Abstract
Paeoniflorin is an active ingredient derived from Paeonia, which has an anti-inflammatory effect. However, the potential role and basis of paeoniflorin in rheumatoid arthritis (RA) are indistinct. Cell viability, cycle distribution, migration, and invasion were evaluated via Cell Counting Kit-8 (CCK-8), flow cytometry, and transwell assays. The contents of inflammatory cytokines were examined using enzyme-linked immunosorbent assay (ELISA). RNA expression levels were determined via qRT-PCR and western blot. The targeting relationship between miR-671-5p and circ-FAM120A (hsa_circ_0003972) or murine double minute 4 (MDM4) was validated via dual-luciferase reporter assay. Paeoniflorin restrained proliferation, migration, invasion, and inflammation and accelerated cell cycle arrest in RA fibroblast–like synoviocytes (RA-FLSs). Circ-FAM120A was boosted in RA synovial tissues and RA-FLSs. Circ-FAM120A upregulation, miR-671-5p knockdown, or MDM4 augmentation reversed the repressive effect of paeoniflorin on RA-FLS progression. Moreover, paeoniflorin attenuated RA-FLS progression by regulating the circ-FAM120A/miR-671-5p/MDM4 axis. Paeoniflorin inhibited RA-FLS proliferation, mobility, and inflammation and triggered cell cycle arrest via mediating the circ-FAM120A/miR-671-5p/MDM4 pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Rheumatoid arthritis (RA) is a common chronic autoimmune disease characterized by joint inflammation, as well as the destruction of articular cartilage and bone [1]. The incidence of RA is about 0.5%, which can lead to increased disability and mortality [2]. Fibroblast-like synoviocytes (FLSs) are strongly related to the pathogenesis of RA, including the production of pathogenic factors [3]. FLSs, as non-immune cells, cause joint damage through multiple mechanisms [4]. Therefore, seeking new drugs targeting FLSs is essential for RA treatment.
Paeoniflorin is the primary active ingredient extracted from Paeonia, which has anti-inflammatory and immunomodulatory effects [5]. The mechanism of paeoniflorin in inflammation and immunity is to inhibit the activation of immune cells and synovial cells, thereby regulating the release of inflammatory cytokines and abnormal signal transduction of synovial cells [6]. In a collagen-induced RA rat model, paeoniflorin inhibits RA progression by reducing the release of pro-inflammatory factors [7]. Despite this, the molecular mechanism of paeoniflorin in RA still needs further elucidation.
Accumulating evidence has corroborated that circular RNAs (circRNAs) are implicated in various biological functions and exert essential roles in diverse diseases [8]. Besides, recent studies have manifested that circRNAs participate in regulating autoimmune diseases, including systemic lupus erythematosus (SLE), dermatomyositis, and RA [9]. According to RNA sequencing in peripheral blood mononuclear cells (PBMCs) from RA, many circRNAs aberrantly expressed in RA may be diagnostic biomarkers for RA [10]. In addition, Wen et al. revealed that circ-FAM120A (hsa_circ_0003972) derived from family with sequence similarity 120A (FAM120A) was overtly upregulated in RA patients through RNA Sequencing and RT-qPCR [11]. Nonetheless, the potential function and basis of circ-FAM120A in RA remain unknown.
Extensive literatures have confirmed that circRNAs serve as microRNA (miRNA) sponges to compete for miRNA binding sites, thereby indirectly modulating miRNA targets [12]. Furthermore, a large number of miRNAs dysregulated in RA are intimately related to RA occurrence and development [13]. For example, miR-22 suppressed the growth and inflammation of synovial fibroblasts from RA by inhibiting SIRT1 expression [14]. Additionally, augmentation of miR-410-3p restrained FLS proliferation and facilitated apoptosis in RA via repressing YY1 [15]. In this research, bioinformatics analysis discovered that circ-FAM120A might have a targeting relationship with miR-671-5p. Nonetheless, the association between circ-FAM120A and miR-671-5p remains unknown.
Hence, we confirmed the anti-proliferative and anti-inflammatory effects of paeoniflorin in RA-FLSs. Further, we illuminated the association between paeoniflorin and circ-FAM120A/miR-671-5p/MDM4 axis in RA-FLS progression, which might provide new biomarkers for RA treatment.
MATERIALS AND METHODS
Clinical Samples
RA synovial tissues (n = 23) were collected from RA patients undergoing knee synovectomy or knee replacement surgery. Healthy synovial tissues (n = 21) were obtained from volunteers with traumatic knee condition. Among them, volunteers as normal controls had no history of joint abnormalities or systemic diseases. All patients were recruited from Henan Province Hospital of Traditional Chinese Medicine. All participants signed a written informed consent. This research was endorsed by the Ethics Committee of Henan Province Hospital of Traditional Chinese Medicine. RA synovial tissues and normal synovial tissues were stored at −80 °C for subsequent study.
