Abstract
The region of Qingdao, China, experienced the world's largest green tide from May to July 2008. More than one million tons of fresh algal biomass of the green alga Ulva prolifera was harvested, while more was suspected to have sunk to the bottom. The original source of this seaweed was suspected to be from the south as revealed by satellite images. The floating biomass drifted with the water current northward and flourished in nearshore waters around Qingdao. However, direct biological evidence for “seed” source is lacking. It is still unclear whether this alga could survive the Qingdao local coastal environment and pose future danger of potential blooming. Systematic and seasonal sampling of waters in the intertidal zone at six collection sites along the Qingdao coast was conducted from December 2008 to April 2009. Forty-eight water samples were analyzed. From these, nine different morphotypes of Ulva were grown in the laboratory under standard temperature and light regimes. Growth of Ulva was observed in all water samples. However, molecular phylogenetic analyses revealed that the dominant U. prolifera strain of the 2008 bloom was absent in all the water-derived cultures during the sampling period. These results provide evidence that the dominant bloom-forming alga was unlikely able to survive the coastal waters (the minimal surface water temperature in February is 2°C) in winter conditions in Qingdao, even though all the sampling locations were heavily covered by this alga in June 2008.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Green tides are vast accumulations of unattached green macroalgae associated with eutrophic marine environments (Shimada et al. 2003; Charlier et al. 2007). They have had major ecological and economic impacts (Hiraoka et al. 2003; Sun et al. 2008). In nutrient-rich habitats, Ulva species grow rapidly and are often among the dominant species causing green tides and marine fouling (Leskinen et al. 2004). In May to July 2008, before the Olympic sailing competition, the Qingdao coastline experienced what was believed to be the world's largest green tide (Liu et al. 2009), composed of more than one million tons of drifting biomass covering an area of 13,000 to 30,000 km2 (Sun et al. 2008; Leliaert et al. 2008). The dominant green alga, which was included in the Ulva linza–procera–prolifera complex (LPP clade; Shimada et al. 2008; Leliaert et al. 2009), was identified to be Ulva prolifera (Müller) J. Agardh (Chlorophyta, Ulvophyceae; formerly known as Enteromorpha prolifera) through morphological and molecular analyses (Sun et al. 2008; Ye et al. 2008; Leliaert et al. 2009). The drifting biomass was believed to have been transported across the Yellow Sea offshore from Jiangsu province by seasonal winds and surface currents (Liu et al. 2009). The large-scale occurrence of this alga in the Yellow Sea was thought to be related to a series of complex coupled processes (Sun et al. 2008). This large-scale inter-province green tide bloom in the Yellow Sea reminded people of coastal management issues and brought intense attention from the local and national governments because of the potentially damaging consequences on the environment and local economy.
Species in the genus Ulva are widely distributed in marine and brackish water environments all over the world (Hayden and Waaland 2002; Shimada et al. 2003; McAvoy and Klug 2005). Recently, molecular phylogenetic analyses and culture data provided convincing evidence that Ulva and Enteromorpha are not distinct evolutionary entities and should be recognized as one genus of the Ulvophyceae, Ulva (Tan et al. 1999; Hayden et al. 2003; Shimada et al. 2003). Ulva species are highly tolerant of variations in salinity, temperature, irradiance, and water quality (Taylor et al. 2001; Dan et al. 2002; Cohen and Fong 2006; Conley et al. 2009). They show great intraspecific morphological and cytological plasticity in growth phases with season and environmental conditions (Blomster et al. 1998, 1999). This high level of plasticity often leads to difficulties in species identification when only based on morphological and cytological features (Blomster et al. 2002; Leskinen et al. 2004). In view of these facts, molecular methods were introduced on the basis of sequence comparisons of the nuclear encoded internal transcribed spacer DNA (ITS nrDNA) region, including the 5.8S gene, the more conserved plastid encoded large subunit of the ribulose-1,5-bisphosphate carboxylase/oxgenase (rbcL) gene, and the 5S spacer region (Marks and Cummings 1996; Leskinen and Pamilo 1997; Blomster et al. 1998; Woolcott and King 1999; Shimada et al. 2003, 2008; Hayden and Waaland 2004).
