Abstract
The signaling system of phosphoinositides (PI) is involved in a variety of cell and tissue functions, including membrane trafficking, ion channel activity, cell cycle, apoptosis, differentiation, and cell and tissue polarity. Recently, PI and related molecules, such as the phosphoinositide-specific phospholipases C (PI-PLCs), main players in PI signaling were supposed to be involved in inflammation. Besides the control of calcium levels, PI-PLCs contribute to the regulation of phosphatydil-inositol bisphosphate metabolism, crucial in cytoskeletal organization. The expression of PI-PLCs is strictly tissue specific and evidences suggest that it varies under different conditions, such as tumor progression or cell activation. In a previous study, we obtained a complete panel of expression of PI-PLC isoforms in human umbilical vein endothelial cells (HUVEC), a widely used experimental model for endothelial cells. In the present study, we analyzed the mRNA concentration of PI-PLCs in lipopolysaccharide (LPS)-treated HUVEC by using the multiliquid bioanalyzer methodology after 3, 6, 24, 48, and 72 h from LPS administration. Marked differences in the expression of most PI-PLC codifying genes were evident.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
When inflammation occurs, endothelial cells (EC) in the vessels are the first elements exposed to mediators circulating in the bloodstream [1]. EC react with finely tuned responses mediated by different pathways, including the phosphoinositide (PI) signal transduction system. The PI pathway contributes to a variety of cell functions, including hormone secretion, neurotransmitter signal transduction, cell growth, membrane trafficking, ion channel activity, cytoskeleton regulation, cell cycle control, apoptosis, embryonic development, organogenesis, and cell/tissue polarity [2–7]. A combination of compartmentalized and temporal changes in the levels of phosphorylated derivatives of PI elicits different cellular responses, including regulation of gene expression, DNA replication, and chromatin degradation [2–7].
The phosphoinositide-specific phospholipase C (PI-PLC) enzymes contribute to the regulation of the spatiotemporal balance of molecules belonging to the PI system. After activation by different stimuli, PI-PLC rapidly cleaves the polar head group of phosphatydil inositol bisphosphate (PIP2), a structural component of the cell membrane. Decrease in PIP2 levels affects many activities, such as cytoskeleton reorganization, cytokinesis, membrane dynamics, nuclear events, and channels activity [8–10]. Cleavage of PIP2 produces two second messengers inositol trisphosphate (IP3) and diacylglycerol (DAG) (Berridge). IP3 induces the release of calcium, a crucial mediator in many cell and tissue activities, such as progression through the cell cycle, actin polymerization and subsequent cell migration [2–4]. DAG can be further cleaved to release arachidonic acid, which either acts as a messenger, or can activate the serine/threonine calcium dependent protein kinases C [8–12].
Thirteen mammalian PI-PLC enzymes have been identified, divided into six subfamilies on the basis of amino acid sequence, domain structure, and mechanism of recruitment: β(1–4), γ(1, 2), δ(1, 3, 4), ε(1), ζ(1), and η(2) [8]. Isoforms within subfamilies share sequence similarity, common domain organization, and general regulatory mechanism [8–10].
The distribution of PI-PLC enzymes is strictly tissue specific. Probably, each isoform bears a unique function in the modulation of physiological responses [8–12]. In our previous study, the panel of PI-PLC expression was delineated in quiescent human umbilical vein EC (HUVEC), an experimental model for human vascular differentiated, mature EC [13].
Functional studies based on the measurement of PI-PLC activation products indicated the involvement in the inflammatory activation of macrophages [14]. The PI-PLC γ subfamily isoforms are activated in the presence of arachidonic acid that serves as a common link between receptor activation of phospholipase A2 and the PI pathway [8].
The lipopolysaccharide (LPS), a glicolipid structural component of the outer membrane of Gram-negative bacteria, is widely used as inflammatory stimulus [15–18]. In rat astrocytes, PI-PLC enzymes resulted differently expressed after LPS treatment compared to the untreated counterpart [19], suggesting that the PI-PLC pathway might be involved in the inflammatory activation.
In the present study, we treated cultured HUVEC with LPS in order to investigate whether the inflammatory stimulus might modify the expression of the genes that codify for PI-PLC isoforms.
