Abstract
The liver is one of the most vulnerable organs during sepsis. Current studies have proven that microRNAs play important roles in injury and inflammation. The current study aimed to investigate the role of miR-103 in septic liver injury. The sepsis model was established by cecal ligation and puncture in mice. Then, the mice were divided into four groups: normal group, sepsis group, sepsis + miR-103a-3p agomir group, and sepsis + negative control group. Liver injury was observed by hematoxylin-eosin staining and electron microscopic studies. The sepsis-induced apoptosis in liver tissues was assessed by TUNEL staining. The levels of inflammatory cytokines in liver tissues were determined by enzyme-linked immunosorbent assay kits. The targeted gene of miR-103a-3p in cells was predicted by bioinformatics algorithm and confirmed by dual-luciferase reporter assay. The expression of miR-103a-3p, HMBG1, and the apoptosis-relative proteins was examined by quantitative real-time polymerase chain reaction and Western blotting. miR-103a-3p was downregulated in liver tissues of sepsis animals. miR-103a-3p agomir could alleviate liver injury including the tissue injury and mitochondrial damage, inhibit the secretion of inflammatory factors, and decrease the apoptosis of liver cells. The high-mobility group B1 (HMGB1) was overregulated in sepsis, and it was a downstream target gene of miR-103a-3p. The results of the rescue assay confirmed that miR-103a-3p had a protection role in septic liver injury by targeting HMGB1. In summary, HMGB1 was one of the genes targeted by miR-103a-3p, which played roles in septic injury. These data may provide novel insight for the identification of new target and treatment strategies for septic liver injury.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Sepsis was the leading cause of mortality in the intensive care unit, which kills more than 750,000 patients annually. Severe sepsis is characterized by overwhelming inflammatory responses that cause multiple organ failure [1,2,3]. As an important organ in host-defense homeostasis, the liver is often harmed by sepsis [4]. However, the mechanism of septic liver injury is still unclear. Thus, it is of great value to investigate the mechanism and seek effective treatment methods for septic liver injury.
MicroRNAs (miRNAs) are small noncoding RNAs that bind target mRNAs by complementary base pairing. Changes in miRNA expression can influence the expression of hundreds of proteins directly or indirectly [5, 6]. Previous studies proved that miR-103a-3p was involved in inflammation, immunity, and cancer [7,8,9]. Lipopolysaccharide (LPS) is a heat-stable toxin associated with the outer membranes of gram-negative bacteria, which is widely used to build sepsis models. Recently, Li et al. reported miR-103a-3p was downregulated in LPS-treated lung cells, and overexpression of miR-103a-3p weakened the LPS-induced inflammation [9, 10]. Thus, we speculated that miR-103a-3p is involved in septic injury. However, the potential functions of miR-103a-3p in septic liver injury remain unclear. Thus, we examined the expression of miR-103a-3p in the liver of septic mice and found that miR-103a-3p decreases in septic liver, which implies that miR-103a-3p may play an important role in septic liver injury.
High-mobility group B1 (HMGB1) is a DNA-associated nuclear protein that can stabilize the nucleosome and take part in gene transcription. Previous study proved HMGB1 is a critical mediator in sepsis that increases in sepsis animals and mediates the systemic inflammatory response and death [11]. However, whether miR-103a-3p regulates HMGB1 in sepsis has not been reported. It is predicted that miR-103a-3p has putative binding sites with HMGB1. Thus, we speculated that sepsis could induce the abnormal low expression of miR-103a-3p, which might activate the HMGB1 and then promoted the inflammatory reactions in the liver.
In this study, the role of miR-103a-3p in septic liver injury and its potential mechanisms were investigated in septic mice by injection of miR-103a-3p agomir. The results demonstrated that miR-103a-3p agomir could alleviate septic liver injury by repressing inflammatory response and cell apoptosis via targeting HMGB1 for the first time. This study may supply one of the probable pathogenesis of septic liver injury and the possible therapeutic target for septic liver injury.
MATERIALS AND METHODS
Animals and Experimental Protocol
The procedures in this study were performed in accordance with the guidelines for the use of experimental animals from the National Institutes of Health. Male BALB/c mice (7 weeks old) were purchased from Shanghai SLAC Laboratory Animal Center. The mice were randomly divided into four groups (n = 10 per group): normal group, sepsis group, sepsis group + negative control, and sepsis + miR-103a-3p agomir group. Sepsis was induced by sepsis, as previously described by Wisnoski et al. with minor modifications [12].
miR-103a-3p agomir, negative control group (GenePharma, Shanghai, China), or HMGB1 protein (Sigma) was injected into mice through the tail vein 24 h before GLP. At 0, 24, and 48 h after sepsis, the mice were killed; peripheral blood and liver tissues were collected.