Localization of the source of the green tide bloom and analyses of the outbreak potential are two prerequisites before any management decisions could be made (Blomster et al. 2002; Ye et al. 2008). Species of Ulva release microscopic gametes or spores which rapidly grow into adult thalli. These sporulation events were found to be accelerated at elevated light and temperature levels and can be facilitated by thallus fragmentation (Dan et al. 2002; Lüning et al. 2008). The microscopic stages of the germlings can be free-floating in seawater and grow into adult filamentous thalli when a more stagnant water environment is encountered (Dan et al. 2002; Shimada et al. 2008). These microscopic gametes, zygotes, spores, or germlings constitute the “seed bank” in nature for future blooms when favorable light, temperature, and pulse of nutrient enrichment occur (Hoffmann and Santelices 1991; Santelices et al. 1995; Zhang et al. 2009).
A very pertinent question after the large bloom in Qingdao in 2008 is whether this considerable biomass left behind germlings in the seawater along the Qingdao coastline, which could become the direct source of future blooms besides the annual arrival of drifting biomass from the south. The goal of this study was to survey whether the dominating algal strain of the 2008 bloom was still present in the coastal waters of Qingdao in the next winter and, if so, investigate the phylogenetic relationship of the bloom-forming strain with native isolates.
Materials and methods
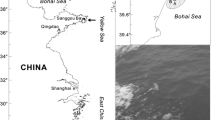
From December 2008 to April 2009, seawater was randomly sampled eight times at six locations (Ferry, Tuandao, Zhanqiao, Institute of Oceanology Chinese Academy of Sciences, Olympic, and Shilaoren) along the Qingdao coast where the green tide was observed in June 2008 (Fig. 1). Seawater was subsequently aerated after adding NO −3 (823 μmol L−1) and PO 3−4 (73 μmol L−1) to reach the levels of Provasoli-enriched seawater medium (PES; Berges et al. 2001) in 1 L glass beakers (two per sampling location) placed in temperature-controlled rooms at 15°C under 80 μmol photons m−2 s−1 with a 12-h light per day light-dark regime. Five hundred microliters of saturated GeO2 aqueous solution was added to the medium to prevent growth of diatoms, and the medium was renewed every 7 days. After 4 weeks, the green algal germlings, attaching to the wall and bottom of the glass beakers, grew up to 1–3 cm. These germlings were derived from microscopic spores, zygotes, gametes or those that were attached to microscopic particles invisible to the naked eye at the time of sampling. After counting, the algae were removed from the beakers and further grown in suspended culture under the same conditions until the morphology of individual plants could be analyzed (6–7 weeks). Twenty-six Ulva strains were preliminarily classified into nine groups according to their gross morphological characters including thallus morphology and the degree of branching (Table 1). Eight strains of Ulva spp. from the 2008 Qingdao green tide were also analyzed in this study.
DNA extraction, ITS nrDNA, and rbcL gene amplification and sequencing
The unialgal samples of 34 Ulva strains were washed three times with sterilized seawater and dried with filter paper. Then, 100 mg of each material was ground to fine powder in liquid nitrogen with a mortar and pestle and transferred to a 2-mL tube. DNA was extracted using the CTAB method (Wang et al. 2006). The concentration and the quality of isolated DNA were assessed by electrophoresis on 1.0% agarose gel. DNA concentration of each sample was adjusted to 50 ng μL−1. Primers sequences and polymerase chain reaction (PCR) amplification of ITS nrDNA and rbcL gene were described by Leskinen and Pamilo (1997) and Hayden et al. (2003). Total genomic DNA (30–40 ng) was added to 50 μL PCR reactions containing final concentrations of 1 × PCR buffer (Takara, Japan), 2 mM MgCl2 (Takara), 0.8 mM dNTPs (Takara), 25 μM of each primer, and 1.6 U Taq polymerase (Takara). Amplification products were separated by 1.0% agarose gel electrophoresis, and fragments of an expected length were cut from the gel and purified by use of a DNA gel extraction kit (Bio Basic Inc, Canada) according to the manufacturer's instructions. ITS nrDNA and rbcL gene were sequenced on both strands using ABI 3730 XL automated sequencers (Shanghai Biosune Biotechnology Co., Ltd.). Each sequencing reaction was repeated twice.
