and prove the significance of the parameters evaluated in this study, towards the enhancement in plant vigour (using fresh weight and ARA as indices). In general, path coefficient analysis has been employed mainly in plant breeding or selection of varieties or yield analyses, but is not used for analyses of microbial inoculation, as evidenced from published literature. The observations from the analyses in this study serve to highlight the significant role of activity of hydrolytic and defense enzymes in enhancing growth, ARA and thereby colonisation of cyanobacteria in wheat roots.
This study is a novel report which illustrates the mode of entry and colonisation in wheat plants by cyanobacteria through stimulating the activity of plant defense and pathogenesis enzymes and is further confirmed using DNA-based profiles. The colonisation by cyanobacteria led to improved plant growth, nitrogen fixation and nutrient mobilisation in such associations. This can also be considered a step forward in the dream of developing nitrogen-independent cereals, without the need for genetic engineering and prove more time and cost effective. Such plants need to be tested under different agro-climatic conditions to evaluate their agronomic efficiency.
References
Buchanan M, Burton R, Dhugga K, Rafalski A, Tingey S, Shirley N, Fincher G (2012) Endo-(1,4)-beta-glucanase gene families in the grasses: temporal and spatial Co-transcription of orthologous genes. BMC Plant Biol 12(1):235
Çakmakçi R, Dönmez F, Aydın A, Şahin F (2006) Growth promotion of plants by plant growth-promoting rhizobacteria under greenhouse and two different field soil conditions. Soil Biol Biochem 38:1482–1487
Chaudhary V, Prasanna R, Bhatnagar AK (2012) Modulation of fungicidal potential of Anabaena strains by light and temperature. Folia Microbiol 57:199-208
Cox WJ, Reisenauer HM (1973) Growth and ion uptake by wheat supplied nitrogen as nitrate, or ammonium, or both. Plant Soil 38:363–380
Daizy R, Kohli RK (1991) Fresh matter is not an appropriate relation unit for chlorophyll content: experience from experiments on effect of herbicide and allelopathic substance. Photosynthetica 25:655–657
Dobbelaere S, Vanderleyden J, Okon Y (2003) Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22:107–149
Dommergues Y, Balandreau J, Rinaudo G, Weinhard P (1973) Non-symbiotic nitrogen fixation in the rhizospheres of rice, maize and different tropical grasses. Soil Biol Biochem 5:83–89
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Gantar M (2000) Mechanical damage of roots provides enhanced colonization of the wheat endorhizosphere by the dinitrogen-fixing cyanobacterium Nostoc sp. strain 2S9B. Biol Fertil Soils 32:250–255
Gantar M, Elhai J (1999) Colonization of wheat para-nodules by the N2-fixing cyanobacterium Nostoc sp. strain 2S9B. New Phytol 141:373–379
Gantar M, Kerby NW, Rowell P, Obreht Z (1991) Colonization of wheat (Triticum vulgare L.) by N2-fixing cyanobacteria: I. A survey of soil cyanobacterial isolates forming associations with roots. New Phytol 118:477–483
Gantar M, Kerby NW, Rowell P, Obreht Z, Scrimgeour C (1995) Colonization of wheat (Triticum vulgare L.) by N2-fixing cyanobacteria. New Phytol 129:337–343
Gupta V, Prasanna R, Chaudhary V, Nain L (2012) Biochemical, structural and functional characterization of two novel antifungal endoglucanases from Anabaena laxa. Biocatal Agric Biotechnol 1:338–347
Gupta V, Ratha SK, Sood A, Chaudhary V, Prasanna R (2013) New insights into the biodiversity and applications of cyanobacteria (blue-green algae)—prospects and challenges. Algal Res 2:79–97
Hageman RH (1984) Ammonium versus nitrate nutrition of higher plants. In: Nitrogen in crop production. Am Soc Agron, Madison, WI, p. 804
Hiroki M, Shimizu A, Li R, Watanabe M, Watanabe MM (1998) Development of a database system useful for identification of Anabaena spp. (Cyanobacteria). Phycol Res 46:85–93
