Abstract
Increasing evidence indicates that the biosynthesis and accumulation of the keto-carotenoid astaxanthin represents a chronic (in days or weeks) molecular defense mechanism in the green alga, Haematococcus pluvialis, to protect cells from abiotic stress. However, information with regard to the acute (in minutes or hours) response of the cells to stress is scarce. In this study, two cDNAs encoding manganese superoxide dismutase (MnSOD) were cloned and their full length nucleotide sequences were obtained. Quantitative real-time RT-PCR analysis revealed a general increase in MnSOD transcriptions in response to various abiotic stressors, e.g., high light, excessive amounts of iron or sodium acetate, either singly or in combination. Meanwhile, four isoforms of MnSOD were detected by a nondenaturing polyacrylamide gel electrophoresis approach. In addition, an iron-containing superoxide dismutase (FeSOD) was detected in the cells using the same technique. Differential expressions of SOD isoenzymes under different abiotic stressors were also investigated and discussed, at both mRNA and protein levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photosynthetic microalgae generate reactive oxygen species (ROS), including the superoxide anion (O −2 ), hydrogen peroxide (H2O2), singlet oxygen (O2 (1∆g)) and the hydroxyl radical (OH), through photosynthesis and cellular respiration, particularly under photo-oxidative stress conditions (Asada 1994; Perl-Treves and Perl 2002). Excess amounts of ROS can damage macromolecules (e.g., proteins, lipids, and DNA) and cell membranes (Asada 1994) while at the same time ROS may serve as signaling molecules to trigger and mediate cell defense systems to improve resistance and adaptation of microalgae to an unfavorable environment (Lu et al. 1998; Boussiba et al. 1999; Alscher et al. 2002; Zhekisheva et al. 2002; Wang et al. 2004b; 2009; Li et al. 2008). Cells have developed a variety of defensive mechanisms against the harmful effects of ROS. These may include low molecular weight compounds, such as glutathione, phenolics, ascorbate, flavonoids, tocopherols, and carotenoids, as well as enzymatic catalysts of high molecular weight, such as superoxide dismutase (SOD), catalase, and glutathione peroxidase, etc. (McCord and Fridovich 1969; Tsang et al. 1991; Shaish et al. 1993; Karpinski et al. 1997; Erturk 1999; Geigenberger et al. 2005; Buchanan and Luan 2005; Kwon et al. 2006; Mur et al. 2006; Grun et al. 2006). SODs (EC 1.15.1.1) are ubiquitous metalloenzymes in aerobic organisms and are an essential part of the first line of the antioxidant defense system (Blokhina et al. 2003). It removes O −2 , prevents generation of OH−, which is highly toxic, and catalyses the dismutation of superoxide radical into molecular oxygen and hydrogen peroxide (Fridovich 1975). Hydrogen peroxide can be further detoxified into water by ascorbate peroxidases (Blokhina et al. 2003).
Four forms of SOD are known based on their metal active sites. These forms are Cu/Zn SOD (in the cytosol of eukaryotes), manganese superoxide dismutase (MnSOD; in the mitochondria of both eukaryotes and prokaryotes), iron superoxide dismutase (FeSOD; in both eubacteria and archaebacteria) (Fridovich 1986), and NiSOD (in Streptomyces and predicted to be present in some cyanobacteria) (Youn et al. 1996; Wuerges et al. 2004). Damage from ROS generated during photosynthesis can also be minimized by several isoforms of the same SOD. Usually there is at least one class of SODs or more within each species. In rice, there are three classes of SODs, including two Cu/Zn, one Mn, and one Fe SODs (Kanematsu and Asada 1990; Sakamoto et al. 1993; Kaminaka et al. 1999). A total of seven SOD genes were identified in Arabidopsis, including three CuZnSODs, three FeSODs, and one MnSOD (Kliebenstein et al. 1998). Most eukaryotic algae contain either FeSOD or MnSOD or both. From the first complete red algal genome (Cyanidioschyzon merilae), three MnSODs were detected (Matsuzaki et al. 2004). In the genome of the diatom Thalassiosira pseudonana, two FeSODs, and two MnSODs were found (Armbrust et al. 2004). The green alga Chlamydomonas reinhardtii contains at least three MnSODs and one FeSOD (Chlamy center, http://www.chlamy.org).