Phylogenetic analysis
ITS nrDNA and rbcL genes were sequenced from 34 Ulva strains representing 11 different morphotypes of Ulva spp. (Table 1). Sequences were aligned using Clustal W (Thompson et al. 1994). Evolutionary relationship was inferred with the neighbor-joining (NJ) and maximum likelihood (ML) methods using Mega 4.0 (Saitou and Nei 1987) and PhyML 3.0 (Guindon and Gascuel 2003), respectively. Ulvaria fusca and Umbraulva amamiensis served as outgroup taxa. For the NJ and ML trees, the bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980). All positions containing gaps and missing data were eliminated from the dataset (complete deletion option; Tamura et al. 2007). Thirty-five closely related ITS nrDNA sequences of Ulva spp. in the U. linza–procera–prolifera (LPP) clade were used to construct a parsimony network with TCS 1.21 (Clement et al. 2000). Final results are presented using gaps as fifth character states for bases in DNA sequences.
Relative growth rate and photosynthetic activity of Ulva linza and U. prolifera at different temperatures
For testing the growth performance of U. linza and U. prolifera, 6 × 0.1 g of healthy biomass of each species without epiphytic algae were weighed and cultured in separate 500-mL glass beakers at 5, 10, 15, 20, and 25°C under 100 μmol photons m−2 s−1 cool fluorescent white light in a 12-h light per day light-dark regime, in a GXZ-260C incubator (Ningbo Jiangnan Instrument, China). Temperature fluctuation was less than 0.5°C. PES medium was used and renewed every 2 days in the 7-day culture experiment. At the end of the experiment, the biomass and the maximal photochemical efficiency of PS II (F v/F m) were measured. F v/F m was determined using a Portable Chlorophyll Fluorometer (Mini PAM, Walz, Germany) by the method of Fleming et al. (2007). Before measurement, samples were dark adapted for 20 min. Optimal chlorophyll fluorescence quantum yield was calculated according to the following equation: \( {F_{\text{v}}}/{F_{\text{m}}} = \left( {{F_{\text{m}}} - {F_{\text{o}}}} \right)/{F_{\text{m}}} \). F o and F m refer to the minimal fluorescence and the maximal fluorescence from dark adapted samples, respectively. F v is the difference between F m and F o. The culture experiments were repeated four times.
Results
Quantitative and morphological analyses of the culture-derived algal samples
The total number of culturable Ulva individuals from 1 L surface seawater collected at the six nearshore locations increased over the study period (Fig. 2). Each time, the most algal individuals were found in the seawater from Zhanqiao, ranging from the lowest at 113 ± 24 strains L−1 on December 30, 2008 to the maximum at 1,840 ± 160 strains L−1 on April 17, 2009. Seawater from Ferry gave the lowest number of Ulva strains each time. According to gross morphological characters of Ulva spp. derived from the water samples, nine groups of Ulva were preliminarily classified (Table 1). The occurrence percentage of each group at each location and at each time was determined (Fig. 3). Ulva spp. in group 1 was the dominating species along the Qingdao coasts, representing more than 50% of total amount of Ulva strains at each location and at each sampling time, with the exception of Shilaoren. Based on morphological characters, Ulva spp. in group 6 (Ulva strains 6a, b, c, and d) and group 8 (Ulva strains 8a and b), both with branches, were the most similar to the dominating alga of the 2008 green tide.
Phylogenetic analysis and morphology comparisons
Alignment of ITS nrDNA and rbcL gene sequences included about 522 and 1,288 aligned characters, respectively. According to both phylogenetic trees based on ITS nrDNA and rbcL gene sequence, all 34 Ulva strains were resolved into five distinct clades: the U. linza–procera–prolifera (LPP) clade (19 samples), Ulva sp. (1–2) clade (six samples), Ulva rigida clade (three samples), Ulva compressa clade (three samples), and Ulva pertusa clade (three samples). The LPP clade included the strains of U. prolifera from the Qingdao bloom in 2008 (Ulva strains 2008a, b, c, d, e, f, and g), native Ulva strains (Ulva strains 1a, 1c, 3a, 4, 5, 6a, 6b, 6c, 6d, 7, 8a, and 8b), and Genbank sequences for U. linza, Ulva procera, and U. prolifera (Figs. 4 and 5). The LPP clade was highly supported in both ITS and rbcL phylogenies. The other 15 Ulva strains were included in the remaining four clades with high bootstrap values (96–100%) in NJ and ML analyses.