MATERIALS AND METHODS
Cell Culture
HUVEC (Cambrex Corporation) were cultured following manufacturer’s instructions. Endothelial Cell Growth Supplement (Endogrow, Millipore) was added to the culture medium. Confluent monolayer was obtained after 6–12 days. Cells were cultured from passage 2 to 7. Cells were stimulated by adding to the medium of culture 100 ng/ml LPS (Sigma, St. Louis, MO, USA). Cultures were stopped at the following times: 3 and 24 h after LPS stimulation for western blot and 3, 6, 24, 48, and 72 h after LPS stimulation for RT-PCR.
Western Blot
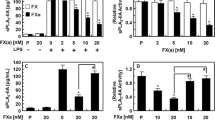
Whole-cell lysates were prepared by lysing cells in RIPA buffer (50 mM Tris pH = 7.5, NP-40, 0.1 % SDS, 100 mM NaCl, 50 mM NaF, 1 mM EDTA) supplemented with a set of protease inhibitors: 10 μg of leupeptin per milliliter, 10 μg of aprotinin per milliliter, 1 mM sodium benzamidine, and 1 mM phenylmethylsulfonyl fluoride. Proteins (50 μg) were separated on 12 % polyacrylamide, 0.1 % SDS gel. Then, incubation with a monoclonal antibody specific for each PI-PLC isoform (Santa Cruz, CA, USA) followed. Immunoreactive bands were visualized using the enhanced chemiluminescence method (Fig. 1).
RT-PCR and Multiliquid Bioanalysis
Both treated cells and untreated negative controls, plated on six-well plates, were suspended in TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA). Total RNA was isolated from samples following the manufacturer’s instructions. The obtained RNA purity was assessed by using a UV/visible spectrophotometer (SmartSpec 3000, Bio-Rad Laboratories, Hercules, CA, USA). One microgram of total RNA was reverse transcribed by using High Capacity cDNA Reverse Transcription Kit (Applied Byosystems, Carlsbad, CA, USA) according to manufacturer’s instructions. Briefly, RT Buffer, dNTP Mix, RT Random Primers, Multiscribe Reverse Transcriptase, RNase Inhibitor and DEPC-treated distilled water were added in RNase free tubes on ice. RNA sample was added. Thermal cycler was programmed as follows: 25 °C for 10 m and 37 °C for 120 m; the reaction was stopped at 85 °C for 5 m. The final volume was 20 μl. For PCR reactions, the following primer pairs (Bio Basic Inc., Amherst- NY, USA) were used: for PI-PLC β1 (gene PLCB1; OMIM ID *607120) forward 5′-AGCTCTCAGAACAAGCCTCCAACA-3′ and reverse 5′-ATCATCGTCGTCGTCACTTTCCGT-3′; for PI-PLC β2 (gene PLCB2; OMIM ID *604114) forward 5′-AAGGTGAAGGCCTATCTGAGCCAA-3′ and reverse: 5′-CTTGGCAAACTTCCCAAAGCGAGT-3′; for PI-PLC β3 (gene PLCB3; OMIM ID *600230) forward 5′-TATCTTCTTGGACCTGCTGACCGT-3′ and reverse 5′-TGTGCCCTCATCTGTAGTTGGCTT-3′; for PI-PLC β4 (gene PLCB4; OMIM ID *600810) forward 5′-GCACAGCACACAAAGGAATGGTCA-3′ and reverse 5′-CGCATTTCCTTGCTTTCCCTGTCA-3′; for PI-PLC γ1 (gene PLCG1; OMIM ID *172420) forward 5′-TCTACCTGGAGGACCCTGTGAA-3′ and reverse 5′-CCAGAAAGAGAGCGTGTAGTCG-3′; for PI-PLC γ2 (gene PLCG2; OMIM ID *600220) forward 5′-AGTACATGCAGATGAATCACGC-3′ and reverse 5′-ACCTGAATCCTGATTTGACTGC-3′; for PI-PLC δ1 (gene PLCD1; OMIM ID *602142) forward 5′-CTGAGCGTGTGGTTCCAGC-3′ and reverse 5′-CAGGCCCTCGGACTGGT-3′; for PI-PLC δ3 (gene PLCD3; OMIM ID *608795) forward 5′-CCAGAACCACTCTCAGCATCCA-3′ and reverse 5′-GCCATTGTTGAGCACGTAGTCAG-3′; for PI-PLC δ4 (gene PLCD4; OMIM ID *605939) forward 5′-AGACACGTCCCAGTCTGGAACC-3′ and reverse 5′-CTGCTTCCTCTTCCTCATATTC-3′; for PI-PLC ε (gene PLCE; OMIM ID *608414) forward 5′-GGGGCCACGGTCATCCAC-3′ and reverse 5′-GGGCCTTCATACCGTCCATCCTC-3′; for PI-PLC η1 (gene PLCH1; OMIM ID *612835) forward 5′-CTTTGGTTCGGTTCCTTGTGTGG-3′ and reverse 5′-GGATGCTTCTGTCAGTCCTTCC-3′; for PI-PLC η2 (gene PLCH2; OMIM ID *612836) forward 5′-GAAACTGGCCTCCAAACACTGCCCGCCG-3′ and reverse 5′-GTCTTGTTGGAGATGCACGTGCCCCTTGC-3′. The specificity of the primers was verified by searching in National Center for Biotechnology Information database possible homology to cDNAs of unrelated proteins. Each PCR tube contained the following reagents: 0.2 μM of both sense and antisense primers, 1–3 μl (about 1 μg) template cDNA, 0.2 mM dNTP mix, 2.5 U REDTaq Genomic DNA polymerase (Sigma-Aldrich) and 1× reaction buffer. MgCl2 was added at variable (empirical determination by setting the experiment) final concentration. The final volume was 50 μl. The amplification was started with an initial denaturation step at 94 °C for 2 min and was followed by 35 cycles consisting of denaturation (30 s) at 94 °C, annealing (30 s) at the appropriate temperature for each primer pairs and extension (1 min) at 72 °C. The PCR products were analyzed by 1.5 % TAE ethidium bromide stained agarose gel electrophoresis (Agarose Gel Unit, BIO-Rad Laboratories Inc., UK). PC-assisted CCD camera UVB lamp (Vilber Lourmaret, France) was used for gel documentation. Gel electrophoresis of the amplification products revealed single DNA bands with nucleotide lengths as expected for all primer pairs. To exclude possible DNA contamination during the RT-PCR, RNA samples were amplified by PCR without reverse transcription. No band was observed, suggesting that there was no DNA contamination in the RNA preparation procedure (data not shown). The PCR products were analyzed and quantified with the Agilent 2100 bioanalyzer using the DNA 1000 LabChip kit (Agilent Technologies, Germany). All the experiments and concentration dosages were repeated at least three times.
RESULTS
Western Blot
Accordingly to previous literature data [20], the treatment of cells with LPS induced reduction in the levels of PI-PLC β1 and PI-PLC β2 proteins. Accordingly to previous literature data, the treatment of cells with LPS did not significantly modify the levels of PI-PLC β3, PI-LPC β4, PI-PLC γ1, and PI-PLC δ1. No further data were available regarding the other isoforms. In the present analysis, the levels of PI-PLC γ2 and PI-PLC δ3 proteins were reduced. The levels of PI-PLC η1 were not significantly modified (Fig. 1).