Haematococcus pluvialis is a flagellated unicellular green alga (Chlorophyta) that produces the red pigment astaxanthin under oxidative stress conditions, such as high light, high salts, or deprivation of nutrients, either singly or in combination (Steinbrenner and Linden 2001; Wang et al. 2004b). It has been shown that Haematococcus has two antioxidative mechanisms, antioxidative enzymes in vegetative cells and antioxidative astaxanthin in cyst cells (Kobayashi et al. 1997; Wang et al. 2004b) and that the accumulation of astaxanthin is a survival strategy of this organism and other related organisms in response to stressful environments (Boussiba 2000). Previously, Kobayashi et al. (1997) conducted an assay for SOD using H. pluvialis whole-cell and cell-free extract samples. Our previous work (Wang et al. 2004b) identified a mitochondrion-associated MnSOD and a cytosol-localized Cu/ZnSOD-like SOD. The expression of MnSOD was constitutively high in vegetative cells and progressively down-regulated after onset of stress, and the protein content was reduced to one tenth of its basal level after 72 h of stress. Cu/ZnSOD like SOD expression, however, was up-regulated and reached the highest level (ca. 8-fold) at 48 h, and then was reduced to only a trace amount after 72 h of stress (Wang et al. 2004b). None of studies mentioned above, however, have performed gene expression analysis of SODs at both mRNA and protein levels. Information with regard to the acute (in minutes or hours) response of the algal cells to stress is scarce. These issues can only be resolved by further work to examine the expression of SODs at both mRNA and enzymatic activity levels.
In this study, we cloned and characterized two MnSOD cDNAs and investigated the regulation of SOD isoenzymes at transcriptional and translational levels in H. pluvialis in response to various abiotic stressors.
Materials and methods
H. pluvialis Flotow NINE144 was obtained from the National Institute for Environmental Studies in Tsukuba, Japan. An acetate basal medium described by Kobayashi et al. (1991) was used: 14.6 mM sodium acetate, 2.7 mM l-asparagine, 2 g L–1 yeast extract, 0.985 mM MgCl2, 0.135 mM CaCl2, 0.036 mM FeSO4, and pH 6.8. The cells were grown in 500-mL Erlenmeyer flasks each containing 100-mL culture medium. Cultures were incubated in a growth chamber at 22°C and 20 μmol photons m–2 s–1 of light under a 12:12-h light/dark cycle. Cultures were shaken manually once each day. For stress induction, exponentially growing cultures (cell density of approximately 5 × 105cells mL–1) were exposed to continuous illumination of 150 μmol photons m–2 s–1 in the absence or presence of 45 mM sodium acetate and 450 μM ferrous sulfate.
Genomic DNA and RNA isolations
For genomic DNA isolation, a simplified protocol (Wang et al. 2004a) was followed. Briefly, 50 mL of green cells in log phase were collected by centrifugation and resuspended in 700 μL DNA lysis buffer (0.25 M Tris-HCl, pH 8, 60 mM Na2EDTA, and 4% SDS), then followed by phenol extraction and isopropanol precipitation at room temperature. After a 70% ethanol wash and air drying, the DNA pellet was dissolved in dH2O. RNA was then isolated according to the miniprep RNA extraction procedure (Wang et al. 2008) with minor modifications. Nuclear acids were quantified by NanoDrop 3.0 (Coleman Technologies Inc., USA). Both DNA and RNA solutions were aliquoted and stored at −80°C, if not immediately used.
Degenerated PCR for SOD genes from H. pluvialis
Both Mn/Fe and Cu/Zn SODs from higher plants and green alga C. reinhardtii were retrieved from the GenBank database and were aligned with Clustal W software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Based on the alignment of SOD amino acid sequences, several highly conserved domains (GGGHWNHS and DVWEHAYY) were used for degenerated PCR primer design (FSOD: 5′-GGGGGCGGCCACTGGAAYCA-3′; RSOD: 5′-TARTANGCGTGY TCCCANCRTC-3′). Using 50 ng of genomic DNA or 20 ng cDNA as template, PCR was programmed at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 58°C for 55 s, and 72°C for 55 s with 72°C extension for 7 min.
PCR, TA cloning, sequencing, and homologues searching
TA Cloning®Kit with One Shot®TOP chemically competent Escherichia coli (Invitrogen, Carlsbad, USA) was used for PCR and rapid amplification of cDNA ends (RACE) products cloning. PCR and RACE products were purified with the Geneclean Kit (QBIOgene, USA). A 3-μL aliquot of the DNA recovered from PCR or RACE was ligated to a pCR21 vector (Invitrogen) overnight at 14°C. Transformation of the ligation was carried out following instructions from Invitrogen. Positive clones were confirmed by PCR method with M13 primers. Plasmid was isolated using the Plasmid MiniPrep kit (QIAGEN, USA) and five clones were selected for DNA sequencing using the dideoxy-nucleotide terminator method with an ABI Prism Dye-terminator system (PE Biosystems, USA). Both strands were sequenced with an overlapping scheme throughout the whole gene fragment. Sequences were analyzed using DNAclub (Xiongfong Chen, Cornell Univ., Ithaca), and homologue searching was done by translated query vs. protein database (blastx) and nucleotide–nucleotide BLAST (blastn) in the GenBank database.