Cell Culture and Paeoniflorin Treatment
Human normal FLSs and RA-FLSs were commercially obtained from Shanghai and Shanghai Zhen Biotechnology Co., Ltd. (Shanghai, China). All cells were maintained in Dulbecco’s modified Eagle medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone) at 37 °C with 5% CO2. For paeoniflorin treatment, RA-FLSs were exposed to various doses (0, 25, 50 and 100 µM) of paeoniflorin (Sigma-Aldrich, St Louis, MO, USA) for 48 h.
Cell Transfection
Circ-FAM120A overexpression vector (circ-FAM120A), miR-671-5p mimics (miR-671-5p), circ-FAM120A or MDM4 siRNA (si-circ-FAM120A or si-MDM4), miR-671-5p inhibitor (anti-miR-671-5p), MDM4 overexpression vector (MDM4), and negative controls (pCD5-ciR, miR-NC, si-NC, anti-miR-NC, and pcDNA) were purchased from GeneChem (Shanghai, China). These vectors and oligonucleotides were introduced into RA-FLSs via Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Cell Viability Assay
The treated RA-FLSs (1 × 104) were transferred to 96-well plates and cultured for 24 h. Afterwards, DMEM medium containing 10% Cell Counting Kit-8 (CCK-8) reagent (Solarbio, Beijing, China) was injected into each well. After reaction for 4 h, the absorbance was examined via a microplate reader (BioTek, Burlington, VT, USA).
Flow Cytometry
The treated RA-FLSs were trypsinized, and the pellets were rinsed in phosphate-buffered saline (PBS; Solarbio). Next, cells were reacted with 80% ethanol and washed in PBS. Subsequently, the pellets were exposed to RNase A (Solarbio). Afterwards, cells were incubated with propidium iodide (Sigma-Aldrich). At last, cell cycle distribution was monitored via flow cytometry (BD Biosciences, San Diego, CA, USA).
Transwell Assay
Cell migration and invasion abilities were assessed via transwell chambers (Corning, Corning, NY, USA) pre-coated without or with Matrigel. The treated RA-FLSs were injected into the upper chamber. In the meantime, the lower chamber was filled with 10% FBS. Then, RA-FLSs were treated with crystal violet. At last, the migrated and invaded cells were calculated under a microscope at 100 × magnification.
Enzyme-Linked Immunosorbent Assay
The concentrations of interleukin 1 beta (IL-1β), interleukin-6 (IL-6), and interleukin-8 (IL-8) in cell culture medium were examined using ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the supplier’s requirements.
Quantitative Real-Time PCR
RNA was extracted with TRIzol reagent (Leagene, Beijing, China). Next, cDNA was obtained using the specific RT-PCR kit (Takara, Dalian, China). For circular determination, 2 μg of RNA was treated with RNase R (Epicentre, Madison, WI, USA). For circRNA localization assay, the nucleus and cytoplasm of RA-FLSs were separated with Nuclear/Cytosol Fractionation Kit (BioVision, Palo Alto, CA, USA). Subsequently, RNA levels were measured by SYBR Green PCR Master Mix (Takara) and counted via the 2−ΔΔCt method. The primers included circ-FAM120A-F: 5′-CCACCACATTACTTAGGAAATCACA-3′, circ-FAM120A-R: 5′-GGCTTTGATCACTACGTCGC-3′; FAM120A-F: 5′-GGCGGAGTCCAACCTATACC-3′, FAM120A-R: 5′-CTGGTCCTCCGACAGAAACC-3′; miR-671-5p-F: 5′-GCCCGCAGGAAGCCCUGGAGGGGC-3′, miR-671-5p-R: 5′-GTGCAGGGTCCGAGGT3-3′; MDM4-F: 5′-CAGCAGGTGCGCAAGGTGAA-3′, MDM4-R: 5′-CTGTGCGAGAGCGAGAGTCTG-3’; GAPDH-F: 5’-GGGAAACTGTGGCGTGAT-3′, GAPDH-R: 5′-GAGTGGGTGTCGCTGTTGA-3′; U6-F: 5′-CTCGCTTCGGCAGCACA-3′, U6-R: 5′-AACGCTTCACGAATTTGCGT-3′. GAPDH or U6 (miR-671-5p) was regarded as an internal control.