RT-PCR
The following median concentrations were obtained for untreated cells (measured at 24 and 48 h): PLCB1 (6.5 ng/ml), PLCB2 (6.98 ng/ml), PLCB3 (5.35 ng/ml), PLCB4 (1.67 ng/ml), PLCG1 (9.07 ng/ml), PLCG2 (0.87 ng/ml), PLCD1 (6.19 ng/ml), PLCD3 (7.88 ng/ml), PLCD4 (0), PLCE (0), PLCH1 (3.21 ng/ml), PLCH2 (0), and PLCZ (0). After stimulation with LPS, the following median concentrations were obtained: 3-h treatment PLCB1 (3.8 ng/ml), PLCB2 (2.12 ng/ml), PLCB3 (3.81 ng/ml), PLCB4 (0), PLCG1 (5.41 ng/ml), PLCG2 (0 ng/ml), PLCD1 (1 ng/ml), PLCD3 (3.32 ng/ml), PLCD4 (0), PLCE (0), PLCH1 (0), PLCH2 (0), PLCZ (0); 6-h treatment PLCB1 (4.36 ng/ml), PLCB2 (0), PLCB3 (3.59 ng/ml), PLCB4 (0), PLCG1 (5.84 ng/ml), PLCG2 (0), PLCD1 (3.75 ng/ml), PLCD3 (2.41 ng/ml), PLCD4 (0), PLCE (0), PLCH1 (0), PLCH2 (0), PLCZ (0); 24-h treatment PLCB1 (6.06 ng/ml), PLCB2 (0), PLCB3 (3.77 ng/ml), PLCB4 (0), PLCG1 (4.36 ng/ml), PLCG2 (0), PLCD1 (1.88 ng/ml), PLCD3 (0), PLCD4 (0), PLCE (0), PLCH1 (0), PLCH2 (0), PLCZ (0); 48-h treatment PLCB1 (7.33 ng/ml), PLCB2 (0), PLCB3 (4.61 ng/ml), PLCB4 (0.64 ng/ml), PLCG1 (1.78 ng/ml), PLCG2 (0), PLCD1 (3.4 ng/ml), PLCD3 (4.11 ng/ml), PLCD4 (0), PLCE (0), PLCH1 (0), PLCH2 (0), PLCZ (0); 72-h treatment PLCB2 (0), PLCG1 (0.64 ng/ml), PLCG2 (0), PLCD3 (7.55 ng/ml) (Table 1).
Statistical analysis of the results did not show significant differences in three dosages obtained for each isoform. Results in different experiments were comparable. The standard deviation of the compared three dosages was not statistically significant for each isoform.
DISCUSSION
The endothelium acts dynamically, producing and modifying vasoactive molecules [20]. The morphology, molecular expression, and function of EC are influenced by the surrounding environment, leading the endothelium to adapt [21–23].
EC are exposed to mediators acting during the inflammation cascade [1]. Then, cells react by activating different signal transduction pathways. Efforts addressed to identify and investigate the signal transduction pathways might contribute to highlight the cascade of molecular events occurring in EC during inflammation [18, 24].
Evidences suggested that LPS induces activation of selected PI-PLC isoforms [19]. In our previous study, we delineated in HUVEC the expression panel of PI-PLC enzymes. In quiescent HUVEC, the following isoforms were detected: PI-PLC β2, PI-PLC β3, and PI-PLC β4; the isoforms of γ subfamily (PI-PLC γ1, and PI-PLC γ2); the isoforms of δ subfamily (PI-PLC δ1, PI-PLC δ3, and PI-PLC δ4); and the isoforms of η subfamily (PI-PLC η1 and PI-PLC η2). The remaining PI-PLC β1, PI-PLC ε, and PI-PLC ζ resulted in unexpressed isoforms [13]. In order to verify whether in HUVEC LPS might act upon gene expression of PI-PLC enzymes, we analyzed the mRNA transcript of all isoforms after LPS stimulation.
In the present study, the isoforms PI-PLC β1, PI-PLC δ4, and PI-PLC η2 were differently expressed in untreated HUVEC with respect to our previous study [13]. PI-PLC β1 was unexpressed in the previous study, while in the present experiments we found it to be expressed [13]. By contrast, PI-PLC δ4 and PI-PLC η2 resulted expressed in quiescent HUVEC in the previous study [13]. In the present experiments, they are not expressed in untreated HUVEC as well as in LPS treated counterpart. At the moment, no explanation is possible for these different findings. However, the primary HUVEC lines we processed in our two studies were obtained from different batches. Literature data showed a number of genetic differences in HUVEC primary lines [25]. That might partially explain this controversial finding.
According to previous literature data [20], the treatment of cells with LPS induced reduction in the levels of PI-PLC β1 and PI-PLC β2 proteins. Accordingly to previous literature data, the treatment of cells with LPS did not significantly modify the levels of PI-PLC β3, PI-LPC β4, PI-PLC γ1, and PI-PLC δ1. No further data were available regarding the other isoforms. In the present analysis, the levels of PI-PLC γ2 and PI-PLC δ3 proteins were reduced. The levels of PI-PLC η1 were not significantly modified.