SMART cDNA synthesis, 5′- and 3′-rapid amplification of cDNA ends
SMART™ RACE cDNA Amplification Kit (Clontech, BD Bioscience) was applied to perform both 5′- and 3′-RACE with the gene specific primers (M1SOD5, 5′-CGACGCATCGACCGCGGCC-3′, M1SOD3, 5′-GGGCAT CACCTCCACCCCCA-3′, for MnSOD1; M2SOD5, 5′-CTCGCCAGAGGCTGGCGC GG-3′, M2SOD3, 5′-GGACCCACTGAGCACGCTGGG-3′ for MnSOD2) and the universal primer (UPM) provided with the kit. 2 μg of total RNA was used for 5′- and 3′-RACE cDNA synthesis. Then the first-strand cDNA was diluted with 100 μL of Tricine-EDTA buffer. The dilutions, 5′- and 3′-RACE cDNA, after heated at 72°C for 7 min, were used for RACEs. The program for 5′-end RACE was a touch-down program 1 (five cycles of 94°C for 30 s and 72°C for 3 min; five cycles of 94°C for 30 s, 70°C for 30 s, and 72°C for 3 min; 25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min), and a 50-fold dilution of product was used for another RACE program for 25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min).
Quantitative real-time RT-PCR
For real-time reverse transcriptase-polymerase chain reaction (RT-PCR) analysis, first strand synthesis was carried out using the Taqman Reverse Transcription system according to the manufacturer’s instructions (Applied Biosystems, USA). 250 ng of total RNA were used in a 10 μL synthesis mixture. M1SODf, 5′-ACCAGGCCTACGTCACAAACC-3′, M1SODr, 5′-GCTCAGGGAACTTGTC CAG-3′, for MnSOD1; M2SODf, 5′-GCTGTTGCAGCTGATTGAGG-3′, M2SODr, 5′-GCTTTGAAAGCGTCCATGCT for MnSOD2) for real-time RT-PCR were designed by using the Primer Express® Software Version 2.0 (Applied Biosystems, USA) to produce 110 bp PCR product. The experiments were repeated at least twice independently. Transcription levels of the target genes were calculated from the threshold cycle by interpolation from the standard curve. To standardize the results, the relative abundance of 18S rRNA was also determined and used as the normalization standard. All calculations and statistical analyses were performed as described in the ABI 7900 sequence detection system User Bulletin 2 (Applied Biosystems, USA)
Preparation of H. pluvialis cell-free extracts
Culture suspensions (100 mL) were taken at various time intervals and the cells were harvested by centrifugation (3,000×g, 5 min at 4°C) and washed three times with cold phosphate buffered saline. Cell-free extracts were prepared by motor and pestle in the presence of liquid N2, and cell lysates were resuspended in 300 μL of Native Solution (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% NP-40) with Protease Inhibitor Cocktail (Sigma, USA), followed by centrifugation (14,000×g for 20 min at 4°C). Protein concentrations were measured using a Bio-Rad Protein Assay Dye Reagent Concentration (Bio-Rad, USA) in microtitre plates containing 2 μL of sample and 100 μL of dye per well. The measurement was performed at 595 nm with BSA as a standard.
Nondenaturing polyacrylamide gel electrophoresis
Electrophoresis was carried out at room temperature according to a modified procedure (Gabriel 1971) with 1.5 mm of 10% polyacrylamide mini-slab gel in standard tris-glycine buffer (pH 8.3). Samples were loaded into each well and then electrophoresed at 60 V through the 5% stacking gel for 15 min and 100 V through the separating gel for 80 min. After electrophoresis, a modified photochemical method based on Beauchamp and Fridovich (1971) and Elstner and Heupel (1976) was used to locate SOD activities on gels. The gel was first soaked in 25 mL of 1.23 mM NBT for 5 min, briefly washed, then soaked in the dark in 30 mL of 50 mM potassium phosphate buffer (pH 7.0) containing 28 mM TEMED and 0.028 mM riboflavin for another 10 min. The gel was briefly washed again, and then illuminated on a light box with an intensity of 30 μmol photons m−2 s−1 for 20 min to initiate the photochemical reaction.
Cell-free extracts SOD assay
For determination of SOD activity of cell-free extracts, SOD assay Kit-WST (Dojindo Molecular Technologies, Inc., USA) was applied, where the presence of SODs inhibited formation of a colored compound. Assays were followed as the protocol described, and measurements was performed with a SPECTRAmax 340PC384 (Molecular Devices Co., USA) at 450 nm in 96-well microtitre plates. In 1 unit of SOD, the 50% inhibition activity of SOD was determined by dilution of samples at different concentrations.
Nucleotide sequences and accession numbers
The nucleotide sequences of cDNAs of MnSOD1 and MnSOD2 have been deposited in the GenBank database under the accession numbers AY744704 and DQ533711, respectively.