Dual-luciferase Reporter Assay
Circ-FAM120A sequence harboring miR-671-5p wild-type or mutant binding site was inserted into the pmirGLO vector (LMAI Bio, Shanghai, China) to form the WT-circ-FAM120A or MUT-circ-FAM120A reporter. Simultaneously, MDM4 3′UTR containing the miR-671-5p wild-type or mutant binding site was inserted into the pmirGLO vector (LMAI Bio) to construct the MDM4 3′UTR-WT or MDM4 3′UTR-MUT reporter. Next, the constructed reporter was introduced into RA-FLSs with miR-671-5p or miR-NC. The luciferase ability was measured via Dual-Lucy Assay Kit (Solarbio).
Western Blot Assay
Protein samples were extracted with RIPA buffer (Solarbio) and quantified via the BCA method. Afterwards, the protein was separated by 10% SDS-PAGE and transferred to PVDF membranes (Corning). Next, the membranes were incubated with 5% non-fat milk, followed by primary antibodies against MDM4 (1:1000, ab16058, Abcam, Cambridge, UK), p53 (1:500, ab131442, Abcam) or GAPDH (1:2500, ab9485, Abcam) overnight at 4 °C. Subsequently, the membranes interacted with a secondary antibody (1:25,000, ab205718, Abcam). The protein signal was examined using an ECL system (Beyotime, Shanghai, China).
Statistical Analysis
All experimental data were displayed as mean ± standard deviation via GraphPad Prism 7.0 software (GraphPad, San Diego, CA, USA). Student’s t-test and one-way analysis of variance were employed to analyze the differences. The association among circ-FAM120A, miR-671-5p, and MDM4 was tested using Spearman’s correlation coefficient. When P < 0.05, it was considered statistically significant.
RESULTS
Paeoniflorin Inhibits Proliferation, Migration, Invasion, and Inflammation and Induces Cell Cycle Arrest in RA-FLSs
To ascertain the role of paeoniflorin in RA progression, RA-FLSs were stimulated with different doses of paeoniflorin (0, 25, 50, and 100 µM) for 48 h. The CCK-8 method illustrated that paeoniflorin suppressed the viability of RA-FLSs dose-dependently (Fig. 1A). Therefore, 50 µM and 48 h were selected as treatment conditions for paeoniflorin in the following research. Next, flow cytometry showed that paeoniflorin increased the percentage of cells in the G0/G1 phase and decreased the percentage of cells in the S phase, indicating that paeoniflorin promoted cell cycle arrest in RA-FLSs (Fig. 1B). Transwell revealed that paeoniflorin suppressed the migration and invasion of RA-FLSs (Fig. 1C–F). In addition, IL-1β, IL-6, and IL-8 levels were strikingly reduced after paeoniflorin treatment, suggesting that paeoniflorin hindered inflammation in RA-FLSs (Fig. 1G). Taken together, paeoniflorin hindered the proliferation, migration, invasion, and inflammation of RA-FLSs and triggered cell cycle arrest.
Effects of paeoniflorin on proliferation, cell cycle arrest, migration, invasion, and inflammation in RA-FLSs. A RA-FLSs were stimulated with different doses of paeoniflorin (0, 25, 50, and 100 µM) for 48 h, and cell viability was determined by using CCK-8 analysis. B Cell cycle distribution was evaluated by flow cytometry in RA-FLSs treated with paeoniflorin (50 µM) for 48 h. C–F After RA-FLSs were treated with paeoniflorin (50 µM) for 24 h, the migration and invasion of RA-FLSs were measured by transwell analysis. G The concentration of IL-1β, IL-6, and IL-8 was examined by ELISA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Circ-FAM120A Is Upregulated in RA and Promotes the Development of RA
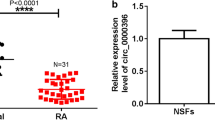
Next, we detected the expression trend of circ-FAM120A in RA patients. As exhibited in Fig. 2A, the circ-FAM120A level in synovial tissues from RA patients was remarkably increased relative to healthy synovial tissues. Similarly, the circ-FAM120A level in RA-FLSs was markedly higher than that in FLSs (Fig. 2B). Besides, circ-FAM120A was not affected by RNase R, indicating that circ-FAM120A was highly stable in RA-FLSs (Fig. 2C). Moreover, circ-FAM120A was principally distributed in the cytoplasm (Fig. 2D). Thus, circ-FAM120A is a novel circRNA and is upregulated in RA.
circ-FAM120A is upregulated in RA synovial tissues and RA-FLSs. A The expression of circ-FAM120A in synovial tissues from RA patients (n = 23) and normal control (n = 21) was detected by RT-qPCR. B Circ-FAM120A level was examined in RA-FLSs and the control FLSs. C The levels of circ-FAM120A and linear FAM120A were measured by RT-qPCR after treatment with RNase R. D The expression level of circ-FAM120A in cytoplasm and nucleus was detected using RT-qPCR. ***P < 0.001, ****P < 0.0001.