Cell Culture and Treatment
293FT cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Beijing, People’s Republic of China) and maintained in Dulbecco modified eagle medium (DMEM) containing 10% fetal bovine serum, at 37 °C in 5% CO2 atmosphere.
Kupffer cells (KCs) from mice were isolated according to previously published methods [13]. Briefly, mice were anesthetized effectively, and the liver was excised and chopped. The liver tissues were further digested in RPMI-1640 (Life Technology, Grand Island, NY) containing 0.1% type IV collagenase (Roche, Indianapolis, IN) for 30 min at 37 °C. The liver homogenate was filtered and centrifuged at 50×g for 5 min. The top aqueous phase was reserved and centrifuged at 1400×g for 10 min; the cell sediment mainly contained KCs. The cells were suspended and maintained in a 6-well plate at a density of 2 × 106/well in DMEM (Life Technology) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Sigma). After culturing for 2 to 3 h, the non-KCs were removed followed by washing with phosphate-buffered saline (PBS). The remaining adherent cells, which proved to be mainly KCs, were continued to culture for 12 to 24 h before different treatment. LPS (10 ng/mL) was used to activate KC inflammatory response.
For miRNA agomir transfection, the KCs were transfected using the Amaxa mouse macrophage Nucleofector kit using the Y-001 program (Lonza, Cologne, Germany), as previously reported [14, 15].
Enzyme-Linked Immunosorbent Assay
After the mice in different groups were killed, the liver was excised quickly, washed with buffer solution, and homogenized, and then, ultrasound was performed for 10 min. Tissue homogenate was centrifuged at 12,000×g for 10 min; the concentration of proteins in liver homogenate was measured by BCA assay. The levels of tumor necrosis factor α (TNF-α), interleukin 6 (IL-6), and IL-10 in the supernatant were measured by commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems Inc., USA) following the manufacturer’s instructions. Then, the quantity of each cytokine in liver homogenate was divided by the protein content in the sample and regarded as the content of cytokines per gram of protein (in ng/g).
Detection of Serum Alanine Aminotransferase Level
The serum level of alanine aminotransferase (ALT) was examined using Alanine Aminotransferase Assay Kit (Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer’s instructions.
Luciferase Assays
The miR-103a-3p sequence was amplified and cloned into pcDNA3.1 constructs (Invitrogen) to generate the pcDNA3.1-miR-103a-3p expression vector. We also constructed mutated Luci-HMGB1 reporter vectors bearing deletions of the UTR target regions and named them Luci-ΔHMGB1. The vectors were cotransduced with control or pcDNA3.1-miR-103a-3p expression vectors into 293 FT cells. Lysates were prepared 48 h after transduction. Luciferase activity was measured using a dual-luciferase reporter assay system (Promega). All experiments were performed in triplicate at least three independent times.
Histopathological and Immunohistochemical Analysis
Liver tissues were fixed in 10% buffered formalin, embedded in paraffin wax after dehydration by ascending grades of ethanol, and then sectioned. The sections were stained with hematoxylin and eosin. Then, they were observed by light microscopy for histological analysis.
For immunohistochemistry staining, sections were incubated with anti-mouse HMGB1 and caspase-3 antibodies (Santa Cruz Biotechnology USA) at 4 °C overnight, washed with PBS for three times, and incubated with fluorescein isothiocyanate–conjugated immunoglobulin G for 1 h.
For electron microscopic studies, the liver tissues were immediately dissected into 60 nm and placed into 4% glutaraldehyde (Solarbio, USA) for 5 min at 4 °C. The samples then were washed with PBS buffer 0.1 M for three times, dehydrated, and embedded in epoxy resin. Further processing was carried out according to the standard technique. The material was studied and documented in electron micrographs by using a TEM-125K electronic microscope.