Results
MnSOD sequence analysis
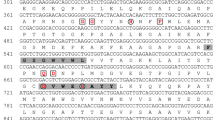
Using genomic DNA and cDNA as templates, each degenerated primer pairs yielded one or several specific fragments from H. pluvilias (data not shown). The molecular size of the amplicon was about 300 bp from primers FSOD/RSOD. Sequence analyses of the PCR confirmed that two partial sequences of a SOD gene were obtained. Based on the confirmed sequences of 300 bp fragment of the SOD gene, specific 5′-RACE and 3′RACE primers were designed and synthesized. Using cDNA from total RNA of green cells, 5’-RACE and 3’-RACE were performed to clone the 5′- and 3′-end of SOD mRNA. For both 5′-RACE and 3′-RACE, more than one band was amplified. For cloning and sequencing, the bands with the highest molecular weight were selected for subsequent steps. After TA cloning, sequencing and database searching, two full lengths of cDNAs were obtained by overlapping of 5′- and 3′-end sequences. Based on the amino acid residues that are specific for MnSOD and FeSODs (Parker et al. 1987), e.g., Gly79, Gly 80, Phe87, Gln153, and Asp154 unique for MnSOD (numbers were based on SOD1) in H. pluvilias and residues that are ligands to the Mn cofactor, e.g., His31, His84, Asp172, and His176, these two SOD genes were determined to encode Mn-containing SOD. These two genes contained an open reading frame of 654 bp coding for 217 amino acids (MnSOD1) and 660 bp for 220 amino acids (MnSOD2) with the stop codon of TGA (Fig. 1).
The alignment of MnSODs from green algae, human, fruitfly, higher plant, bacterium, and blue green alga. MnSOD1 and MnSOD2, from this study. MnSODChlam, MnSOD from Chlamydomonas (GenBank accession ID: XP_001700058.1); Human, MnSOD from Human (GenBank accession ID: AAP34409.1); Fruitfly, MnSOD from Drosophila melanogaster (GenBank accession ID: NP_476925.1); Arabi, MnSOD from Arabidopsis thaliana (GenBank accession ID: AAM62550.1); E.coli, MnSOD from Escherichia coli ED1a (GenBank accession ID: YP_002400405.1), and Cyano, MnSOD from the cyanobacterium Anabaena variabilis ATCC 29413 (Genbank accession ID: YP_321963.1). The conserved residues that may distinguish Mn- and FeSODs are indicated with dots. Asterisks denote the ligands to the Mn cofactor. The residues conserved in all MnSODs are highlighted in a grey color
Real-time RT-PCR analyses of MnSODs
In order to understand the biological roles of the two MnSODs in H. pluvialis, we analyzed the transcriptional expression of the MnSOD genes under different oxidative stresses, e.g., high light of 150 μmol photons m–2 s–1 (HL), HL in the presence of 450 μM iron (HL + Fe), HL in the presence of 45 mM sodium acetate (HL + SA), and HL in the presence of both Fe and SA (HL + Fe + SA) using the quantitative real-time RT-PCR technique. The gene expression levels were normalized to the housekeeping gene 18S rRNA. Overall, MnSOD1 was induced by all treatments and reached the highest levels at around 6 h (HL, HL + Fe or HL + SA) and 12 h (HL + Fe + SA) (Fig. 2a). Oxidative stresses increased transiently the mRNA level of MnSOD1 gene to 287% under HL after 6 h and 325% under HL + Fe + SA after 12 h, as compared to the onset of stress. Afterwards, the transcripts of MnSOD decreased. Under HL, it decreased to ca. 200% after 24 h and to 160% after 72 h of the onset level. However, under HL + Fe + SA, MnSOD mRNA decreased rapidly after 12 h, and reached a relatively stable level (140% of the onset level) after 24 h. At 144 h, its level was still 1.26 times higher than that at the onset. As a control, the mRNA level of LL treated cells remained relatively low and stable for the first 72 h. Afterwards, MnSOD mRNA copies increased slightly, and remained at a slightly higher level (130% of the onset level) until 144 h (Fig. 2a). The transcriptional expression of MnSOD2 was also up-regulated under all the above treatments; however, its expression patterns were somewhat different from those of MnSOD1. As such, the MnSOD2 mRNA levels were induced steadily and reached the highest levels at 12 h (all the treatments except for HL + Fe + SA) or at 24 h under HL + Fe + SA, and then remained at a relatively stable level (Fig. 2b).
Transcriptional expression of MnSOD1 and MnSOD2 and growth curves of H. pluvialis under different treatment conditions. A, Expression of MnSOD1; B, Expression of MnSOD2. C. Growth curves from different treatments. Relative mRNA levels were calculated and normalized with respect to the 18S rRNA level. Values represent the mean and standard deviation of triplicates. LL (■) low light of 20 µmol photons m–2 s–1, HL (●) high light of 150 µmol photons m–2 s–1, LL + Fe (□) low light with addition of 450 µM ferrous sulfate (FeSO4), HL + Fe (○) high light with addition of 450 µM FeSO4, LL + SA (◊) low light with addition of 45 mM sodium acetate (NaAc), HL + SA (▼) high light with addition of 45 mM NaAc, HL+ Fe + SA (Δ) high light with addition of 450 µM FeSO4 and 45 mM NaAc
Nondenaturing polyacrylamide gel electrophoresis
The SODs in H. pluvialis was characterized at the protein level by a nondenaturing polyacrylamide gel electrophoresis (PAGE). The SOD activity of cell-free extracts from green vegetative cells grown under favorable culture conditions was resolved into at least three bands, bands I, II, and III (Fig. 3a–c). These SODs were tested for their susceptibility to KCN and H2O2. None of them were sensitive to 2 mM KCN, indicating the absence of Cu/ZnSOD in accordance with previous reports (Asada et al. 1977; Sakurai et al. 1993). Band I was sensitive to H2O2, indicating that it is a FeSOD. Bands II and III were insensitive to both KCN and H2O2, indicating that they are MnSODs (Asada et al. 1975). Among them, band III covered around 50% of the total SOD activity in the cell-free extracts (Fig. 3).