Next, we further investigated the function of circ-FAM120A in RA. RA-FLSs were transfected with si-circ-FAM120A, and the results indicated that circ-FAM120A was suppressed by half in RA-FLSs (Fig. S1A). Besides, circ-FAM120A knockdown suppressed cell proliferation, but induced cell cycle arrest in contrast with the si-NC group (Fig. S1B and S1C). Transwell assay indicated that the migration and invasion abilities of RA-FLSs were significantly suppressed by circ-FAM120A knockdown (Fig. S1D and S1E). In addition, circ-FAM120A knockdown also reduced inflammation in RA-FLSs, as identified by the decreased levels of pro-inflammatory cytokines (IL-1β, IL-6, and IL-8) in the si-circ-FAM120A group (Fig. S1F). Thus, circ-FAM120A promotes the development of RA.
Overexpression of Circ-FAM120A Inhibits the Role of Paeoniflorin in RA-FLSs
To explore whether circ-FAM120A is regulated by paeoniflorin in RA, RA-FLSs were introduced with pCD5-ciR or circ-FAM120A before paeoniflorin administration. Firstly, paeoniflorin reduced the expression level of circ-FAM120A dose-dependently (Fig. 3A). As shown in Fig. 3B, circ-FAM120A transfection restored the decrease of the circ-FAM120A level caused by paeoniflorin stimulation. Moreover, paeoniflorin suppressed cell proliferation and induced cell cycle arrest in RA-FLSs, while these impacts were abolished by upregulating circ-FAM120A (Fig. 3C and D). Additionally, transwell assay and ELISA suggested that paeoniflorin stimulation restrained the migration, invasion, and inflammatory response of RA-FLSs, whereas introduction of circ-FAM120A alleviated these effects (Fig. 3E–G). Overall, these data evidenced that paeoniflorin suppressed RA-FLS growth via decreasing circ-FAM120A.
Overexpression of circ-FAM120A inhibits the role of paeoniflorin in RA-FLSs. A RA-FLSs were treated with various concentrations of paeoniflorin for 48 h, and circ-FAM120A level was examined by RT-qPCR. After RA-FLSs transfected with pCD5-ciR or circ-FAM120A were exposed to 50 µM paeoniflorin, appropriate methods were used to detect circ-FAM120A expression (B), cell viability (C), cell cycle distribution (D), cell migration (E), invasion (F), and inflammation (G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Circ-FAM120A Directly Interacts with miR-671-5p
The prediction software CircInteractome revealed that the putative binding site of miR-671-5p was located in the circ-FAM120A sequence (Fig. 4A). Dual-luciferase reporter assay showed that miR-671-5p upregulation prominently decreased the luciferase activity of WT-circ-FAM120A, but had no effect on MUT-circ-FAM120A (Fig. 4B). In addition, miR-671-5p levels in RA synovial tissues and RA-FLSs were overtly reduced relative to the control group (Fig. 4C and D). Spearman’s correlation analysis exhibited that circ-FAM120A and miR-671-5p in RA synovial tissues were negatively correlated (Fig. 4E). Subsequently, the knockdown efficiency and overexpression efficiency of circ-FAM120A were determined by RT-qPCR analysis (Fig. 4F). Furthermore, silencing of circ-FAM120A accelerated miR-671-5p expression, and augmentation of circ-FAM120A suppressed miR-671-5p expression (Fig. 4G). These data evidenced that circ-FAM120A was a decoy of miR-671-5p in RA-FLSs.
circ-FAM120A directly interacts with miR-671-5p. A The online database CircInteractome displayed the binding site between circ-FAM120A and miR-671-5p. B Luciferase activity was tested in RA-FLSs co-transfected with WT-circ-FAM120A or MUT-circ-FAM120A and miR-NC or miR-671-5p. C MiR-671-5p level was detected in RA patients (n = 23) and non-RA patients (n = 21). D MiR-671-5p expression was measured in RA-FLSs and the control FLSs. E Spearman’s correlation coefficient was used to assess the relationship between circ-FAM120A and miR-671-5p. F, G The levels of circ-FAM120A and miR-671-5p were examined in RA-FLSs transfected with si-NC, si-circ-FAM120A, pCD5-ciR, or circ-FAM120A. **P < 0.01, ****P < 0.0001.