TUNEL Assays
TUNEL assays were performed to evaluate cell apoptosis in different groups. After rehydration, slides were treated with 20 mg/mL proteinase K for 8 to 10 min at room temperature, washed with PBS, and refixed in 4% paraformaldehyde. The fragmented DNA of apoptotic cells was labeled using Nucleotide Mix and rTdT enzyme (Dead End Fluorometric TUNEL System; Promega) at 37 °C for 60 min, and the reaction stopped by incubation in 2 SSC for 15 min. After washing with deionized water, the slides were covered with glass coverslips and analyzed under a fluorescence microscope. Images were acquired using the fluorescence microscope. The percentage of apoptotic cells was calculated as the ratio of TUNEL-positive cells to all cells in 10 high-power microscopic fields.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA isolation, reverse transcription, and the quantification of target gene expression were performed as previously described [16]; GAPDH was used as an internal control of the expression of HMGB1. miR-103a-3p expression levels were quantified using U6 as the internal control (GenePharma). The fold change in expression was calculated using the following primer pairs for the amplification of target RNAs: miR-103a-3p F: 5′-AGCAGCATTGTACAGGGCTA TGAA-3′, R: 5′-TGGTGTCGTGGAGTCG-3′; U6 F: 5′-TCGCACAGACTTGTGGGAGAA-3′, R: 5′-CGCACATTAAGCCTCTATAGTTACTAGG-3′; HMGB1 F: 5′-TATGGCAAAAGCGGACAAG G-3′, R: 5′-CTTCGCAACATCACCAATGGA-3′; TNF-α F: 5′-CAGGCGGTGCCTATGTCTC-3′, R: 5′-CGATCACCCCGAAGTTCAGTAG-3′; IL-1β F: 5′-GAAATGCCACCTTTTGACAGTG-3′, R: 5′-TGGATGCTCTCATCAGGACAG-3′; IL-6 F: 5′-CTGCAAGAGACTTCCATCCAG-3′, R: 5′-AGTGGTATAGACAGGTCTGTTGG-3′; GAPDH F: 5′-GGAGCGAGATCCCTCCAAA A T-3′, R: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western Blotting
Tissues of mice from different groups were washed by PBS solution and added into cell lysate containing moderate protease inhibitor. The mixture was shaken at 4 °C for 5 min and centrifuged at 12,000×g for 10 min at 4 °C. The protein concentration was determined and trimmed. Fifty micrograms of protein was separated on polyacrylamide gels, transferred onto nitrocellulose membrane by iBlot (Invitrogen), and detected using horseradish peroxidase–conjugated secondary antibodies and chemiluminescence (Santa Cruz) exposure of BioMax film (Kodak). The following antibodies were used: anti-mHMGB1, Bcl-2, Bax, caspase-3, and GAPDH (Santa Cruz).
Statistical Analysis
All data were expressed as the mean ± standard deviation (SD). Statistical evaluation was performed using a Student t test between two groups or one-way analysis of variance for multiple comparisons. All statistical analyses were carried out using GraphPad Prism 5 software. p < 0.05 (*) was considered statistically significant.
RESULTS
Effect of miR-103a-3p on Liver Injury in Septic Mice
First, we measured the expression of miR-103a-3p by quantitative real-time polymerase chain reaction (qRT-PCR) in liver tissues from different groups at 24 h after cecal ligation and puncture (CLP). We found that the expression of miR-103a-3p was significantly downregulated in liver tissues from septic mice when compared with the normal group, whereas miR-103a-3p agomir treatment could promote the expression of miR-103a-3p in the liver tissue of septic mice (Fig. 1a).
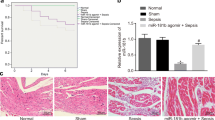
Effect of miR-103a-3p on liver injury in septic mice. a The expression of miR-103a-3p in liver tissues from different groups was measured by qRT-PCR at 24 h after sepsis (n = 10). U6 served as an internal control. miR-103a-3p expression in tissue from normal mice 1 was set as a standard. *P < 0.05 when compared with standard. b The level of serum ALT from various groups was detected by ELISA kit at 12 or 24 h after sepsis (n = 5 at each time point). c The morphological changes of liver tissues were assessed by H&E staining. Bar = 50 μm. d Ultrastructural imaging of the hepatocyte in different groups at 24 h after sepsis. The arrow directed the mitochondria in liver tissues from the different groups. Bar = 250 nm.
Then, the level of serum liver function marker ALT could be promoted by sepsis, whereas miR-103a-3p agomir treatment led to approximately a third or fourth less of ALT level when compared with negative control at 12 or 24 h after CLP (Fig. 1b).
Moreover, hematoxylin-eosin (H&E) staining was performed to evaluate the effect of miR-103a-3p on the liver injury at 24 h after CLP. As shown in Fig. 1c, the inflammatory cell infiltration and cell necrosis were obvious in livers from the sepsis group and the sepsis group + negative control group. However, there were no obvious differences in the histological morphology of liver tissues from normal and miR-103a-3p agomir groups. These results implied that septic liver injury induced by sepsis could be alleviated by miR-103a-3p agomir treatment.