Hisbergues M, Christiansen G, Rouhiainen L, Sivonen K, Börner T (2003) PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch Microbiol 180:402–410
Jaiswal P, Prasanna R, Nayak S, Sood A, Suseela MR (2008) Characterization of rhizo-cyanobacteria and their associations with wheat seedlings. Egypt J Biol 10:20–27
Karthikeyan M, Radhika K, Mathiyazhagan S, Bhaskaran R, Samiyappan R, Velazhahan R (2006) Induction of phenolics and defense-related enzymes in coconut (Cocos nucifera L.) roots treated with biocontrol agents. Braz J Plant Physiol 18:367–377
Karthikeyan N, Prasanna R, Lata N, Kaushik BD (2007) Evaluating the potential of plant growth promoting cyanobacteria as inoculants for wheat. Eur J Soil Biol 43:23–30
Karthikeyan N, Prasanna R, Sood A, Jaiswal P, Nayak S, Kaushik BD (2009) Physiological characterization and electron microscopic investigation of cyanobacteria associated with wheat rhizosphere. Folia Microbiol 54:43–51
Kaushik BD (2004) Use of blue green algae and Azolla biofertilizer in rice cultivation and their influence on soil properties. In: Jain PC (ed) Microbiology and Biotechnology for sustainable development. CBS, New Delhi, pp 166–184
Kloepper J, Ryu C-M (2006) Bacterial endophytes as elicitors of induced systemic resistance. In: Schulz BE, Boyle CC, Sieber T (eds) Microbial root endophytes, vol 9. Soil biology. Springer, Berlin, pp 33–52
Knight CD, Adams DG (1996) A method for studying chemotaxis in nitrogen fixing cyanobacterium-plant symbioses. Physiol Mol Plant Pathol 49:73–77
Kovtunovych G, Lar O, Kleiner D, Kozyrovska N (1999) Enhancing the internal plant colonization rate with endophytic nitrogen-fixing bacteria. Biopolimery i Kletka 15(4):300–305
Loon LCV, Bakker PAHM, Pieterse CMJ (1998) Systematic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Lucas García JA, Probanza A, Ramos B, Colón Flores JJ, Gutiérrez Mañero FJ (2004) Effects of plant growth promoting rhizobacteria (PGPRs) on the biological nitrogen fixation, nodulation, and growth of Lupinus albus l. cv. Multolupa. Eng Life Sci 4:71–77
Mackinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140(2):315–322
Mäder P, Kaiser F, Adholeya A, Singh R, Uppal HS, Sharma AK, Srivastava R, Sahai V, Aragno M, Wiemken A, Johri BN, Fried PM (2011) Inoculation of root microorganisms for sustainable wheat–rice and wheat–black gram rotations in India. Soil Biol Biochem 43:609–619
Manjunath M, Prasanna R, Nain L, Dureja P, Singh R, Kumar A, Jaggi S, Kaushik BD (2009) Biocontrol potential of cyanobacterial metabolites against damping off disease caused by Pythium aphanidermatum in solanaceous vegetables. Arch Phytopathol Plant Protect 43:666–677
McCowen S, MacArthur L, Gates J (1986) Azolla fern lectins that specifically recognize endosymbiotic cyanobacteria. Curr Microbiol 14:329–333
Nain L, Rana A, Joshi M, Jadhav S, Kumar D, Shivay YS, Paul S, Prasanna R (2010) Evaluation of synergistic effects of bacterial and cyanobacterial strains as biofertilizers for wheat. Plant Soil 331:217–230
Nayak S, Prasanna R, Pabby A, Dominic TK, Singh PK (2004) Effect of urea, blue green algae and Azolla on nitrogen fixation and chlorophyll accumulation in soil under rice. Biol Fertil Soils 40:67–72
Nayak S, Prasanna R, Prasanna BM, Sahoo D (2009) Genotypic and phenotypic diversity of Anabaena isolates from diverse rice agro-ecologies of India. J Basic Microbiol 49:165–177
Obreht Z, Kerby N, Gantar M, Rowell P (1993) Effects of root-associated N2-fixing cyanobacteria on the growth and nitrogen content of wheat (Triticum vulgare L.) seedlings. Biol Fertil Soils 15:68–72
Patil HJ, Srivastava AK, Singh DP, Chaudhari BL, Arora DK (2011) Actinomycetes mediated biochemical responses in tomato (Solanum lycopersicum) enhances bioprotection against Rhizoctonia solani. Crop Prot 30:1269–1273
Piromyou P, Buranabanyat B, Tantasawat P, Tittabutr P, Boonkerd N, Teaumroong N (2011) Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur J Soil Biol 47:44–54