Inhibition of miR-671-5p Abolishes the Effect of Paeoniflorin on the Progression of RA-FLSs
To elucidate the effect of miR-671-5p on RA development, RA-FLSs transfected with anti-miR-NC or anti-miR-671-5p were exposed to 50 µM paeoniflorin. First of all, paeoniflorin treatment elevated the miR-671-5p level in a concentration-dependent manner (Fig. 5A). Moreover, anti-miR-671-5p transfection undermined the increase of the miR-671-5p level mediated by paeoniflorin administration (Fig. 5B). As illustrated in Fig. 5C, miR-671-5p depletion partially abolished the suppressive effect of paeoniflorin treatment on RA-FLS proliferation. In addition, miR-671-5p downregulation partially reversed the inductive effect of paeoniflorin on cell cycle arrest (Fig. 5D). Furthermore, downregulation of miR-671-5p alleviated the repressive effect of paeoniflorin on the migration and invasion of RA-FLSs (Fig. 5E and F). Also, paeoniflorin stimulation led to a significant decrease in IL-1β, IL-6, and IL-8 levels, while miR-671-5p depletion partially abolished the effect of paeoniflorin on inflammation (Fig. 5G). Collectively, these data evidenced that paeoniflorin restrained RA-FLSs progression via elevating miR-671-5p.
Inhibition of miR-671-5p abolishes the effect of paeoniflorin on the progression of RA-FLSs. A The expression of miR-671-5p was measured in RA-FLSs stimulated with different doses of paeoniflorin. MiR-671-5p level (B), cell viability (C), cell cycle distribution (D), cell migration (E), invasion (F), and inflammation (G) were examined in RA-FLSs transfected with anti-miR-NC or anti-miR-671-5p following 50 µM paeoniflorin treatment. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
MiR-671-5p Directly Targets MDM4
TargentScan online database suggested that miR-671-5p had multiple target mRNAs. Among these mRNAs, three mRNAs (TLR4, IGF-1R, and MDM4) are of concern. TLR4 expression is elevated in chondrocytes, osteoblasts, and synoviocytes, and has been confirmed to be a promising therapeutic target for RA [16]. IGF-1R is activated in RA, and the activation of IGF-1R signaling contributes to T cell–dependent inflammation in rheumatoid arthritis [17]. MDM4 is highly expressed in OA, and was reported to be closely related to the hyperplasia phenotype in RA [18]. In the present research, we found that MDM4 was the most downregulated mRNA in RA-FLSs with miR-671-5p mimic transfection (Fig. S2). Hence, MDM4 was chosen for further research. As displayed in Fig. 6A, MDM4 3′UTR contained the possible miR-671-5p binding site. Next, dual-luciferase reporter assay showed that co-transfection of miR-671-5p and MDM4 3′UTR-WT resulted in a marked decrease in luciferase activity (Fig. 6B). Compared with the normal group, MDM4 mRNA and protein levels in RA synovial tissues were conspicuously increased (Fig. 6 C and D). Simultaneously, the MDM4 protein level in RA-FLSs was overtly elevated compared to the control group (Fig. 6E). Besides, the miR-671-5p level in RA synovial tissues was inversely correlated with MDM4 level (Fig. 6F). Then, RT-qPCR assay confirmed that miR-671-5p overexpression and inhibition efficiency were significant (Fig. 6G). In the meantime, miR-671-5p upregulation markedly reduced the MDM4 expression and knockdown of miR-671-5p remarkably increased the MDM4 expression (Fig. 6H). Taken together, miR-671-5p directly targeted MDM4 in RA-FLSs.
MiR-671-5p directly targets MDM4. A The predicted miR-671-5p binding site in MDM4 3′UTR was shown. B Luciferase activity was detected in RA-FLSs introduced with MDM4 3′UTR-WT or MDM4 3′UTR-MUT and miR-NC or miR-671-5p. C, D The mRNA and protein levels of MDM4 were measured in RA patients (n = 23) and non-RA patients (n = 21). E MDM4 protein level was determined in RA-FLSs and the control FLSs. F The correlation between miR-671-5p and MDM4 was evaluated by Spearman’s correlation coefficient. After RA-FLSs were transfected with miR-NC, miR-671-5p, anti-miR-NC, or anti-miR-671-5p, miR-671-5p expression (G) and MDM4 protein expression (H) were examined by RT-qPCR and western blot, respectively. **P < 0.01, ***P < 0.001, ****P < 0.0001.
MDM4 Overexpression Reverses the Role of Paeoniflorin in RA-FLSs
Next, to investigate the biological function of MDM4 in RA progression, RA-FLSs were introduced with pcDNA or MDM4 and then stimulated with 50 µM paeoniflorin. As depicted in Fig. 7A, paeoniflorin suppressed the MDM4 expression dose-dependently. In addition, introduction of MDM4 restored the decrease of the MDM4 level caused by paeoniflorin treatment (Fig. 7B). Besides, western blot showed that suppression of MDM4 promoted p53 expression (Fig. S3). Furthermore, overexpression of MDM4 mitigated the repressive effects of paeoniflorin on cell proliferation, migration, invasion, and inflammation, and the inductive effect of paeoniflorin on cell cycle arrest (Fig. 7C–G).