Effect of miR-103a-3p Agomir on the Ultrastructure of Liver
To further elucidate the effect of miR-103a-3p agomir treatment on the sepsis-induced liver injury, the ultrastructure of liver tissues in different groups was recorded by a transmission electron microscope. Then, we observed abundant mitochondria in liver tissues from normal and miR-103a-3p agomir groups with clear and wide mitochondrial cristae and intact membrane. However, the number of mitochondrial in liver tissues from the sepsis group and the sepsis group + negative control group decreased with irregular shapes and degenerative mitochondrial crest (normal vs. agomir vs. sepsis vs. sepsis group + negative 8 ± 0.707 vs. 2.8 ± 0.58 vs. 2.62 ± 0.50 vs. 8.4 ± 0.7/field), as shown in Fig. 1d and Supplementary Fig. 1.
Effect of miR-103a-3p Agomir on the Release of Inflammatory Cytokines
To evaluate septic liver injury–induced inflammatory responses, the secretion of inflammatory factors including TNF-α, IL-1β, and IL-6 in the supernatant of liver homogenate was measured using ELISA. As presented in Fig. 2, the expression of inflammatory factors in the liver could be induced by sepsis. At 12 h following sepsis, the quantity of TNF-α, IL-1β, and IL-6 in the sepsis group and sepsis group + negative control group was more than two times those in the normal group, whereas treatment with miR-103a-3p agomir could inhibit the secretion of TNF-α, IL-1β, and IL-6. At 24 h following LPS treatment, the quantity of IL-1β and IL-6 was further increased, and pretreatment with miR-103a-3p agomir also alleviates the sepsis-induced inflammatory factor secretion. These results suggested that miR-103a-3p could restrain septic liver injury–induced inflammatory responses by regulating the release of inflammatory cytokines.
Effect of miR-103a-3p agomir on the release of inflammatory cytokines. The secretion of inflammatory factors including TNF-α, IL-1β, and IL-6 in the supernatant of liver homogenate from different groups was measured using ELISA. The results of three independent experiments were presented as means ± SD, *P < 0.05 as compared with the sepsis group.
Effect of miR-103a-3p Agomir on Septic Liver Injury–Induced Cell Apoptosis
To assess the effect of miR-103a-3p on injury-induced apoptosis, sepsis-induced apoptosis in liver tissues was assessed by TUNEL staining at 24 h after sepsis (Fig. 3a, b). The results showed that the ratio of apoptosis could be obviously induced by sepsis treatment and greatly alleviated by miR-103a-3p agomir.
Effect of miR-103a-3p agomir on septic liver injury–induced cell apoptosis. a The apoptosis of liver cells in different groups was measured by TUNEL assay at 24 h after sepsis. Bar = 50 μm. b Statistical analysis for the apoptosis ratio of liver cells in different groups at 24 h after sepsis. The percent of apoptotic cells was calculated as the ratio (TUNEL-positive cells/all cells in a field). Data represent the mean percentage of positive cells ± SD. *P < 0.05 when compared with the sepsis group. c, d The protein levels of Bcl-2, Bax, and caspase-3 in livers from different groups (normal, sepsis, sepsis + negative: sepsis + N, sepsis + agomir: sepsis + A) were examined by Western blotting and analyzed by grayscale scanning at 12 or 24 h after sepsis. The data represent the mean ± SD from three experiments. *P < 0.05 as compared with sepsis group.
Furthermore, the protein levels of Bcl-2, Bax, and caspase-3 in livers from different groups were examined by Western blotting. As shown in Fig. 3c and d, sepsis could induce a remarkable increase in the levels of Bax and caspase-3 and a decrease in the levels of Bcl-2 in liver tissues at 12 and 24 h after sepsis, whereas miR-103a-3p agomir treatment could reverse the alterations of the above proteins at 12 and 24 h after sepsis, which was in accordance with the results of TUNEL staining.
miR-103a-3p Targets HMGB1
Next, we explored the mechanism of miR-103a-3p in sepsis-induced injury. According to the prediction of the miRNA target gene database TargetScan, we identified HMGB1 as a potential target of miR-103a-3p (Fig. 4a). To confirm the bounding between miR-103a-3p and the 3′ UTR region of HMGB1, we transduced 293FT cells by control or miR-103a-3p expression vector along with either the full-length 3′ UTR of HMGB1(Luci-HMGB1) or Luci-HMGB1 vectors without 3′ UTR target regions (ΔLuci-HMGB1) (Fig. 4a). Then, we examined the luciferase activity of 293FT cells in different groups. The results showed that after cotransduction of the miR-103a-3p expression vector and vector carrying the HMGB1/miR-103a-3p target sequence, the luciferase activity of cells significantly decreased when compared with other groups (Fig. 4b).