The non-denaturing PAGE gel images revealing different isoenzymes of SOD in H. pluvialis. 100 mg protein sample was loaded into each well of non-denaturing PAGE gel. a. control; b. incubated with KCN; c. incubated with H2O2; d and e, cell free extracts obtained from HL- and LL-treated algal cells, respectively. Each well was loaded with 10 mg protein extracts. S, cell free extracts from H. pluvialis at LL, Fe, control FeSOD protein. The different bands (I-VI) were described in the text
The nondenaturing PAGE analyses indicated that SOD enzymatic activity remained high, even after 144 h of HL treatment (Fig. 3). In this study, MnSOD and FeSOD displayed different alterations during stress induction. In contrast to the relatively stable level of FeSOD, MnSOD activity began to decrease after 72 h under LL (Fig. 3e) and after 120 h under HL (Fig. 3d).
More bands of SOD activities were observed in the nondenaturing PAGE of cell-free extracts of red cyst cells under HL (Fig. 3d). There were at least three additional bands induced after 48 h of HL treatment. Among them, there were two bands (V and VI) that migrated slightly faster than band III. The other one (band IV) migrated slightly faster than band I but slower than band II. Susceptibility test of these bands with KCN and H2O2 indicated that they were probably all MnSODs. It suggests that additional isoenzymes of MnSODs were expressed or activated under stress conditions. Although unlikely, however, we could not rule out the possibility of the additional bands being the proteolytic products of the isoemzymes of band I or III.
Cell-free extract-based SOD assays
In order to determine the SOD activity, several direct and indirect methods have been developed (McCord and Fridovich 1969; Beauchamp and Fridovich 1971; Elstner and Heupel 1976). SOD Assay Kit-WST allows sensitive and reliable SOD assaying by utilizing Dojindo’s highly water-soluble tetrazolium salt, WST-1 (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) that produces a water-soluble formazan dye upon reduction with a superoxide anion (Ukeda et al. 1999).
Figure 4 shows the alteration in the total SOD activity during oxidative stress under high light. Total enzyme activity increased gradually from 55 to 110 U mg−1 after induction for 12 h. SOD activity reached the highest level of 150 U mg−1 at 120 h and decreased slight thereafter.
Discussion
MnSOD gene sequence analysis
Comparative sequence analysis revealed a similar amino acid composition of MnSOD in H. pluvialis to that of other organisms. For example, the amino acid sequence of MnSOD1 had the highest homology (73% identity and 84% similarity) with MnSOD in the green alga Chlamydomonas reinhartii and shared 52% identity and 66% similarity with MnSOD in the diatom Thalassiosira weissflogii. An identity of 47% and a similarity of 64% were found with the higher plant Arabidopsis thaliana.
Transcriptional expression of MnSODs
As shown in Fig. 2a, MnSOD1 underwent transient up-regulation under HL and the maximum transcript level occurred at 6 h, followed by rapid reduction at 12 h and remained more or less constant for 48 h. Under HL + Fe or HL + SA, the gene expression followed the same trend as that occurring under HL. However, the maximum gene expression level was lower under HL + Fe or HL + SA than under HL. The gene expression under HL + Fe + SA exhibited a delayed pattern compared to HL, HL + Fe or HL + SA, and the maximum transcript level did not occur until 12 h, followed by a rapid decrease in the transcriptional expression. As a control, the mRNA level of MnSOD1 under LL remained relatively low and stable for the first 72 h and increased slightly thereafter (data not shown).
The transcriptional expression of MnSOD2 was also up-regulated under all the above conditions. However, its expression pattern was somewhat different from that of MnSOD1. Under LL, the MnSOD2 mRNA level remained low and unchanged. A small but noticeable increase in MnSOD2 transcripts occurred under LL + Fe or LL + SA, suggesting that the presence of 45 mM sodium acetate or 450 μM ferrous sulfate imposed mild stress to H. pluvialis cells under LL, and resulted in modest up-regulation of MnSOD2 to cope with the stress.
A rapid increase in MnSOD2 transcripts occurred under HL, reaching the maximum transcript level at 12 h and the expression level remained stable for 48 h (Fig. 2b). The gene expression under HL + Fe or HL + SA exhibited the same pattern of that under HL only. However, the maximum transcript level of MnSOD2 was lower under HL + Fe or HL + SA than under HL only. Under HL + Fe + SA, the maximum transcript level did not occur until 24 h and yet the value of the maximum transcripts was the lowest among all the treatments under HL.