In the present experiments, PLCB1 transcript decreases about 50 % after 3 h from LPS treatment, increases after 6 h, and reaches the initial levels after 24 h. Therefore, the expression of PI-PLC β1 is rapidly and briefly modified by LPS. In rat astrocytes, Pi-Plc β1, unexpressed in untreated cells, was expressed after LPS treatment [19]. PI-PLC β1 seemed to be downregulated in endometriosis, both in eutopic and ectopic endometrium, compared to the endometrium of unaffected women [26]. Endometriosis is a multifactorial disease, with a strong inflammatory component. Therefore, although the present results remain to be confirmed, our data might suggest that under inflammatory stimulus the transcription of the PLCB1 gene is modified with respect to untreated counterpart. However, the regulation of PLCB1 is controversial. Further studies are required in order to investigate the regulation of PI-PLC β1 expression during the inflammation cascade.
In the present experiments, the transcription of PLCB2 gene decreases about 40 % after 3 h from LPS treatment. The observation was prolonged to 72 h, and no transcript was detected during the whole period. Unfortunately, it was not possible to further prolong the observation for more than 72 h, as HUVEC primary lines die after eight to nine passages [25]. By the present results, LPS treatment seems to efficiently silence the expression of PI-PLC β2 in HUVEC. PI-PLC β2 isoform is strictly expressed in hematopoietic cell lines [27, 28]. In fact, in our previous studies, PI-PLC β2 was not expressed in quiescent cell lines as well as in the pathological counterpart [13, 19, 26, 29]. PI-PLC β2 is known to be involved in the pathogenesis of some autoimmune diseases [30]. Moreover, interleukin-8, a pro-inflammatory chemokine, induces activation of G-protein which results in the activation of PI-PLC β2 [31]. However, further studies are required in order to investigate the role of PI-PLC β2 and the relationship with the elements of the immune system [31].
In the present experiments, the transcription of PLCB3 gene is uniformly reduced about 40 % after LPS treatment since 3–72 h. Therefore, the effect of LPS treatment seems to stably reduce the expression of PLCB3 in HUVEC. PI-PLC β3 findings are controversial compared to our previous studies. We described downregulation during astrocyte activation after LPS stimulation [19]. By contrast, we found upregulation of PI-PLC β3 expression in the ectopic endometrium [27]. Further studies are required in order to investigate the differences in PLCB3 expression. In fact, the different regulation of PLCB3 transcription might only partially be explained with the different features of the analyzed cytotypes (rat astrocytes versus human endometrium and EC).
In the present experiments, after LPS treatment, PLCB4 transcript decreases since 6–24 h. PI-PLC β4 isoform is highly expressed in the nervous system, and is involved in the nociceptive pathway in mouse [32]. In our previous studies, Pi-Plc β4 was expressed both in untreated and in LPS-treated rat astrocytes [19]. Unfortunately, we had not quantified the transcript concentrations. Therefore, it is not possible to evaluate whether in rat astrocytes Plcb3 as well as genes codifying for other isoforms were downregulated.
In the present experiments, the expression of PI-PLC δ1 is reduced about 80 % after LPS treatment, then stably increases about 50 % since 6 h. Therefore, the downregulating effect of LPS acts rapidly upon PLCD1 transcription and continues, although less evident, for a prolonged period. These observations accord to our previous studies. In rat astrocytes, LPS stimulation acted rapidly and briefly downregulating the expression of Plcd1. Moreover, PI-PLC δ1 seemed to be downregulated in the ectopic endometrium [26]. Interestingly, evidences suggested a critical role for PI-PLC δ1 in the progression the cell cycle from G(1)-to-S-phase [33]. Further literature data indicated that lack of Plcδ1 in knock-out (KO) mice induces skin inflammation [33]. Further studies are required in order to investigate the role played by PI-PLC δ1 during inflammation.
The transcription of PLCD3 decreases more than 50 % after 3 h from LPS treatment of HUVEC. It further decreases after 6 h (60 %) and no mRNA was detected after 24 h. The expression begins after 48 h from LPS treatment. In addition to its role in the cell cycle control and osmotic response, recent evidences indicate that PI-PLC δ1 might act as an antioncogene [33, 34]. PI-PLC δ isozymes, the most primitive and evolutionary conserved, are known to be the most sensitive to calcium. Our previous studies described the variation of PI-PLC δ1 and PI-PLC δ3 expression during activation after LPS stimulation and in endometriosis [26]. Both isoforms seem to be downregulated in a time-dependent manner under inflammatory conditions [13, 16]. That suggested that both PI-PLC δ1 and PI-PLC δ3 might be involved in the fine tuning and regulation of the inflammation cascade. However, their role, timing of action, and possible interplay during the complex inflammation mechanism remains to be elucidated.