MDM4 overexpression reverses the role of paeoniflorin in RA-FLSs. A MDM4 protein expression was measured in RA-FLSs treated with different doses of paeoniflorin. After RA-FLSs were introduced with pcDNA or MDM4 and exposed to 50 µM paeoniflorin, MDM4 protein level (B), cell viability (C), cell cycle distribution (D), cell migration (E), invasion (F), and inflammation (G) were detected using the appropriate methods. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Paeoniflorin Regulates RA Development via the Circ-FAM120A/miR-671-5p/MDM4 Axis
To illuminate the association between paeoniflorin and the circ-FAM120A/miR-671-5p/MDM4 axis, MDM4 protein expression was measured in RA-FLSs treated with different combinations. As displayed in Fig. 8A, si-circ-FAM120A transfection markedly decreased MDM4 protein level, while this change was abolished via repressing miR-671-5p. Additionally, overexpression of circ-FAM120A abrogated the reduction of MDM4 protein level mediated by paeoniflorin administration (Fig. 8B). Similarly, depletion of miR-671-5p alleviated the suppression of paeoniflorin on MDM4 protein expression (Fig. 8C). These results hinted that paeoniflorin ameliorated RA via regulating the circ-FAM120A/miR-671-5p/MDM4 axis.
Paeoniflorin regulates RA development via circ-FAM120A/miR-671-5p/MDM4 axis. A MDM4 protein level was detected in RA-FLSs transfected with si-NC, si-circ-FAM120A, si-circ-FAM120A + anti-miR-NC, or si-circ-FAM120A + anti-miR-671-5p. B, C RA-FLSs transfected with pCD5-ciR, circ-FAM120A, anti-miR-NC, or anti-miR-671-5p were exposed to 50 µM paeoniflorin for 48 h, and MDM4 protein expression was examined using western blot. **P < 0.01, ***P < 0.001, ****P < 0.0001.
DISCUSSION
RA is characterized by the activation of FLSs, leading to synovial inflammation and joint destruction [19]. In RA progression, FLSs possess tumor-like phenotypes, including increased aggressiveness and reduced apoptosis [20]. IL-1β, IL-6, and IL-8 are critical pro-inflammatory cytokines and contribute to the activation of RA-FLSs [21]. Besides, substantial studies have demonstrated paeoniflorin plays an essential role in various inflammatory disorders including RA by virtue of its anti-inflammatory effect [22]. In the present research, paeoniflorin hindered proliferation, migration, invasion, and inflammation and triggered cell cycle arrest in RA-FLSs, suggesting that paeoniflorin might be an effective drug for treating RA.
Compelling evidence has revealed that circRNAs are crucial regulators in the immune system and are inextricably linked to the occurrence and development of autoimmune diseases [23, 24]. For example, circFADS2 expedited proliferation and restrained apoptosis in lipopolysaccharide-triggered RA chondrocytes via sponging miR-498 and upregulating mTOR [25]. Furthermore, depletion of hsa_circ_0001859 attenuated inflammatory activity in RA by competitively combining with miR-204/211 [26]. In this report, the circ-FAM120A level was prominently increased in RA synovial tissues and RA-FLSs. Subsequently, circ-FAM120A upregulation abolished the effect of paeoniflorin in RA-FLSs.
Plentiful literatures have corroborated that circRNAs regulate a series of biological processes through functioning as competing endogenous RNAs (ceRNAs) for miRNAs [27]. Herein, miR-671-5p was selected as a potential target for circ-FAM120A based on prediction software and previous research. Many investigations have certified that miR-671-5p exerts an anti-tumor property in various cancers, including osteosarcoma [28], gastric cancer [29], and esophageal squamous cell carcinoma [30]. In osteoarthritis, miR-671-5p overexpression alleviated IL-1β-resulted chondrocyte injury [31]. Tang et al. unveiled that miR-671 was strikingly downregulated in RA patients [32]. In the present research, we validated that miR-671-5p was conspicuously downregulated in RA synovial tissues and RA-FLSs. Importantly, circ-FAM120A directly targeted miR-671-5p and miR-671-5p depletion relieved the suppressive effect of paeoniflorin on RA-FLSs development.
Moreover, extensive evidence has highlighted that miRNAs directly modulate gene expression through combining with 3′UTR of mRNAs [33]. Herein, we verified the targeted binding of MDM4 to miR-671-5p based on bioinformatics analysis and dual-luciferase reporter analysis. MDM4 (alias MDMX) is a negative regulator of tumor suppressor p53 [34]. A previous study indicated that MDM4 facilitated RA-FLS proliferation in RA via inactivating p53 [35]. Furthermore, Hou et al. showed that miR-34a-3p mimics blocked RA-FLS growth and reduced pro-inflammatory cytokines in RA by targeting MDM4 [36]. In the present research, upregulation of MDM4 undermined the effect of paeoniflorin on RA-FLS progression. In terms of mechanism, paeoniflorin ameliorated RA via the circ-FAM120A/miR-671-5p/MDM4 axis.