Considering that sodium acetate and ferrous sulfate at the final concentration of 45 mM and 450 μM, respectively, induced stress and thus resulted in up-regulation of MnSOD under LL (Fig. 2b) and of several genes encoding for carotenoid synthesis in H. pluvialis (Steinbrenner and Linden 2001, 2003; Wang et al. 2004b; Li et al. 2008), it was expected that the maximum transcript level of MnSODs would be higher when H. pluvialis cells were exposed to HL in the presence of sodium acetate and/or ferrous sulfate than exposed to HL alone. However, our real-time RT-PCR results indicated that the maximum transcript level of MnSOD1 was lower when two stressors (i.e., HL + FE or HL + SA) were combined compared with HL alone, and the maximum gene expression was delayed from 6 h under HL to 12 h under HL + FE + SA (Fig. 2a). Likewise, two or three stressors (i.e., HL + FE, HL + SA, or HL + Fe + SA) resulted in low maximum transcripts of MnSOD2 than HL alone, and the maximum gene expression delayed from 12 h under HL + Fe + SA to 24 h under HL alone (Fig. 2b).
This phenomenon was previously observed in a study of carotenoid gene expression in H. pluvialis under stress. Li et al. (2008) reported that transcripts of the carotenoid genes were lower when two or three stressors were combined (i.e., HL + FE, HL + SA or HL + FE + SA) compared with HL. Our results supported the interpretation proposed by Li et al. (2008) that each individual stressor (e.g., HL, FE, or SA), may simultaneously trigger specific stress-dependent protective mechanism(s) (e.g., specific enzymatic defense pathways) in addition to inducing common, shared protective mechanism(s) (e.g., carotenogenesis, storage lipid biosynthesis, or secondary wall formation). When HL in combination with SA or FE or both is applied to H. pluvialis cells, more individual specific defense pathways could be activated or up-regulated than those triggered by HL alone, with each contributing to overall protection and thus, may to a lesser extent, depend upon SOD or carotenogenesis.
Presence of FeSOD and multiple MnSODs in H. pluvialis
SODs in H. pluvialis were characterized by nondenaturing PAGE, which detected various types of SODs and quantified the enzymatic activity (not the amount of SOD protein) of individual SODs at the same time. These SODs could be distinguished for their susceptibility to KCN and H2O2. For instance, Cu/ZnSOD and FeSOD are sensitive to KCN and H2O2 respectively, while MnSOD is not sensitive to either of the chemicals (Asada et al. 1975). In our study, at least three MnSODs and one FeSOD were detected. The gel analysis suggests that additional isoenzymes of MnSODs were induced or up-regulated under stress conditions. We speculated that there might be one major constitutively expressed and several stress-responsive MnSOD isoforms in H. pluvialis to augment the SOD-based protection against stress. If true, then our finding agrees with the reports of multiple forms of MnSOD in C. reinhardtii (Chlamy center, http://www.chlamy.org) and several other species of algae (Matsuzaki et al. 2004; Armbrust et al. 2004), and in higher plants such as maize (Zhu and Scandalios 1993), as well as in humans (Church 1990) and rat tissues (Ho et al. 1991).
We also used a SOD Assay Kit-WST to determine the enzymatic activity of SODs in H. pluvialis. Figure 4 shows a transient increase in the total SOD activity under HL. It increased gradually after induction for 12 h and reached the highest level at 120 h and decreased slightly thereafter. By adding different SOD-sensitive agents, e.g., KCN and H2O2, it is possible to determine MnSOD, Cu/ZnSOD, or FeSOD enzyme activity separately in cell-free extracts. In our study, the SOD activities of cell-free extracts were not altered by adding KCN during incubation (data not shown). FeSOD activity could not be quantitatively determined by substracting total enzyme activity from H2O2 addition assays because even 10 mM of H2O2 could not inhibit 100% of the FeSOD activity in our study (data not shown).
In an effort to confirm the presence of FeSOD in H. pluvilais, we designed several degenerated FeSOD primers for PCR detection using both cDNA and genomic DNA as templates. However, we failed to detect any FeSOD gene in H. pluvilais using RNA from green cells or genomic PCR. This cloning failure may be caused by very low expression of FeSOD in green cells or the PCR condition was not optimal to obtain FeSOD gene amplification. In further investigations, RNA from cyst cells or genomic southern blot will be required to verify the presence of FeSOD. We also designed several degenerated Cu/ZnSOD primers for PCR detection of any Cu/ZnSOD gene. However, we did not detect any Cu/ZnSOD in H. pluvilais. The latter was in consistent with the previous finding that Cu/ZnSODs are absence in most eukaryotic algae examined (Asada et al. 1977; Sakurai et al. 1993; Kitayama 1994), although Cu/ZnSOD seems to be the predominant species of SOD in higher plant cells (Bowler et al. 1994).