miR-103a-3p targets HMGB1. a Bioinformatics analysis of the predicted interactions of miR-103a-3p and its binding sites within the 3′ UTR of HMGB1. b Luciferase analysis in 293 FT cells. Data represent the mean ± SD from three experiments. *P < 0.05 relative to control. c, d The mRNA expression levels of miR-103a-3p or HMGB1 in 293FT cells were analyzed by qPCR after they were transfected with miR-103a-3p agomir or antagomir. U6 or GAPDH served as an internal control. *P < 0.05 relative to control or negative control. e The protein expression of HMGB1 in 293FT cells was examined by Western blotting and analyzed by grayscale scanning after they were transfected with miR-103a-3p agomir or antagomir. GAPDH served as an internal control. The data represent the mean ± SD from three experiments. *P < 0.05 as compared with control.
Furthermore, we transduced miR-103a-3p agomir or antagomir into 293FT cells and examined the effect of miR-103a-3p on the expression of HMGB1 at the mRNA and protein level 48 h after transfection. The results showed that the level of miR-103a-3p was promoted by miR-103a-3p agomir and inhibited by antagomir (Fig. 4c), whereas the mRNA and protein level of HMGB1 significantly decreased by the treatment of miR-103a-3p agomir and vice versa (Fig. 4d, e).
miR-103a-3p Inhibits Sepsis-Induced Liver Injury by Suppressing HMGB1 Expression
Then, we further evaluate whether miR-103a-3p regulated septic liver injury by targeting HMGB1.
First, we examined the effect of the expression of miR-103a-3p on HMGB1. As illustrated in Fig. 5a–c, the mRNA and protein expression of HMGB1in liver tissues significantly increased at 12 or 24 h after sepsis, whereas miR-103a-3p agomir could inhibit the expression of HMGB1. Furthermore, we found that HMGB1 protein injection could abolish the effect of miR-103a-3p agomir in inhibiting sepsis-induced cell apoptosis and apoptosis-relative protein expression in livers (Fig. 5d).
miR-103a-3p inhibits sepsis-induced liver injury by suppressing HMGB1 expression. a The mRNA expression of HMGB1 in liver tissues from different groups was measured by qPCR at 12 or 24 h after sepsis. GAPDH served as an internal control. HMGB1 expression in tissue from normal mice 1 was set as a standard. b The protein expression of HMGB1 in liver tissues was examined by Western blotting and analyzed by grayscale scanning after 12 or 24 h after sepsis. GAPDH served as an internal control. c The protein levels of Bcl-2, Bax, and caspase-3 in livers from different groups were examined by Western blotting and analyzed by grayscale scanning at 12 or 24 h after sepsis. d The apoptosis of liver cells in different groups was measured by TUNEL assay at 24 h after sepsis. Bar = 50 μm. e Statistical analysis for the apoptosis ratio of liver cells in different groups at 24 h after sepsis. The percentage of apoptotic cells was calculated as the ratio (TUNEL-positive cells/all cells in a field). The data represent the mean ± SD from three experiments. *P < 0.05 when compared with the sepsis group.
DISCUSSION
Liver injury is a common complication of sepsis, which would lead to multiple organ dysfunction syndrome and cause of death of patients [17]. The ideal targets for the therapies of septic liver injury are inhibiting the inflammation reaction, preventing the cell apoptosis, and maintaining the function of the liver [18]. However, the molecular mechanism of septic liver injury remains unclear, which greatly limits the screening for septic liver injury diagnostic markers. Many studies have highlighted the participation of miRNAs in pathological and physiological processes including sepsis [19, 20]. miR-103a-3p was located in 5q34 and associated with the tumor, Parkinson disease, and pregnancy-related complications [21,22,23]. As a key multifunctional miRNA, miR-103a-3p could exert biological functions by negatively regulating the target genes. However, the physiological function of miR-103a-3p and its underlying molecular mechanisms in sepsis have never been reported. Thus, in the present study, we explored the functions of miR-103a-3p in sepsis-induced septic liver injury in mice. First, we examined the expression levels of miR-103a-3p in liver of mice after sepsis treatment. And we found that the expression of miR-103a-3p decreased after sepsis treatment, and downregulation of miR-103a-3p was accompanied by the liver injury of septic mice including the tissue and mitochondrial injury. Mitochondria were one of the most important organelles in hepatocytes. The damage of mitochondria can impair the respiratory function and energy metabolism of hepatocytes.
miRNA agomir could simulate the effect of miRNA and be administered by local or tail vein injection in animal experiments. The previous study reported that miRNA agomir possessed long-term activity and miRNA specificity under in vivo conditions [24]. Here, we observed that miR-103a-3p agomir could alleviate mitochondria injury of liver in sepsis by maintaining the integrity of the mitochondrial structure.