Phylogenetic tree constructed by neighbor-joining (NJ) method from the analysis of the nuclear encoded internal transcribed spacer DNA (ITS nrDNA) region, including the 5.8S gene, of Ulva species. The tree was rooted with Ulvaria fusca and Umbraulva amamiensis. Bootstrap values (percentage of 1,000 replicates) for neighbor-joining (above) and maximum likelihood (below) are indicated at nodes. The numbers under the branches represent full heuristic bootstrap values (1,000 replicates) greater than 50%. Branch lengths are proportional to the amount of sequence change, which are indicated by the scale bar below the tree
Phylogenetic tree constructed by neighbor-joining (NJ) method from the analysis of the rbcL gene sequences of Ulva species. The tree was rooted with Ulvaria fusca and Umbraulva amamiensis. Bootstrap values (percentage of 1,000 replicates) for neighbor-joining (above) and maximum likelihood (below) are indicated at nodes. The numbers under the branches represent full heuristic bootstrap values (1,000 replicates) greater than 50%. Branch lengths are proportional to the amount of sequence change, which are indicated by the scale bar below the tree
The ribotype network of ITS nrDNA from samples in the LPP clade showed that there were at least 14 distinct ITS ribotypes in this clade (Fig. 6). The ITS nrDNA sequences of Ulva strains 2008a, b, c, d, e, f, and g from 2008 green algal bloom were identical with U. linza (EU888138), U. prolifera (FJ426613), U. prolifera (AB298314), Ulva sp. HT7 (FJ194958), and U. prolifera (FJ026732). The dominant Ulva species forming the Qingdao bloom had at least two mutational steps with the native Ulva strains in the LPP clade, which were included in four ribotypes in the network (Fig. 6).
Relative growth rate and photosynthetic activity of U. linza and U. prolifera at different temperatures
At 5°C, both U. linza and U. prolifera grew slowly (t test, P > 0.05. Fig. 7a). When the temperature was raised to 10, 15, 20, and 25°C, U. prolifera grew faster than U. linza (two-way analysis of variance, species effect: P < 0.001, temperature effect: P < 0.001, species × temperature effect: P < 0.001). Both species had the highest growth rates between 10°C and 20°C. However, the photosynthetic activity of U. prolifera was significantly impacted at 5°C compared to that of U. linza (t test, P < 0.01. Fig. 7b). F v/F m of U. prolifera was reduced to 0.48 ± 0.08, while that of U. linza was at 0.61 ± 0.07, indicating that U. linza was better adapted to lower temperature than the stain of U. prolifera from the 2008 green tide in Qingdao. There was no significant difference in F v/F m between U. linza and U. prolifera at all the other temperatures tested (t test, P > 0.05).
Discussion
This study demonstrates the absence of the dominant U. prolifera strain from the 2008 bloom at all six locations near Qingdao from December 2008 to April 2009. This conclusion is based on the phylogenetic analyses, the ribotype network analyses in the LPP clade, and unique features of ramification. These results indicated that Ulva strains 2008a, b, c, d, e, f, and g collected from different areas around Qingdao during the bloom represented a unique algal strain with intensive ramifications, different from Ulva strains 1a, 1c, 3a, 4, 5, 6a, 6b, 6c, 6d, 7, 8a, and 8b. The latter belong to native Ulva species in Qingdao.
Ulva strains 2008 a, b, c, d, e, f, and g with dense branches, as the representative of the dominating Ulva strain in the 2008 green tide, were collected from different sites during the summer of 2008. The sequence divergence within the seven samples was between 0% and 0.2% according to the combined ITS nrDNA and rbcL sequences (data not given). The low sequence divergence was striking because of the large covering area of the green tide in the Qingdao region. These results confirm a previous study (Jiang et al. 2008) in which the sequence divergences among all free-floating samples collected along the Qingdao coastline in summer 2007 were between 0% and 2.5% based on the sequencing of the ITS nrDNA including the 5.8S gene. It was reported that data from molecular analyses based on ITS nrDNA showed a clade representing the U. linza–procera–prolifera (LPP) complex in the phylogenetic trees (Shimada et al. 2008; Leliaert et al. 2009). We also found that the ITS nrDNA and rbcL sequences from the strain of U. prolifera in the 2008 green tide were similar to that of Ulva strains 1a, 1c, 3a, 4, 5, 6a, 6b, 6c, 6d, 7, 8a, and 8b derived from seawater of Qingdao coasts, with just a few changes of a few bases in the ITS nrDNA sequences. However, a parsimony network analysis of the LPP clade revealed that there were at least two mutational steps between the dominating Ulva strain in the 2008 green tide and the native Ulva strains 1a, 1c, 3a, 4, 5, 6a, 6b, 6c, 6d, 7, 8a, and 8b. In this analysis, the position of the species forming the Qingdao bloom was fully consistent with that reported by Leliaert et al. (2009). Additionally, it is easy to distinguish them based on significant differences in the morphological characteristics under identical culture conditions (data not given). The exception was Ulva strain 2008h, with few branches and similar to U. compressa based on their gross morphology (Tseng et al. 1962). The molecular phylogenetic analysis showed that it was closer to Ulva strains 2b, 3b, and U. compressa (Figs. 4 and 5).