In the present experiments, the transcription levels of PLCG1 progressively decrease. The transcript is reduced about 45–50 % since 3 h, about 75 % at 48 h and 95 % at 72 h. Therefore, LPS acts rapidly and progressively. Moreover, LPS effects last until 72 h of observations. In our previous study, Pi-Plc γ1 was expressed both in untreated and in LPS-treated astrocytes. However, results are not comparable to the present data. In fact, in our previous study, we did not quantify the transcript. Literature data demonstrated that PI-PLC γ1 is essential for T cell development, activation and tolerance. In KO mice, deficiency of Plcg1 impairs the functionality of regulatory T cells, causing inflammatory/autoimmune symptoms [35].
In the present experiments, no transcript for PLCG2, low in quiescent HUVEC, was detected until 48 h after LPS treatment. PI-PLC γ2 transcript was detected exclusively after 72 h, although reduced about 25 % compared to untreated levels. PI-PLC γ2 isoform is tissue restricted to the hematopoietic lineage [8, 36]. In our previous study, PI-PLC γ2 was unexpressed in untreated and LPS-treated astrocytes, and in endometrium, according to literature data. PI-PLC γ2 was also identified as a promising target in inflammatory arthritis. In fact, recent studies suggested that PI-PLC γ2 isoform might represent a critical regulator of the cellular and molecular mechanisms occurring in bone and immune cells during autoimmune inflammation [37].
In the present experiments, PI-PLC η1, expressed in quiescent HUVEC, resulted unexpressed after LPS treatment, and no transcript was detected after 72 h from stimulation. LPS seems to act rapidly and definitely upon PLCH1 transcription. Unfortunately, no literature data are available about the possible role of PI-PLC η subfamily in the inflammation cascade. The remaining isoforms, PI-PLC δ4, ε, and η2 remained unexpressed both in untreated controls and LPS treated HUVEC.
Resuming, our data suggest that some PI-PLC isoforms might be involved in the metabolic pathways activated by LPS-induced inflammatory stimulation of HUVEC. For some isoforms, namely PI-PLC β2 and γ2, our results accord to literature data suggesting the involvement of PI-PLC in the regulation of the immune system. The present data partially contrast with previous results obtained in quiescent HUVEC, remarkably with regard to PI-PLC β1, δ4, and η2. In rat astrocytes, LPS seems to upregulate PI-PLC β1 expression [19]. In the present experiment, LPS seems to downregulate the transcription of PLCB1 gene in HUVEC. The differences we observed might be due to the peculiar role played by PI-PLC β1 in the nervous system [32]. The remaining findings partially or completely accord to literature data.
The present results suggest that the treatment with LPS affects the gene expression of PI-PLC enzymes, and thus contribute to regulate the production of the enzymes. LPS seems to downregulate the overall expression of most PI-PLC isoforms. That is an interesting long-term effect of LPS, otherwise known to induce the activation of the corresponding enzymes, probably followed by depletion of the intracellular stores. Although all to demonstrate, in HUVEC the effects of LPS upon PLC genes expression might be the attempt to self-limit the inflammatory reaction or might contribute to the regulation of the cell cycle. In this perspective, the role of the PI-PLC enzymes remains to be highlighted.
To understand the complex network that regulates the inflammatory process and the ability to manipulate the EC might offer a powerful tool of considerable practical and clinical importance, providing useful insights for potential therapeutic strategies.
However, although HUVEC represent a widely used, useful experimental model for human macrovascular EC, limitations in the present study remain in that they cannot fully represent the metabolic properties and interactions of the EC distributed in the living organism. Further studies are required to analyze the role and the crosstalk among the PI-PLC isoforms and the reciprocal regulation in order to tune the cell responses.
References
Gonzalez, M.A., and A.P. Selwyn. 2003. Endothelial function, inflammation, and prognosis in cardiovascular disease. Am J Med 115(Suppl 8A): 99S–106S.
Berridge, M.J. 1981. Phosphatidylinositol hydrolysis: a multifunctional transducing mechanism. Mol Cell Endocrinol 24(2): 115–140.
Berridge, M.J., and R.F. Irvine. 1989. Inositol phosphates and cell signalling. Nature 341(6239): 197–205.