In conclusion, paeoniflorin impeded RA-FLS progression via mediating the circ-FAM120A/miR-671-5p/MDM4 pathway. These findings demonstrated that the paeoniflorin/FAM120A/miR-671-5p/MDM4 axis might provide new therapeutic targets for RA. Thereafter, in vivo experiments are needed to confirm the conclusion of this research.
Availability of Data and Materials
The data that support the findings of this study are available on request from the corresponding author.
References
Sparks JA. Rheumatoid arthritis[J]. Ann Intern Med 2019,170(1): ITC1-ITC16.
Aletaha, D., and J.S. Smolen. 2018. Diagnosis and management of rheumatoid arthritis: A Review[J]. JAMA 320 (13): 1360–1372.
Nygaard, G., and G.S. Firestein. 2020. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes[J]. Nature Reviews Rheumatology 16 (6): 316–333.
Yoshitomi, H. 2019. Regulation of immune responses and chronic inflammation by fibroblast-like synoviocytes[J]. Frontiers in Immunology 10: 1395.
Zhang, L., and W. Wei. 2020. Anti-inflammatory and immunoregulatory effects of paeoniflorin and total glucosides of paeony[J]. Pharmacol Ther 207: 107452.
Zhang, L., J. Yu, C. Wang, et al. 2019. The effects of total glucosides of paeony (TGP) and paeoniflorin (Pae) on inflammatory-immune responses in rheumatoid arthritis (RA)[J]. Functional Plant Biology 46 (2): 107–117.
Zhai, W., Z. Ma, W. Wang, et al. 2018. Paeoniflorin inhibits Rho kinase activation in joint synovial tissues of rats with collagen-induced rheumatoid arthritis[J]. Biomedicine & Pharmacotherapy 106: 255–259.
Kristensen, L.S., M.S. Andersen, L.V.W. Stagsted, et al. 2019. The biogenesis, biology and characterization of circular RNAs[J]. Nature Reviews Genetics 20 (11): 675–691.
Zhou, Z., B. Sun, S. Huang, et al. 2019. Roles of circular RNAs in immune regulation and autoimmune diseases[J]. Cell Death & Disease 10 (7): 503.
Yang, X., J. Li, Y. Wu, et al. 2019. Aberrant dysregulated circular RNAs in the peripheral blood mononuclear cells of patients with rheumatoid arthritis revealed by RNA sequencing: Novel diagnostic markers for RA[J]. Scandinavian Journal of Clinical and Laboratory Investigation 79 (8): 551–559.
Wen, J., J. Liu, P. Zhang, et al. 2020. RNA-seq reveals the circular RNA and miRNA expression profile of peripheral blood mononuclear cells in patients with rheumatoid arthritis[J]. Biosci Rep 40 (4).
Chen, X., T. Yang, W. Wang, et al. 2019. Circular RNAs in immune responses and immune diseases[J]. Theranostics 9 (2): 588–607.
Evangelatos, G., G.E. Fragoulis, V. Koulouri, et al. 2019. MicroRNAs in rheumatoid arthritis: from pathogenesis to clinical impact[J]. Autoimmun Rev 18 (11): 102391.
Zhang, C., L. Fang, X. Liu, et al. 2020. miR-22 inhibits synovial fibroblasts proliferation and proinflammatory cytokine production in RASF via targeting SIRT1[J]. Gene 724: 144144.
Wang, Y., T. Jiao, W. Fu, et al. (2019) miR-410–3p regulates proliferation and apoptosis of fibroblast-like synoviocytes by targeting YY1 in rheumatoid arthritis[J]. Biomed Pharmacother 119: 109426.
Samarpita, S., J.Y. Kim, M.K. Rasool, et al. 2020. Investigation of toll-like receptor (TLR) 4 inhibitor TAK-242 as a new potential anti-rheumatoid arthritis drug[J]. Arthritis Res Ther (in eng) 22 (1): 16.
Erlandsson, M.C., S. Töyrä Silfverswärd, M. Nadali, et al. 2017. IGF-1R signalling contributes to IL-6 production and T cell dependent inflammation in rheumatoid arthritis[J]. Biochim Biophys Acta Mol Basis Dis (in eng) 1863 (9): 2158–2170.
Hou, C., D. Wang, and L. Zhang. 2019. MicroRNA-34a-3p inhibits proliferation of rheumatoid arthritis fibroblast-like synoviocytes[J]. Mol Med Rep (in eng) 20 (3): 2563–2570.
Tristano, A.G. 2009. Tyrosine kinases as targets in rheumatoid arthritis[J]. International Immunopharmacology 9 (1): 1–9.