Species in the genus Ulva are famous for their wide tolerance of temperature, salinity, and nutrient levels (Tan et al. 1999; Dan et al. 2002; Cohen and Fong 2006; Conley et al. 2009). Thus, the absence of the dominant 2008 green tide alga in the following winter is surprising, considering that extremely large biomass which was present along the entire coast half a year before the sampling and that large number of gametes or spores were discharged into coastal waters (based on laboratory releasing experiments, data not shown). A logical explanation seems to be that the microscopic stages of the bloom-forming alga were not able to survive the winter conditions around Qingdao. Another research team found that the microscopic stages of the dominating 2008 green tide alga (thalli or fragments) were also not detected, as investigated by trawling, along the Qingdao coasts from November 2008 onwards (Z.L.Wang, personal communication). This conclusion is further strengthened by the fact that significantly reduced photosynthetic activity was found at 5°C in the bloom-forming U. prolifera strain, in comparison to a Qingdao native species, U. linza.
Ulva spp. in the coastal waters of Qingdao in the winter after the 2008 bloom
It has been reported that Ulva species are morphologically simple but exhibit considerable plasticity in response to environmental factors (Malta et al. 1999; Blomster et al. 1999; Hiraoka et al. 2004; Leskinen et al. 2004). Environmental conditions such as fluctuating salinity, light intensity, and temperature could cause the wide morphological variations and the differences in branching in some Ulva species (Blomster et al. 1998). Based on the above, in our experiments, the Ulva strains were cultured under the same condition to minimize the morphological changes caused by the environmental factors.
Under the described culture condition in this investigation, Ulva strains in groups 1 and 2 were unbranched, ribbon-like, and often had a frilled margin. However, based on molecular data, Ulva strains 1a, 1b, 1c, 1d, 1e, 2a, 2c, and 2d were included in two clades: strains 1a and 1c in the LPP clade and strains 1a, 1b, 1c, 1d, 1e, 2a, 2c, and 2d in the Ulva sp. (1–2) clade, which indicated that they might belong to two different Ulva species. However, further identification is needed. Meanwhile, Ulva strains 1a and 1c were identified as U. linza (data not given), a littoral and sublittoral species commonly found on rocks or in tide pools around Qingdao (Tseng et al. 1962). Ulva linza was found in surprisingly higher numbers in the seawater and rocks along the Qingdao coasts (Figs. 2 and 3), indicating that it could have the potential to form a local small-scale green tide. In a field survey in April and May 2009, U. linza was found to be the most common Ulva species along the Qingdao coasts and was often washed ashore and piling up at a small scale (personal observation). According to the phylogenetic trees, Ulva strains in group 9 could be separated into two species, Ulva strains 9a, 9d, 9e close to U. rigida and Ulva strains 9b, 9c, 9f close to U. pertusa. Further investigations are required on morphological features including cell arrangement, shape, pyrenoid number, etc.
Quantification of the microscopic stages of Ulva spp. in seawater
Field observation showed that the large-scale drifting Ulva biomass in the Yellow Sea in 2008 was gradually accumulating from small patches of drifting algae under the influence of winds and surface currents (Sun et al. 2008; Liu et al. 2009). How the sessile plants started to float and where these small patches of drifting biomass came from are two key issues to be addressed. One hypothesis is that the microscopic gametes or zygotes or spores attached to small particles in the water (high turbidity along the coast of Jiangsu province) and developed into a few cells stage. Adult thalli were derived from these microscopic stages in the water under favorable condition in early spring. However, quantification of culturable Ulva individuals at the microscopic stages (gametes, zygotes, spores, or microscopic germlings) has been difficult. For biomass forecasting estimation in green tide investigations, quantification of conspicuous adult plants only will underestimate the real biomass that could potentially be the entire bloom, since the microscopic plants could rapidly grow into adult plants when light, nutrients, and temperature levels reach the optimal combination (Hoffmann and Santelices 1991; Santelices et al. 1995; Worm et al. 2001; Zhang et al. 2009). In practice, quantifying the microscopic stages in the water through filtration at 0.45 or 0.22 μm is not reliable because Ulva gametes, zygotes, and spores often are confused with microalgal cells. It is also not possible to obtain enough live material for DNA analyses. In this investigation, large numbers of individuals were detected and determined from different sampling locations, indicating the ubiquitous existence of Ulva in seawater along the Qingdao coastline. This culture method will help to accurately delineate species structure of Ulva populations in the water and thus help understanding, and possibly forecasting, potential blooms in the investigated region.