Berridge, M.J. 2009. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta 1793(6): 933–940.
Comer, F.I., and C.A. Parent. 2007. Phosphoinositides specify polarity during epithelial organ development. Cell 128(2): 239–240.
Crooke, C.E., A. Pozzi, and G.F. Carpenter. 2009. PLC-gamma1 regulates fibronectin assembly and cell aggregation. Exp Cell Res 315(13): 2207–2214.
Mi, L.Y., D.S. Ettenson, and E.R. Edelman. 2008. Phospholipase C-delta extends intercellular signalling range and responses to injury-released growth factors in non-excitable cells. Cell Prolif 41(4): 671–690.
Suh, P.G., J. Park, L. Manzoli, L. Cocco, J.C. Peak, M. Katan, K. Fukami, T. Kataoka, S. Yuk, and S.H. Ryu. 2008. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Reports 41: 415–434.
Bunney, T.D., and M. Katan. 2011. PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem Sci 36(2): 88–96.
Katan, M. 2005. New insights into the families of PLC enzymes: looking back and going forward. Biochem J 391(Pt 3): e7–e9.
Nakamura, Y., and K. Fukami. 2009. Roles of phospholipase C isozymes in organogenesis and embryonic development. Physiology 24: 332–341.
Hisatsune, C., K. Nakamura, Y. Kuroda, T. Nakamura, and K. Mikoshiba. 2005. Amplification of Ca2+ signaling by diacylglycerol-mediated inositol 1,4,5-trisphosphate production. J Biol Chem 280(12): 11723–11730.
Lo Vasco, V.R., L. Pacini, T. Di Raimo, D. D’Arcangelo, and R. Businaro. 2011. Expression of phosphoinositide-specific phospholipase C isoforms in HUVEC. J Clin Pathol 64(10): 911–915.
Lo, C.J., H.G. Cryer, and R.V. Maier. 1996. Prostaglandin E2 production by endotoxin-stimulated alveolar macrophages is regulated by phospholipase C pathways. J Trauma. 40(4): 557–62. discussion 563.
Tang, X., E.M. Edwards, B.B. Holmes, J.R. Falck, and W.B. Campbell. 2006. Role of phospholipase C and diacylglyceride lipase pathway in arachidonic acid release and acetylcholine-induced vascular relaxation in rabbit aorta. Am J Physiol Heart Circ Physiol 290(1): H37–H45.
Loppnow, H., and P. Libby. 1989. Comparative analysis of cytokine induction in human vascular endothelial and smooth muscle cells. Lymphokine Res 8(3): 293–299.
Reyes, C.L., and G. Chang. 2005. Structure of the ABC transporter MsbA in complex with ADP.vanadate and lipopolysaccharide. Science. 308(5724): 1028–31. Retraction in: Chang G, Roth CB, Reyes CL, Pornillos O, Chen YJ, Chen AP. Science. 2006. 22;314(5807):1875.
Zhang, X.M., A. Morikawa, K. Takahashi, G.Z. Jiang, Y. Kato, T. Sugiyama, M. Kawai, M. Fukada, and T. Yokochi. 1994. Localization of apoptosis (programmed cell death) in mice by administration of lipopolysaccharide. Microbiol Immunol 38(8): 669–671.
Lo Vasco, V.R., C. Fabrizi, L. Fumagalli, and L. Cocco. 2010. Expression of phosphoinositide specific phospholipase C isoenzymes in cultured astrocytes activated after stimulation with lipopolysaccharide. J Cell Biochem 109(5): 1006–1012.
Grinberg, S., G. Hasko, D. Wu, and S.J. Leibovich. 2009. Suppression of PLCbeta2 by endotoxin plays a role in the adenosine A(2A) receptor-mediated switch of macrophages from an inflammatory to an angiogenic phenotype. Am J Pathol 175(6): 2439–2453.
Gloe, T., H.Y. Sohn, G.A. Meininger, and U. Pohl. 2002. Shear stress-induced release of basic fibroblast growth factor from endothelial cells is mediated by matrix interaction via integrin alpha(v)beta3. J Biol Chem 277(26): 23453–23458.
Christ, M., and M. Wehling. 1998. Cardiovascular steroid actions: swift swallows or sluggish snails? Cardiovasc Res 40(1): 34–44.