Moreover, the results from TUNEL assays confirmed that overexpression of miR-103a-3p could prevent sepsis-induced apoptosis in liver tissue. It has been documented that apoptosis was induced by inhibiting the expression of antiapoptotic proteins and promoting the expression of proapoptotic proteins [25, 26]. In this study, we observed that the apoptosis in liver was induced by sepsis through upregulating the levels of proapoptotic proteins Bax and caspase-3 and downregulating the levels of Bcl-2, whereas miR-103a-3p antagomir could inhibit sepsis-induced apoptosis by reversing the expressions of the above proteins in liver tissues. These results first proved that miR-103a-3p could protect liver cells against inflammatory-induced injury in sepsis by regulating apoptosis-relative protein.
Then, we tested whether overexpression of miR-103a-3p affected the inflammatory response in sepsis-induced liver injury. The previous studies reported that KCs were responsible for producing inflammatory cytokines in sepsis. And the KC-derived proinflammatory cytokines IL-1β and IL-6 were directly or indirectly responsible for hepatocellular dysfunction. Moreover, TNF-α was an inducer of IL-1β and IL-6 [27, 28]. So, the secretion of TNF-α, IL-1β, and IL-6 in the supernatant of liver homogenate was measured using ELISA after treatment with miR-103a-3p agomir. The results showed miR-103a-3p greatly prevented the release of inflammatory factors including TNF-α, IL-1β, and IL-6. The previous study proved that overexpression of miR-103a-3p could decrease the expression of IL-6 in lower-extremity deep venous thrombosis mice [29]. Here, we also observed that miR-103a-3p could suppress the secretion of inflammatory factors in sepsis-induced injury, which was in accordance with previous studies.
It is well known that miRNAs exert their physiological functions by regulating their downstream target genes [20, 21]. So, we predicted the potential targets of miR-103a-3p by TargetScan website and found the matched sequence in the target region of HMGB1 and miR-103a-3p, which implied that HMGB1 might be a target gene of miR-103a-3p. Literature studies reported that HMGB1 was associated with sepsis and took part in systemic inflammatory response. However, whether miR-103a-3p can regulate HMGB1 expression during sepsis has not been determined. In this article, we first verified the conserved region in the 3 ′UTR of HMGB1 was binding to miR-103a-3p in 293 cells using the double luciferase reporter gene assay. Besides, the mRNA and protein expression of HMGB1 significantly changed after miR-103a-3p agomir or antagomir transfection. Furthermore, our results showed that the increased protein levels of HMGB1 in the liver induced by sepsis were inhibited by miR-103a-3p agomir. These results indicated that miR-103a-3p agomir might restrain tissue injury by targeting HMGB1 expression in septic liver injury.
To further confirm the underlying mechanisms through which miR-103a-3p alleviated liver damage, the HMGB1 protein was injected into septic mice that were pretreated by miR-103a-3p agomir, and then, the apoptosis ratio of cells in the liver and the expression of apoptosis-relative proteins were assessed. The results proved that proapoptotic proteins Bax and caspase-3 were downregulated by miR-103a-3p agomir in septic liver injury accompanied by the decrease of apoptosis ratio, whereas the inhibition of apoptosis by miR-103a-3p agomir could be canceled by HMGB1 overexpression in septic liver injury. It was the first time to report the relation between miR-103a-3p and HMGB1 in septic liver injury.
Kupffer cells were regarded as the major source of hepatic inflammation, so we examined the effect of miR-103a-3p on KCs in vitro. The results showed that the expression of miR-103a-3p decreased significantly in KCs after LPS treatment. At the same time, the expression level of TNF-α, IL-1β, and IL-6 in KCs increased after LPS treatment. However, miR-103a-3p agomir transfection could obviously inhibit the LPS-induced expression of TNF-α, IL-1β, and IL-6 in KCs (Supplementary Fig. 2). These results implied that miR-103a-3p may play important roles in septic liver injury through KCs.