The biological role of SOD in H. pluvialis stress response
The transient up-regulation of FeSOD and MnSOD at the mRNA and protein level in H. pluvialis in response to the different stress conditions confirmed the previous finding that the majority of the antioxidative enzymes identified in H. pluvialis were boosted under oxidative stress (Wang et al. 2004b). Given that both the transcripts and enzymatic activity of MnSODs decreased to various degrees after their maximum levels within several hours or days under stress, it suggested that SODs may represent a short-term strategy to protect H. pluvialis against stress. As the stress persists, H. pluvialis cells may depend upon a long-term cell protective strategy to survive, which is thought to be the astaxanthin synthesis pathway (Kobayashi et al. 1991, 1997; Hagen et al. 1993; Wang et al. 2004b; 2009; Li et al. 2008).
References
Alscher RG, Erturk N, Lenwood SH (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341
Armbrust E, Berges J, Bowler C, Green B, Martinez D, Putnam H, Zhou S, Allen E, Apt E, Bechner M, Brzezinski M, Chaal B, Chiovitti A, Davis A, Demarest M, Detter J, Glavina T, Goodstein D, Hadi M, Hellsten U, Hildebrand M, Jenkins B, Jurka J, Kapitonov V, Kröger N, Lau W, Lane T, Larimer F, Lippmeier J, Lucas S, Medina M, Montsant A, Obornik M, Parker M, Palenik B, Pazour G, Richardson P, Rynearson T, Saito M, Schwartz D, Thamatrakoln K, Valentin K, Vardi A, Wilkerson F, Rokhsa D (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Asada K (1994) Production and action of active oxygen species in photosynthetic tissues. In: Foyer C, Mullineaux P (eds) Photooxidative Stresses in Plants: Causes and Amelioration. CRC Press, Boca Raton, pp 77–104
Asada K, Yoshikawa K, Takahashi M, Maeda Y, Enmanji K (1975) Superoxide dismutases from a blue-green alga, Plectonema boryanum. J Biol Chem 250:2801–7
Asada K, Kanematsu S, Uchida K (1977) Superoxide dismutases in photosynthetic organisms: absence of the cuprozinc enzyme in eukaryotic algae. Arch Biochem Biophys 179:243–256
Beauchamp C, Fridovich I (1971) Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287
Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91:179–94
Boussiba S (2000) Carotenogenesis in the green alga Haematococcus pluvialis: cellular physiology and stress response. Physiol Plant 108:111–117
Boussiba S, Wang B, Yuan JP, Zarka A, Chen F (1999) Changes in pigments profile in the green alga Haematococcus pluvialis exposed to environmental stresses. Biotechnol Lett 21:601–604
Bowler C, Camp W, Vanmontagu M, Inze D (1994) Superoxide-dismutase in plants. CRC Crit Rev Plant Sci 13:199–218
Buchanan BB, Luan S (2005) Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J Exp Bot 56:1439–1447
Church S (1990) Manganese superoxide dismutase: nucleotide and deduced amino acid sequence of a cDNA encoding a new human transcript. Biochim Biophys Acta 1087:250–252
Elstner EF, Heupel A (1976) Inhibition of nitrite formation from hydroxylammonium chloride: a simple assay for superoxide dismutase. Anal Biochem 70:616–620
Erturk HN (1999) Responses of superoxide dismutases to oxidative stress in Arabidopsis thaliana. In: Biology. Virginia Polytechnic Institute and State University, Blacksburg, VA, p 179
Fridovich I (1975) Superoxide dismutases. Annu Rev Biochem 44:147–159
Fridovich I (1986) Biological effects of the superoxide radical. Arch Biochem Biophys 247:1–11
Gabriel O (1971) Locating enzymes on gels. In: Colowick SP, Kaplan NO (eds) Methods in Enzymology, vol 22. Academic, New York, p 578
Geigenberger P, Kolbe A, Tiessn A (2005) Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56:1469–1479
Grun S, Lindermayr C, Sell S (2006) Nitric oxide and gene regulation in plants. J Exp Bot 57:507–516
Hagen C, Braune W, Vogel K, Hader DP (1993) Functional aspects of secondary carotenoids in Haematococcus lacustris (Girod) Rostafinski (Volvocales) V. Influences on photomovement. Plant Cell Environ 16:991–995
Ho Y, Howard A, Crapo J (1991) Molecular structure of a functional rat gene for manganese-containing superoxide dismutase. Am J Respir Cell Mol Biol 4:278–286
Kaminaka H, Morita S, Tokumoto M, Yokoyama H, Masumura T, Tanaka K (1999) Molecular cloning and characterization of a cDNA for an iron-superoxide dismutase in rice (Oryza sativa L.). Biosci Biotech Biochem 63:302–308
Kanematsu S, Asada K (1990) Characteristic amino acid sequences of chloroplast and cytosol isozymes of CuZn-superoxide dismutase in spinach, rice and horsetail. Plant Cell Physiol 31:99–112
Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9:627–640
Kitayama K (1994) Molecular analysis of photooxidative stress responses in Chlamydomonas reinhardtii. Thesis, Indiana University, Bloomington
Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118:637–650
Kobayashi M, Kakizono T, Nagai S (1991) Astaxanthin production by a green-alga, Haematococcus pluvialis accompanied with morphological-changes in acetate media. J Ferment Bioeng 71:335–339
Kobayashi M, Kakizono T, Nishio N, Nagai S, Kurimura Y, Tsuji Y (1997) Antioxidant role of astaxanthin in the green alga Haematococcus pluvialis. Appl Microbiol Biotechnol 48:351–356
Kwon SJ, Choi EY, Choi YJ (2006) Proteomics studies of post-translational modifications in plants. J Exp Bot 57:1547–1551
Li YT, Sommerfeld M, Chen F, Hu Q (2008) Consumption of oxygen by astaxanthin biosynthesis: a protective mechanism against oxidative stress in Haematococcus pluvialis (Chlorophyceae). J Plant Physiol 165:1783–1797
Lu F, Vonshak A, Zarka A, Boussiba S (1998) Does astaxanthin protect Haematococcus against light damage? Z Naturforsch 53c:93–100
Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
McCord JM, Fridovich I (1969) Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244:6049–6055
Mur LAJ, Tim L, Carver W (2006) No way to live: The various roles of nitric oxide in plant-pathogen interactions. J Exp Bot 57:489–505
Parker M, Blake C, Barra D, Bossa F, Schinina M, Bannister W, Bannister J (1987) Structural identity between the iron- and manganese-containing superoxide dismutases. Protein Eng 1:393–397
Perl-Treves R, Perl A (2002) Oxidative stress: an introduction. In: Van Montagu M, Inzé D (eds) Oxidative stress in plants. Taylor & Francis, London, pp 1–32
Sakamoto A, Nosaka Y, Tanaka K (1993) Cloning and sequencing analysis of a complementary DNA for manganese-superoxide dismutase from rice (Oryza sativa L.). Plant Physiol 103:1477–1478
Sakurai H, Kusumoto N, Kitayama K, Robert TK (1993) Isozymes of superoxide dismutase in Chlamydomonas and purification of one of the major isozymes containing Fe. Plant Cell Physiol 34:1133–1137
Shaish A, Avron M, Pick U, Ben-Amotz A (1993) Are active oxygen species involved in induction of β-carotene in Dunaliella bardawil? Planta 190:363–8
Steinbrenner J, Linden H (2001) Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol 125:810–817
Steinbrenner J, Linden H (2003) Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol Biol 52:343–56
Tsang EW, Bowler C, Hérouart D, Van Camp W, Villarroel R, Genetello C, Van Montagu M, Inzé D (1991) Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3:783–92
Ukeda H, Sarker AK, Kawana D, Sawamura M (1999) Flow-injection assay of superoxide dismutase based on the reduction of highly water-soluble tetrazolium. Anal Sci 15:353–357
Wang JX, Shi ZX, Xu XD (2004a) Residual plastids of bleached mutants of Euglena gracilis and their effects on the expression of nucleus-encoded genes. Prog Nat Sci 14:213–217
Wang JX, Sommerfeld M, Hu Q (2009) Occurrence and environmental stress responses of two plastid terminal oxidases in Haematococcus pluvialis (Chlorophyceae). Planta 230:191–203
Wang JX, Zhang XZ, Chen YS, Sommerfeld M, Hu Q (2008) Toxicity assessment of manufactured nanomaterials using the unicellular green alga Chlamydomonas reinhardtii. Chemosphere 73:1121–1128
Wang S, Chen F, Sommerfeld M, Hu Q (2004b) Proteomic analysis of molecular response to oxidative stress by the green alga Haematococcus pluvialis (Chlorophyceae). Planta 220:17–29
Wuerges J, Lee J, Yim Y, Yim H, Kang S, Carugo K (2004) Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc Natl Acad Sci USA 101:8569–8574
Youn HD, Kim E, Roe J, Hah Y, Kang S (1996) A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem J 318:889–896
Zhekisheva M, Boussiba S, Khozin-Goldberg I, Zarka A, Cohen Z (2002) Accumulation of oleic acid in Haematococcus pluvialis (Chlorophyceae) under nitrogen starvation or high light is correlated with that of astaxanthin esters. J Phycol 38:325–331
Zhu D, Scandalios J (1993) Maize mitochondrial manganese superoxide dismutases are encoded by a differentially expressed multigene family. Proc Natl Acad Sci USA 90:9310–9314
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, J., Sommerfeld, M. & Hu, Q. Cloning and expression of isoenzymes of superoxide dismutase in Haematococcus pluvialis (Chlorophyceae) under oxidative stress. J Appl Phycol 23, 995–1003 (2011). https://doi.org/10.1007/s10811-010-9631-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-010-9631-6