References
Berges JA, Franklin DJ, Harrison PJ (2001) Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J Phycol 37:1138–1145
Blomster J, Maggs CA, Stanhope MJ (1998) Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. J Phycol 34:319–340
Blomster J, Maggs CA, Stanhope MJ (1999) Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed by molecular analysis. J Phycol 35:575–586
Blomster J, Bäck S, Fewer DP, Kiirikki M, Lehvo A, Maggs CA, Stanhope MJ (2002) Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. Am J Bot 89:1756–1763
Charlier RH, Morand P, Finkl CW, Thys A (2007) Green tides on the Brittany coasts. Environ Res Eng Manage 3:52–59
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Cohen RA, Fong P (2006) Using opportunistic green macroalgae as indicators of nitrogen supply and sources to estuaries. Ecol Appl 16:1405–1420
Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, Lancelot C, Likens GE (2009) Controlling eutrophication: nitrogen and phosphorus. Science 323:1014–1015
Dan A, Hiraoka M, Ohno M, Critchley AT (2002) Observations on the effect of salinity and photon fluence rate on the induction of sporulation and rhizoid formation in the green alga Enteromorpha prolifera (Müller) J.Agardh (Chlorophyta, Ulvales). Fish Sci 68:1182–1188
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fleming ED, Bebout BM, Castenholz RW (2007) Effects of salinity and light intensity on the resumption of photosynthesis in rehydrated cyanobacterial mats from Baja California Sur, Mexico. J Phycol 43:15–24
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hayden HS, Waaland JR (2002) Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. J Phycol 38:1200–1212
Hayden HS, Waaland JR (2004) A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia 43:364–382
Hayden HS, Blomster J, Maggs CA, Silva PC, Stanhope MJ, Waaland JR (2003) Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur J Phycol 38:277–294
Hiraoka M, Shimada S, Uenosono M, Masuda M (2003) A new green-tide-forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan. Phycol Res 51:17–29
Hiraoka M, Ohno M, Kawaguchi S, Yoshida G (2004) Crossing test among floating Ulva thalli forming ‘green tide’ in Japan. Hydrobiologia 512:239–245
Hoffmann AJ, Santelices B (1991) Banks of algal microscopic forms: hypotheses on their functioning and comparisons with seed banks. Mar Ecol Prog Ser 79:185–194
Jiang P, Wang JF, Cui YL, Li YX, Lin HZ, Qin S (2008) Molecular phylogenetic analysis of attached Ulvaceae species and free-floating Enteromorpha from Qingdao coasts in 2007. Chin J Oceanol Limnol 26:276–279
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Leliaert F, Malta EJ, Engelen AH, Mineur F, De Clerck O (2008) Quindao algal bloom culprit identified. Mar Pollut Bull 56:1515–1516
Leliaert F, Zhang X, Ye N, Malta EJ, Engelen AH, Mineur F, Verbruggen H, De Clerck O (2009) Identity of the Qingdao algal bloom. Phycol Res 57:147–151
Leskinen E, Pamilo P (1997) Evolution of the ITS sequences of ribosomal DNA in Enteromorpha (Chlorophyceae). Hereditas 126:17–23
Leskinen E, Alstrőm-Rapaport C, Pamilo P (2004) Phylogeographical structure, distribution and genetic variation of the green algae Ulva intestinalis and U. compressa (Chlorophyta) in the Baltic Sea area. Mol Ecol 13:2257–2265
Liu DY, Keesing JK, Xing QG, Shi P (2009) World's largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Mar Pollut Bull. doi:10.1016/j.marpolbul.2009.01.013
Lüning K, Kadel P, Pang SJ (2008) Control of reproduction rhythmicity by environmental and endogenous signals in Ulva pseudocurvata (Chlorophyta). J Phycol 44:866–873
Malta E, Draisma SGA, Kamermans P (1999) Free-floating Ulva in the southwest Netherlands: species or morphotypes? A morphological, molecular and ecological comparison. Eur J Phycol 34:443–454