Taken together, our research proved that miR-103a-3p may participate in septic liver injury by targeting HMGB1. However, a nucleic miRNA can regulate multiple genes, and a gene can also be regulated by multiple miRNAs; these results only proved that HMGB1 was one of the genes targeted by miR-103a-3p, which played roles in septic injury. Our results provided new evidence that miR-103a-3p might serve as a potential therapeutic target for the septic liver. However, further studies were needed to confirm the physiological functions of miR-103a-3p in sepsis.
References
Angus, D.C., and T. van der Poll. 2013. Severe sepsis and septic shock. The New England Journal of Medicine 369: 840–851.
Angus, D.C., W.T. Linde-Zwirble, J. Lidicker, G. Clermont, J. Carcillo, and M.R. Pinsky. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical Care Medicine 29: 1303–1310.
Martin, G.S., D.M. Mannino, S. Eaton, and M. Moss. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. The New England Journal of Medicine 348: 1546–1554.
Li, A., J. Li, Y. Bao, D. Yuan, and Z. Huang. 2016. Xuebijing injection alleviates cytokine-induced inflammatory liver injury in sepsis-induced septic rats through induction of suppressor of cytokine signaling. Experimental and Therapeutic Medicine 12: 1531–1536.
Ambros, V. 2004. The functions of animal microRNAs. Nature 431: 350–355.
Baek, D., J. Villén, C. Shin, F.D. Camargo, S.P. Gygi, and D.P. Bartel. 2008. The impact of microRNAs on protein output. Nature 455: 64–71.
Kim, H., J.M. Yang, Y. Jin, S. Jheon, K. Kim, C.T. Lee, J.H. Chung, and J.H. Paik. 2017. MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget 8: 8484–8498.
Serafin, A., L. Foco, S. Zanigni, H. Blankenburg, A. Picard, A. Zanon, G. Giannini, I. Pichler, M.F. Facheris, P. Cortelli, P.P. Pramstaller, A.A. Hicks, F.S. Domingues, and C. Schwienbacher. 2015. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology 84: 645–653.
Li, R., P. Liang, J. Yuan, and F. He. 2020. Exosomal miR-103a-3p ameliorates lipopolysaccharide-induced immune response in BEAS-2B cells via NF-κB pathway by targeting transducin β-like 1X related protein 1. Clinical and Experimental Pharmacology & Physiology 47 (4): 620–627.
de Pádua, Lúcio K., A.C.S. Rabelo, C.M. Araújo, G.C. Brandão, G.H.B. de Souza, R.G. da Silva, D.M.S. de Souza, A. Talvani, F.S. Bezerra, A.J. Cruz Calsavara, and D.C. Costa. 2018. Anti-inflammatory and Antioxidant properties of black mulberry (Morus nigra L.) in a model of LPS-induced sepsis. Oxidative Medicine and Cellular Longevity 7: 5048031.
Karlsson, S., V. Pettilä, J. Tenhunen, R. Laru-Sompa, M. Hynninen, and E. Ruokonen. 2008. HMGB1 as a predictor of organ dysfunction and outcome in patients with severe sepsis. Intensive Care Medicine 34: 1046–1053.
Wisnoski, N., C.S. Chung, Y. Chen, X. Huang, and A. Ayala. 2007. The contribution of CD4+ CD25+ T-regulatory-cells to immune suppression in sepsis. Shock 27: 251–257.
Li, P.Z., J.Z. Li, M. Li, J.P. Gong, and K. He. 2014. An efficient method to isolate and culture mouse Kupffer cells. Immunology Letters 158 (1–2): 52–56.
Saikia, P., D. Bellos, M.R. McMullen, K.A. Pollard, C. de la Motte, and L.E. Nagy. 2017. MicroRNA 181b-3p and its target importin α5 regulate toll-like receptor 4 signaling in Kupffer cells and liver injury in mice in response to ethanol. Hepatology 66 (2): 602–615.
Mandal, P., S. Roychowdhury, P.H. Park, B.T. Pratt, T. Roger, and L.E. Nagy. 2010. Adiponectin and heme oxygenase-1 suppress TLR4/MyD88-independent signaling in rat Kupffer cells and in mice after chronic ethanol exposure. Journal of Immunology 185: 4928–4937.