Korb-Pap, A., J. Bertrand, J. Sherwood, et al. 2016. Stable activation of fibroblasts in rheumatic arthritis-causes and consequences[J]. Rheumatology (Oxford) 55(suppl 2): ii64-ii67.
Guo, Q., S. Zhang, J. Huang, et al. 2020. Alogliptin inhibits IL-1beta-induced inflammatory response in fibroblast-like synoviocytes[J]. Int Immunopharmacol 83: 106372.
Xin, Q., R. Yuan, W. Shi, et al. 2019. A review for the anti-inflammatory effects of paeoniflorin in inflammatory disorders[J]. Life Sci 237: 116925.
Yang, L., J. Fu, and Y. Zhou. 2018. Circular RNAs and their emerging roles in immune regulation[J]. Frontiers in Immunology 9: 2977.
Xia, X., X. Tang, and S. Wang. 2019. Roles of circRNAs in autoimmune diseases[J]. Frontiers in Immunology 10: 639.
Li, G., W. Tan, Y. Fang, et al. 2019. circFADS2 protects LPS-treated chondrocytes from apoptosis acting as an interceptor of miR-498/mTOR cross-talking[J]. Aging (Albany NY) 11 (10): 3348–3361.
Li, B., N. Li, L. Zhang, et al. 2018. Hsa_circ_0001859 regulates ATF2 expression by functioning as an MiR-204/211 sponge in human rheumatoid arthritis[J]. Journal of Immunology Research 2018: 9412387.
Ng, W.L., T.B. Mohd Mohidin, and K. Shukla. 2018. Functional role of circular RNAs in cancer development and progression[J]. RNA Biology 15 (8): 995–1005.
Xin, C., S. Lu, Y. Li, et al. 2019. miR-671-5p inhibits tumor proliferation by blocking cell cycle in osteosarcoma[J]. DNA and Cell Biology 38 (9): 996–1004.
Qiu, T., K. Wang, X. Li, et al. 2018. miR-671-5p inhibits gastric cancer cell proliferation and promotes cell apoptosis by targeting URGCP[J]. Experimental and Therapeutic Medicine 16 (6): 4753–4758.
Li, X., C. Nie, B. Tian, et al. 2019. miR-671-5p blocks the progression of human esophageal squamous cell carcinoma by suppressing FGFR2[J]. International Journal of Biological Sciences 15 (9): 1892–1904.
Zhang, B., M. Sun, J. Wang, et al. 2019. MiR-671 ameliorates the progression of osteoarthritis in vitro and in vivo[J]. Pathol Res Pract 215 (7): 152423.
Tang, X., J. Wang, X. Xia, et al. 2019. Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients[J]. Diagnostic Pathology 14 (1): 11.
Mo, Y.Y. 2012. MicroRNA regulatory networks and human disease[J]. Cellular and Molecular Life Sciences 69 (21): 3529–3531.
Espadinha, M., V. Barcherini, E.A. Lopes, et al. 2018. An update on MDMX and dual MDM2/X inhibitors[J]. Current Topics in Medicinal Chemistry 18 (8): 647–660.
Xu, N., Y. Wang, D. Li, et al. 2010. MDM4 overexpression contributes to synoviocyte proliferation in patients with rheumatoid arthritis[J]. Biochemical and Biophysical Research Communications 401 (3): 417–421.
Hou, C., D. Wang, and L. Zhang. 2019. MicroRNA34a3p inhibits proliferation of rheumatoid arthritis fibroblastlike synoviocytes[J]. Molecular Medicine Reports 20 (3): 2563–2570.
Funding
This work was supported by the National Natural Science Foundation of China Youth Project (No. 81804050) and Key Subject of Henan Province Traditional Chinese Medicine Scientific Research Project (No. 2018ZY1011).
Author information
Authors and Affiliations
Contributions
Junfu Ma had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Junfu Ma, Qingliang Meng, Junping Zhan, Huilian Wang, and Wei Fan; acquisition of data: Yanqi Wang, Sudan Zhang, Hua Bian, and Fuzeng Zheng; critical revision of the manuscript for important intellectual content: Junfu Ma; administrative, technical, or material support: Junfu Ma, Qingliang Meng, and Junping Zhan; study supervision: Junfu Ma.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Written informed consent was obtained from patients with approval by the Institutional Review Board in Henan Province Hospital of Traditional Chinese Medicine.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, J., Meng, Q., Zhan, J. et al. Paeoniflorin Suppresses Rheumatoid Arthritis Development via Modulating the Circ-FAM120A/miR-671-5p/MDM4 Axis. Inflammation 44, 2309–2322 (2021). https://doi.org/10.1007/s10753-021-01504-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-021-01504-0