Prasanna R, Lata, Tripathi R, Gupta V, Middha S, Joshi M, Ancha R, Kaushik BD (2008) Evaluation of fungicidal activity of extracellular filtrates of cyanobacteria-possible role of hydrolytic enzymes. J Basic Microbiol 48:186–194
Prasanna R, Jaiswal P, Nayak S, Sood A, Kaushik B (2009) Cyanobacterial diversity in the rhizosphere of rice and its ecological significance. Indian J Microbiol 49:89–97
Prasanna R, Joshi M, Rana A, Shivay Y, Nain L (2012) Influence of co-inoculation of bacteria-cyanobacteria on crop yield and C–N sequestration in soil under rice crop. World J Microbiol Biotechnol 28:1223–1235
Prasanna R, Chaudhary V, Gupta V, Babu S, Kumar A, Singh R, Shivay Y, Nain L (2013a) Cyanobacteria mediated plant growth promotion and bioprotection against Fusarium wilt in tomato. Eur J Plant Pathol 136:337–353
Prasanna R, Sharma E, Sharma P, Kumar A, Kumar R, Gupta V, Pal R, Shivay Y, Nain L (2013b) Soil fertility and establishment potential of inoculated cyanobacteria in rice crop grown under non-flooded conditions. Paddy Water Environ 11:175–183
Prasanna R, Triveni S, Bidyarani N, Babu S, Yadav K, Adak A, Khetarpal S, Pal M, Shivay YS, Saxena AK (2013c) Evaluating the efficacy of cyanobacterial formulations and biofilmed inoculants for leguminous crops. Arch Agron Soil Sci 60:1–18
Rai AN (1990) CRC Handbook of Symbiotic Cyanobacteria. CRC Press Inc., Boca Raton
Rai AN, Soderback E, Bergman B (2000) Cyanobacterial plant symbioses. New Phytol 147:449–481
Rasmussen U, Svenning MM (1998) Fingerprinting of cyanobacteria based on PCR with primers derived from short and long tandemly repeated repetitive sequences. Appl Environ Microbiol 64:265–272
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Singh RK, Chaudhary BD (1979) Biometrical methods in quantitative genetic analysis. Kalyani Publishers, Ludhiana, p 303
Singh D, Prabha R, Yandigeri M, Arora D (2011) Cyanobacteria-mediated phenylpropanoids and phytohormones in rice (Oryza sativa) enhance plant growth and stress tolerance. Anton van Leeuw 100:557–568
Spiller H, Stallings W Jr, Woods T, Gunasekaran M (1993) Requirement for direct association of ammonia-excreting Anabaena variabilis mutant (SA-1) with roots for maximal growth and yield of wheat. Appl Microbiol Biotechnol 40:557–566
Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35:171–205
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. In: Current protocols in bioinformatics. John Wiley Sons, Inc.
Venkataraman GS (1972) Algal biofertilizer and rice cultivation. Today and Tomorrow Publications, New Delhi
Venkataraman GS, Neelakantan S (1967) Effect of cellular constituents of nitrogen fixing blue green alga Cylindrospermum on root growth of rice plants. J Gen Appl Microbiol 13:53–62
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Weissman GS (1951) Nitrogen metabolism of wheat seedlings as influenced by the ammonium: nitrare ratio and the hydrogen ion concentration. Am J Bot 38:162–174
Acknowledgments
This study was supported partially by the funds from the Application of Microorganisms in Agricultural and Allied Sectors (AMAAS) Network Project on Microorganisms (Themes: Microbial Genomics; Nutrient Management) granted by Indian Council of Agricultural Research (ICAR), New Delhi. The authors are also grateful to the National Phytotron Facility and Division of Microbiology, IARI, New Delhi, for providing the necessary facilities for undertaking this study. The authors state that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOC 37 kb)
Supplementary Table 2
(DOC 53 kb)
Supplementary Table 3
(DOC 58 kb)
Rights and permissions
About this article
Cite this article
Babu, S., Prasanna, R., Bidyarani, N. et al. Analysing the colonisation of inoculated cyanobacteria in wheat plants using biochemical and molecular tools. J Appl Phycol 27, 327–338 (2015). https://doi.org/10.1007/s10811-014-0322-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-014-0322-6