Liu, Y.X., L. Wang, W.J. Liu, H.T. Zhang, J.H. Xue, Z.W. Zhang, and C.J. Gao. 2016. MiR-124-3p/B4GALT1 axis plays an important role in SOCS3-regulated growth and chemo-sensitivity of CML. Journal of Hematology & Oncology 9: 69.
Chen, Y.L., G. Xu, X. Liang, J. Wei, J. Luo, G.N. Chen, X.D. Yan, X.P. Wen, M. Zhong, and X. Lv. 2016. Inhibition of hepatic cells pyroptosis attenuates CLP-induced acute liver injury. American Journal of Translational Research 8: 5685–5695.
Lv, X., Y. Zhang, Y. Cui, Y. Ren, R. Li, and Q. Rong. 2015. Inhibition of microRNA155 relieves sepsis induced liver injury through inactivating the JAK/STAT pathway. Molecular Medicine Reports 12: 6013–6018.
Han, Y., Y. Li, and Y. Jiang. 2016. The prognostic value of plasma MicroRNA-155 and MicroRNA-146a level in severe sepsis and sepsis-induced acute lung injury patients. Clinical Laboratory 62: 2355–2360.
Gao, X.L., J.Q. Li, Y.T. Dong, E.J. Cheng, J.N. Gong, Y.L. Qin, Y.Q. Huang, J.J. Yang, S.J. Wang, and D.D. An. 2018. Upregulation of microRNA-335-5p reduces inflammatory responses by inhibiting FASN through the activation of AMPK/ULK1 signaling pathway in a septic mouse model. Cytokine 1109: 466–478.
Hu, X., J. Miao, M. Zhang, X. Wang, Z. Wang, J. Han, D. Tong, and C. Huang. 2018. miRNA-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing ATF7 in vitro. Molecules and Cells 41: 390–400.
Tang, H., D. Zhu, G. Zhang, X. Luo, and W. Xie. 2019. AFAP1-AS1 promotes proliferation of pituitary adenoma cells through miR-103a-3p to activate PI3K/AKT signaling pathway. World Neurosurgery 130: e888–e898.
Liguori, M., N. Nuzziello, A. Introna, A. Consiglio, F. Licciulli, E. D’Errico, A. Scarafino, E. Distaso, and I.L. Simone. 2018. Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Frontiers in Molecular Neuroscience 11: 288.
Hou, Z., X. Qin, Y. Hu, X. Zhang, G. Li, J. Wu, J. Li, J. Sha, J. Chen, J. Xia, L. Wang, and F. Gao. 2019. Longterm exercise-derived exosomal miR-342-5p: a novel exerkine for cardio-protection. Circulation Research 124 (9): 1386–1400.
Youle, R.J., and A. Strasser. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nature Reviews. Molecular Cell Biology 9: 47–59.
Liu, Q., T. Si, X. Xu, F. Liang, L. Wang, and S. Pan. 2015. Electromagnetic radiation at 900 MHz induces sperm apoptosis through bcl-2, bax and caspase-3 signaling pathways in rats. Reproductive Health 12: 65.
Koo, D.J., I.H. Chaudry, and P. Wang. 1999. Kupffer cells are responsible for producing inflammatory cytokines and hepatocellular dysfunction during early sepsis. The Journal of Surgical Research 83 (2): 151–157.
Gaddam, R.R., R. Fraser, A. Badiei, S. Chambers, V.C. Cogger, D.G. Le Couteur, and M. Bhatia. 2017. Differential Effects of Kupffer cell inactivation on inflammation and the liver sieve following caecal-ligation and puncture-induced sepsis in mice. Shock 47 (4): 480–490.
Zhang, P., Q. Zhao, K. Gong, Y. Long, J. Zhang, Y. Li, and X. Guo. 2019. Downregulation of miR-103a-3p contributes to endothelial progenitor cell dysfunction in deep vein thrombosis through PTEN targeting. Annals of Vascular Surgery 19: 30894–30895.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81760523 and 81660328), the Project of the Jiangxi Provincial Department of Science and Technology (No. 20171BAB215024), the Project of the Jiangxi Provincial Department of Education (No. GJJ160251), and the Project of the Jiangxi Provincial Health Commission (No. 2017A264).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
All animal experiments were carried out in accordance with the principles and procedures of the National Institute of Animal Health Care Guidelines and approved by the Animal Ethics Committee of Nanchang University.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, L., Lu, Q., Deng, F. et al. miR-103a-3p Could Attenuate Sepsis-Induced Liver Injury by Targeting HMGB1. Inflammation 43, 2075–2086 (2020). https://doi.org/10.1007/s10753-020-01275-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-020-01275-0