Marks JC, Cummings MP (1996) DNA sequence variation in the ribosomal internal transcribed spacer region of freshwater Cladophora species (Chlorophyta). J Phycol 32:1035–1042
McAvoy KM, Klug JL (2005) Positive and negative effects of riverine input on the estuarine green alga Ulva intestinalis (syn Enteromorpha intestinalis) (Linneaus). Hydrobiologia 545:1–9
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Santelices B, Hoffmann AJ, Aedo D, Bobadilla M, Otaíza R (1995) A bank of microscopic forms on disturbed boulders and stones in tide pools. Mar Ecol Prog Ser 129:215–228
Shimada S, Hiraoka M, Nabata S, Lima M, Masuda M (2003) Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycol Res 51:99–108
Shimada S, Yokoyama N, Arai A, Hiraoka M (2008) Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J Appl Phycol 20:979–989
Sun S, Wang F, Li CL, Qin S, Zhou MJ, Ding LP, Pang SJ, Duan DL, Wang GC, Yin BS, Yu RC, Jiang P, Liu ZL, Zhang GT, Fei XG, Zhou M (2008) Emerging challenges: massive green algae blooms in the Yellow Sea. Nature Precedings. hdl:10101/npre.2008.2266.1
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan IH, Blomster J, Hansen G, Leskinen E, Maggs CA, Mann DG, Sluiman HJ, Stanhope MJ (1999) Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Mol Biol Evol 16:1011–1018
Taylor R, Fletcher RL, Raven JA (2001) Preliminary studies on the growth of selected “green tide” algae in laboratory culture: effects of irradiance, temperature, salinity and nutrients on growth rate. Bot Mar 44:327–336
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tseng CK, Zhang DR, Zhang JF, Xia EZ, Xia BM, Dong ML, Yang ZD (1962) Economic seaweeds of China. Science Press, Beijing, pp 41–50 (in Chinese)
Wang D, Wang XL, Li DP, Wang FJ, Duan DL (2006) The genetic analysis and germplasm identification of the gametophytes of Undaria pinnatifida (Phaeophyceae) with RAPD method. J Appl Phycol 18:801–809
Woolcott GW, King RJ (1999) Ulva and Enteromorpha (Ulvales, Ulvophyceae, Chlorophyta) in Eastern Australia: comparisons of morphological features and analyses of nuclear rDNA sequence data. Aust J Bot 12:709–725
Worm B, Lotze HK, Sommer U (2001) Algal propagule banks modify competition, consumer and resource control on Baltic rocky shores. Oecologia 128:281–293
Ye NH, Zhuang ZM, Jin XS, Wang QY, Zhang XW, Li DM, Wang HX, Mao YZ, Jiang ZJ, Li B, Xue ZX (2008) China is on the track tackling Enteromorpha spp. forming green tide. Nature Precedings. hdl:10101/npre.2008.2352.1
Zhang X, Wang H, Mao Y, Liang C, Zhuang Z, Wang Q, Ye N (2009) Somatic cells serve as a potential propagule bank of Enteromorpha prolifera forming a green tide in the Yellow Sea, China. J Appl Phycol. doi:10.1007/s10811-009-9437-6
Acknowledgements
The authors thank Chun Hua Song for her technical assistance in maintaining the algal culture; Yu Rong Zhang for her technical assistance in the collection of water samples; Xian Liang Meng, Wei Lu, Jin Feng Wang, and Xin Yue for their technical assistance in data analyses; Yao Li for his assistance in editing maps; and Shanghai Biosune Biotechnology Co., Ltd for their technical assistance in sequencing. We also would like to thank two reviewers for their valuable comments to improve this article. Financial support came from Scientific and Technical Supporting Programs of China (2008BAC49B01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, F., Pang, S.J., Chopin, T. et al. The dominant Ulva strain of the 2008 green algal bloom in the Yellow Sea was not detected in the coastal waters of Qingdao in the following winter. J Appl Phycol 22, 531–540 (2010). https://doi.org/10.1007/s10811-009-9489-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-009-9489-7