Grienddling, K.K., and W.R. Alexander. 1996. Endothelial control of the cardiovascular system: recent advances. FASEB J 10: 283–292.
Cuschieri, J., J. Billigren, and R.V. Maier. 2006. Endotoxin tolerance attenuates LPS-induced TLR4 mobilization to lipid rafts: a condition reversed by PKC activation. J Leukoc Biol 80(6): 1289–1297.
Unterluggauer, H., E. Hütter, R. Voglauer, J. Grillari, M. Vöth, J. Bereiter-Hahn, P. Jansen-Dürr, and M. Jendrach. 2007. Identification of cultivation-independent markers of human endothelial cell senescence in vitro. Biogerontology 8(4): 383–397.
Lo Vasco, V.R., M. Leopizzi, C. Chiappetta, R. Businaro, P. Polonia, C. Della Rocca, and P. Litta. 2012. Expression of phosphoinositide-specific phospholipase C enzymes in normal endometrium and in endometriosis. Fertil Steril 98(2): 410–414.
Gratacap, M.P., B. Payrastre, C. Viala, G. Mauco, M. Plantavid, and H. Chap. 1998. Phosphatidylinositol 3,4,5-trisphosphate-dependent stimulation of phospholipase C-gamma2 is an early key event in FcgammaRIIA-mediated activation of human platelets. J Biol Chem 273(38): 24314–24321.
Miao, J.Y., K. Kaji, H. Hayashi, and S. Araki. 1997. Inhibitors of phospholipase promote apoptosis of human endothelial cells. J Biochem 121(3): 612–618.
Lo Vasco, V.R., C. Fabrizi, M. Artico, L. Cocco, A.M. Billi, L. Fumagalli, and F.A. Manzoli. 2007. Expression of phosphoinositide-specific phospholipase C isoenzymes in cultured astrocytes. J Cell Biochem 100(4): 952–959.
Jakus, Z., E. Simon, D. Frommhold, M. Sperandio, and A. Mócsai. 2009. Critical role of phospholipase C gamma2 in integrin and Fc receptor-mediated neutrophil functions and the effector phase of autoimmune arthritis. J Exp Med 206(3): 577–593.
Atta-ur-Rahman, Harvey K., and R.A. Siddiqui. 1999. Interleukin-8: an autocrine inflammatory mediator. Curr Pharm Des 5(4): 241–253.
Lo Vasco, V.R. 2012. The phosphoinositide pathway and the signal transduction network in neural development. Neuroscience Bulletin 28(6): 789–800.
Kaproth-Joslin, K.A., X. Li, S.E. Reks, and G.G. Kelley. 2008. Phospholipase C delta 1 regulates cell proliferation and cell-cycle progression from G1- to S-phase by control of cyclin E-CDK2 activity. Biochem J 415(3): 439–448.
Ichinohe, M., Y. Nakamura, K. Sai, M. Nakahara, H. Yamaguchi, and K. Fukami. 2007. Lack of phospholipase C-delta1 induces skin inflammation. Biochem Biophys Res Commun 356(4): 912–918.
Fu, G., Y. Chen, M. Yu, A. Podd, J. Schuman, Y. He, L. Di, M. Yassai, D. Haribhai, P.E. North, J. Gorski, C.B. Williams, D. Wang, and R. Wen. 2010. Phospholipase C{gamma}1 is essential for T cell development, activation, and tolerance. J Exp Med 207(2): 309–318.
Kurosaki, T., A. Maeda, M. Ishiai, A. Hashimoto, K. Inabe, and M. Takata. 2000. Regulation of the phospholipase C-gamma2 pathway in B cells. Immunol Rev 176: 19–29.
Faccio, R., and V. Cremasco. 2010. PLCgamma2: where bone and immune cells find their common ground. Ann N Y Acad Sci 1192: 124–130.
Acknowledgments
The authors thank the Serena Talarico Association for support. The authors thank Dr Liselotte Setter for technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lo Vasco, V.R., Leopizzi, M., Chiappetta, C. et al. Lypopolysaccharide Downregulates the Expression of Selected Phospholipase C Genes in Cultured Endothelial Cells. Inflammation 36, 862–868 (2013). https://doi.org/10.1007/s10753-013-9613-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-013-9613-3